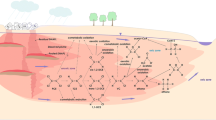
Abstract
Polychlorinated biphenyls (PCBs) are typical lasting organic pollutants. Persistence and recalcitrance to biodegradation of PCBs have hampered the transformation of PCB congeners from the environment. Biological transformation of polychlorinated biphenyls could take place through anaerobic dechlorination, aerobic microbial degradation, and a combination of transformation of anaerobic dechlorination and aerobic degradation. Under anaerobic conditions, microbial dechlorination is an important degradation mode for PCBs, especially high-chlorinated congeners. The low-chlorinated compounds formed after reductive dechlorination could be further aerobically degraded and completely mineralized. This paper reviews the recent advances in biological degradation of PCBs, introduces the functional bacteria and enzymes involved in the anaerobic and aerobic degradation of PCBs, and discusses the synergistic action of anaerobic reduction and aerobic degradation. In addition, the different ways to the microbial remediation of PCBs-contaminated environments are discussed. This review provides a theoretical foundation and practical basis to use PCBs-degrading microorganisms for bioremediation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Polychlorinated biphenyls (PCBs) are chlorinated organic compounds in which the hydrogen atoms in biphenyls are replaced by one or more chlorine atoms. PCBs are often widely used in industrial, commercial, and agricultural fields because of their excellent physical and chemical properties (Jahnke and Hornbuckle 2019). However, environmental and health issues caused by PCBs obsolescence and leakage are increasing due to the widespread use of commercial PCB mixtures (Warenik-Bany et al. 2019; Frederiksen et al. 2020). PCBs can do damage to the health of organisms and even trigger cancer, according to various human and animal studies (Tomza-Marciniak et al. 2019; Klocke and Lein 2020).
Bioremediation is a green economy and promising remediation technology that can make up for the shortcomings of physical and chemical remediation technologies (Lehtinen et al. 2014). Microorganisms are an important part in the remediation of the PCBs-polluted environment. Bacterial degradation of PCB congeners could occur through anaerobic reductive dechlorination, aerobic metabolic degradation (bacterial co-metabolism or bacterial growth on PCBs as sole carbon and energy source), or the coupling of anaerobic dechlorination and aerobic degradation (Field and Sierra-Alvarez 2008). The number of PCB chlorine substitutions and the toxicity of PCB pollutants are reduced via anaerobic dechlorination. In a one-year biodegradation of microcosms containing PCBs-contaminated sediments, high-chlorinated PCBs were observed to decrease, but trichlorobiphenyls (triCBs) and tetrachlorobiphenyls (tetraCBs) increased (Matturro et al. 2016b). This not only reduced the environmental risk but also enabled high-chlorinated PCBs to be more easily degraded by aerobic microorganisms. Then, most aerobic bacterial groups could partially oxidize low-chlorinated PCBs to chlorobenzoic acid, and even complete mineralization. However, only a small quantity of dechlorinating bacteria and related reductive dehalogenases have been isolated or characterized at present, and as a result there is less information available on bacteria responsible for anaerobic reduction of PCBs (LaRoe et al. 2014). On the contrary, the process of aerobic oxidation of PCBs is well understood; numerous bacterial degradation strains have been isolated and functional genes encoding for various enzymes have been published (Wang et al. 2018; Hirose et al. 2019).
There has been a range of research published into the degradation mechanisms of PCBs transformation, intending to promote the understanding of the bioremediation of PCBs-polluted locations. In this paper, we illustrate the aerobic degradation and anaerobic reduction of PCBs that displays the recent advances in PCBs degradation, focusing on specific degradation bacteria and the description of their key active enzymes. In addition, the different ways to the microbial remediation of PCBs-contaminated environments are discussed.
Anaerobic degradation of PCBs
Bacteria involved in anaerobic degradation
Qrganohalide-respiring bacteria (OHRB) which apply PCBs as substrates for organohalide respiration activity mainly are the members of Dehalococcoides mccartyi, belonging to phylum Chloroflexi (Table 1). Non-obligate organohalide respirers capable of PCBs dechlorination have not been reported. There are only a few anaerobic PCBs dechlorinating bacteria that have been isolated to date, and this is largely due to the difficulty in growing these bacteria and the requirements for the maintenance of the anaerobic environment. Dehalococcoides has been divided into three subgroups named Cornell, Victoria, and Pinellas according to specific base differences in variable regions 2 and 6 of 16S rDNA sequence (Hendrickson et al. 2002). The first strain D. mccartyi 195 was isolated from Cornell, so the sequences of rDNA that had the same base substitutions with D. mccartyi 195 were named Cornell subgroup. Likewise, the rDNA sequences that were identical to the representative rDNA sequences from Victoria and Pinellas were divided into members of Victoria group and Pinellas group, respectively (Hendrickson et al. 2002). The majority of PCBs dechlorinating bacteria belong to the Pinellas subgroup, including D. mccartyi CBDB1, JNA, CG5, and 195. D. mccartyi CG1 in subgroup Victoria and D. mccartyi CG4 in subgroup Cornell are also PCBs dechlorinating bacteria.
Dehalococcoides mccartyi strains are considered to be the preferred microorganisms for organochlorine microbial dechlorination. However, most dechlorinated cultures must maintain the dechlorination of PCBs with the help of media such as sediments, due to the extremely low bioavailability of PCBs and the slow growth of PCBs dechlorination bacteria (Bedard et al. 2007). D. mccartyi CBDB1 and some PCB dechlorinated cultures can only dechlorinate Aroclor 1260 under conditions using silicon powder as a sediment substitute (Adrian et al. 2009). The PCB dechlorination culture JN showed highly efficient dechlorination in the presence of sediments, but the dechlorination rate significantly decreased after two consecutive inoculations into the non-particulate medium and there was no activity observed after the third inoculation under the same conditions (Bedard et al. 2007). Because of the large surface area and capacity to absorb the highly hydrophobic PCBs, the particles can be used as a biofilm with a growing medium, with the result that the microbes can more easily contact the PCB congeners.
How to effectively enrich and isolate PCBs dechlorinating bacteria has been a problem for researchers for a long time. In addition to adding sediment substitutes, researchers have used various methods such as primer activation, increasing substrate concentration, and selecting the best PCBs monomer substrate to promote the growth of PCBs dechlorinating bacteria (Bedard et al. 2007; Wang et al. 2014). Increasing the addition concentration of PCBs to 675μM can improve the dechlorination efficiency of JN cultures. After 110 days of culture, the tetrachloride products from JN cultures increased from 2.19 to 65.23 mol% (Bedard et al. 2007). Three D. mccartyi strains CG1, CG4, and CG5 that can respire on Aroclor 1260 were isolated using tetrachloroethylene (PCE) as a substitute electron acceptor (Wang et al. 2014). Finding alternative electron receptors provides a feasible method to isolate other difficult-to-cultivate bacteria.
In addition to Dehalococcoides mccartyi bacteria, members of several other genera, such as the member of Dehalobacter genus, Dehalogenimonas genus, and family Geobacteraceae (phylum δ-Proteobacteria), play roles in PCBs anaerobic degradation. Dehalogenimonas (2.16%), from culture CG-3, is capable of degrading complex Aroclor 1260 and shares the highest full-length 16S rRNA gene sequence identity to Dehalogenimonas lykanthroporepellens BL-DC-9 (Wang and He 2012). Likewise, Mortan et al. (2017) also confirmed that synergistic action of Dehalogenimonas and D. mccartyi bacteria completely detoxicate 1,1,2-trichloroethane to ethene. Furthermore, Yang et al. (2017) and Qiao et al. (2018) demonstrated the importance of the Dehalogenimonas genus in the dechlorination of chlorinated pollutants. Therefore, the function of the Dehalogenimonas genus, closely related to Dehalococcoides, is probably underappreciated and further studies of the halogenated substrate range of Dehalogenimonas are needed.
The possibility of the family Geobacteraceae participating in PCBs degradation was revealed in the sediment-free microcosms treated by 2-bromoethanesulfonate (BES), an inhibitor of methanogenic activity to hinder the growth of Dehalococcoides strains (Praveckova et al. 2016). Previous research showed that lateral gene acquisition could confer dehalogenation activity on some bacteria (Wagner et al. 2012). Praveckova et al. (2016) surmised that Geobacteraceae species acquire the relevant genes responsible for dechlorination of low chlorinated PCBs. Moreover, the authors also found many sequences related to Methanosaeta sp., which were shown to contribute to the growth of Geobacter genus via direct interspecies electron transfer (Rotaru et al. 2015; Yang et al. 2017). Therefore, family Geobacteraceae may make contributions to PCBs dehalogenation and expand the metabolic diversity of PCB congeners.
Reductive dehalogenases responsible for the degradation of PCBs
Each dechlorination microorganism has gene sequences encoding one or more dehalogenases that catalyze the reductive dechlorination process. However, reductive dehalogenases (RDase) genes that participate in PCBs dechlorination remain poorly understood, and the characteristics of RDase still need to be further studied. PCBs dechlorination genes could be inferred by exploring the expression amount of RDase genes in the PCBs dechlorination process and establishing a corresponding quantitative relationship with the amount of cell growth and dechlorination (Ewald et al. 2019).
The overly slow growth of anaerobic PCBs-degradation strains hinders the sequencing of the genomes and the identification of functional genes (Bedard 2014). Researchers applied PCE instead of PCB as an electron acceptor to support the growth of D. mccartyi strains CG1, CG4, and CG5 (Wang et al. 2014). Three RDases from these three strains, named PcbA1, PcbA4, and PcbA5, were preliminary purified from native polyacrylamide gel electrophoresis, and tested to capable of catalyzing the PCB and PCE degradation, indicating that they are bifunctional PCB/PCE RDases (Wang et al. 2014). Three research papers showed that pcbA4 and pcbA5 dechlorinase genes were enriched in PCBs-contaminated sediment, but the pcbA1 sequence abundance was less than the other two or even was not detected, indicating that pcbA4 and pcbA5 genes are feasible biomarkers during PCBs degradation (Matturro et al. 2016a; Matturro et al. 2016b; Ewald et al. 2019). In addition, the rd8 gene and rd11 genes in D. mccartyi strain JNA were highly transcribed using PCE as an electron acceptor (Wang et al. 2014).
Ewald et al. (2019) found that the sequence abundance of the rd14 gene from D. mccartyi CG5 was more than that of the pcbA4 gene and the pcbA5 gene during anaerobic degradation. This may have something to do with the condition that PcbA4 and PcbA5 as bifunctional enzymes grow in sediments without chlorinated ethenes. Therefore, CG5’s rd14 also could be used as a biomarker to monitor PCBs dechlorination.
Aerobic degradation of PCBs
Bacteria capable of aerobic degradation
The aerobic oxidation of PCBs by most microorganisms is a co-metabolic process using biphenyl as a growth substrate, such as in the genera Pseudomonas, Ralstonia, Acinetobacter, and Rhodococcus (Field and Sierra-Alvarez 2008). Newly isolated strains from 2014 are shown on Table 2. The difference in the activity of bacterial metabolized PCBs is attributed to the number and the position of chlorine atoms in different PCB congeners. The degradation effect of PCBs is inversely proportional to the chlorine content, and high-chlorinated PCB congeners are not easily metabolized. Rhodococcus ruber SS1 and Rhodococcus pyridinivorans SS2 can both significantly degrade CB, dichlorobiphenyl (diCB), and triCB, but cannot obviously degrade tetraCB and hexaCB (Wang et al. 2018).
PCBs-degrading bacteria mainly attack 2, 3-carbon bond by biphenyl-2, 3-dioxygenase to make PCBs open the ring and generate chlorobenzoic acid (CBA) and 2-hydroxy-penta-2,4-dienoic acid. Nam et al. (2014) found dihydroxybiphenyl, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid, and benzoic acid during the degradation of biphenyl by a PCBs-degrading bacteria Pseudomonas sp. KM-04, indicating that this strain conformed to the catalytic mode of biphenyl 2, 3-dioxygenase. Paraburkholderia xenovorans LB400 is a high-efficiency PCBs-degrading bacteria and its range of degradation of PCB homologs is relatively wide because strain LB400 could open the benzene ring through 3,4-dioxygenation pathways besides 2,3-dioxygenation (Haddock and Gibson 1995).
The majority of PCBs-degrading bacteria are unable to completely mineralize PCB congeners, resulting in the accumulation of some toxic intermediates such as benzoic acid and different CBAs, which both have significant inhibitory effects on bacterial growth and PCBs degradation (Wójcik et al. 2020; Xing et al. 2020). However, Sphingobium fuliginis strain HC3 could degrade benzoic acid and 3-CBA simultaneously and produced no intermediates (Hu et al. 2015).
In addition, a small number of bacteria could use PCBs with one or two chlorines, mainly with 4-CB as the only carbon source and energy. Achromobacter xylosoxidans strain IR08 utilized all CBs, 2,4’-and 4,4’-diCB, as sole carbon and energy sources (Ilori et al. 2008). At the same time, it also degraded three kinds of CBAs and generated no metabolites, indicating that A. xylosoxidans IR08 could mineralize CBs (Ilori et al. 2008). Although various isomers of diCBs are present in very small amounts in commercial PCB mixtures, researchers have found that they occupied an important position in wastewater samples due to the degradation of other pollutants, and they were accumulated in large quantities after anaerobic reduction dechlorination, thus indicating that the metabolism of diCBs is vital in the bioremediation process of PCBs-polluted environments. Ralstonia spp. SA-3 and SA-4 utilized all CBs, the meta-substituted diCBs 3,3’- and 3,5-diCB, and the ortho-substituted diCBs, 2,2’-, 2,4’- and 2,3-diCB as carbon sources (Adebusoye et al. 2008a; Adebusoye et al. 2008b).
In recent years, several strains responsible for using highly chlorinated PCBs as carbon and energy sources have been isolated. Sinorhizobium meliloti NM degraded coplanar 3,3’,4,4’-tetraCB, possessing the highest toxicity of all tetraCBs, and its degradation effect was remarkable at low concentrations (Wang et al. 2016). Furthermore, a cold-tolerant strain, Comamonas testosteroni strain QL, could use decachlorobiphenyl (decaCB) as a carbon source and it almost completely degraded decaCB at a concentration of 400 μg L-1 (Qiu et al. 2016).
Enzymes and functional genes in aerobic degradation
This series of reactions is known as the upstream pathway for biphenyl degradation (Field and Sierra-Alvarez 2008; Elangovan et al. 2019). The first step of degradation is to catalyze the addition of a molecule of oxygen to the 2,3-position of PCBs under the action of biphenyl 2,3-dioxygenase (BphA) to form cis-2,3-dihydro-2,3-dihydroxychlorobiphenyl. Then this is catalyzed by dehydrogenase (BphB) to generate 2,3-dihydroxychlorobiphenyl. 2,3-Dihydroxychlorobiphenyl undergoes 2,3-dihydroxybiphenyl-dioxygenase (BphC) to catalyze ring opening to form 2-hydroxy-6-ox-6-phenylhexa-2,4-chlorodienoic acid (HOPDA). Finally, HOPDA is hydrolyzed to chlorobenzoic acid and 2-hydroxy-penta-2,4-dienoic acid by HOPDA hydrolase (BphD) (Fig 1). In some strains, the degradation product 2-hydroxypenta-2,4-dienoic acid could be further degraded to pyruvate and acetyl-CoA (Field and Sierra-Alvarez 2008). The biphenyl/polychlorinated biphenyl-degradation locus (bph) of Pseudomonas sp. LB400 encodes four additional metabolic enzymes. BphK, a glutathione-S-transferase, is probably involved in the dechlorination of HOPDA. 2-Hydroxypenta-2,4-dienoic acid is catalyzed by 2-hydroxypenta-2,4-dienoic acid hydratase (BphH), acetaldehyde dehydrogenase (BphI) and 4-hydroxy-2-oxovalerate aldolase (BphJ) to acetyl-CoA.
Polychlorinated biphenyl degrading enzyme genes (bph gene clusters) are situated on chromosomes, plasmids, and transposons. The bph genes of Sphingobium fuliginis HC3 were located on the plasmid (Hu et al. 2015). The bph gene clusters in four KF strains, respectively similar to the genes in Pseudomonas furukawaii KF707 and Tn4371 of Cupriavidus oxalacticus A5, are located on an element which is more than 110 kb (named the ICEbph-sal), and this element could be reintegrated into the host’s chromosome (Hirose et al. 2019). The bph cluster moves between different bacteria through various mobile genetic elements. The different ways of horizontal gene transfer make contributions to bacterial evolution and environment adaptation (Suenaga et al. 2017).
The typical bph gene cluster (bphA1A2A3A4BCKHJID) was observed in P. xenovorans LB400 (Hofer et al. 1994; Chain et al. 2006). Genes encoding enzymes involved in the upstream pathway and lower pathways for biphenyl degradation that transform PCB congeners to acetyl-CoA were located on the same operon. The bph gene organization and the structure of some bacteria are the same, and some other bacteria are quite different, indicating that the bph gene cluster undergoes restructuring. Another type of bph gene cluster was found in Rhodococcus RHA1(bphA1A2A3A4CB) (Masai et al. 1995; Sha’arani et al. 2019). This strain has no bphD, but this is replaced by bphS and bphT which encode a two-component signal transduction system (Takeda et al. 2004). The order of the genes and the sequence in R. globerulus P6 and Rhodococcus sp. TA421 is similar to that in RHA1. However, some other Rhodococcus strains have bph gene clusters different from that in RHA1. In Rhodococcus rhodochrous K37, the bph genes are preceded by bphB, and followed by bphC, bphA1A2A3A4 and bphD (Taguchi et al. 2007). This kind of bph gene cluster evolved separately from the previously known bph gene clusters of PCBs degraders. This strain has eight bphC genes and the bphC8 gene located on a 200-kb linear plasmid was induced by biphenyl, suggesting that it is involved in biphenyl degradation in K37. Interestingly, bphC8 had a sequence identity below 41% with the known bphC involved in biphenyl metabolism but shared the highest identity with phdF from Nocardioides sp. KP7 responsible for degrading phenanthrene. Similarly, Rhodococcus spp. R04 and WAY2 were observed to harbor the bphBCA1A2A3A4D cluster (Yang et al. 2007; Garrido-Sanz et al. 2020).
The genetic organization of the bph gene clusters of multiple degradation strains has been analyzed and partial comparisons are listed in figure 2. In nature, the distribution of bph gene clusters is very wide. The order of bph genes and amino acid sequences in strains of different species are also quite different, but the functions of related genes are highly conserved, indicating that the bph gene clusters are constantly changing in the process of gene transfer and evolution. For the origin of PCBs-degrading genes, aromatic ring dioxygenases are evolved from a common ancestor, as evidenced with toluene dioxygenase, benzene dioxygenase, naphthalene dioxygenase and biphenyl dioxygenase (Harayama et al. 1992). It also could be postulated that many degraders of aromatics might be involved in the degradation of plant lignin, which is massively distributed in the environment (Furukawa 1994).
Genetic organization of the bph gene clusters of Paraburkholderia xenovorans LB400 (Hofer et al. 1994; Chain et al. 2006), Rhodococcus jostii RHA1 (Masai et al. 1995, Takeda et al. 2004), Rhodococcus rhodochrous K37 (Taguchi et al. 2007), Rhodococcus sp. WAY2 (Garrido-Sanz et al. 2020), Acidovorax sp. KKS102 (Kikuchi et al. 1994), and Bacillus sp. JF8 (Mukerjee-Dhar et al. 2005).
Coupling of anaerobic dechlorination and aerobic degradation
High-chlorinated PCBs as electron acceptors could be dechlorinated into low-chlorine PCBs by anaerobic microorganisms, but their benzene ring structures are generally degraded by aerobic strains. PCBs-dechlorination anaerobic consortia also could completely mineralize PCBs (Meckenstock et al. 2016). According to Payne’s opinion (2013)), the combination of anaerobic dechlorination and aerobic degradation could greatly increase the degradation rate of high-chlorinated PCBs and is a promising biodegradation method to completely degrade high-chlorinated PCBs.
Chen et al. (2014) designed an experiment that continuous flooding and drying processes, simulating a natural sequential aerobic-anaerobic environment in the paddy field, degraded more than 40% of PCB congeners, as compared to only 20% degradation under the constant-drying condition. A commercial PCB mixture Aroclor1260, subjected to alternating anaerobic-aerobic degradation under the treatment of three facultative anaerobic microorganisms, resulted in a 49% total reduction of PCBs over two weeks (Pathiraja et al. 2019b). In addition, Payne et al. (2013) showed that remediation of PCBs contaminated sediment with simultaneously added anaerobic dechlorinator Dehalobium sp. DF1 and aerobic Paraburkholderia sp. LB400 significantly enhanced the degradation of PCBs, resulting in 80% total PCBs reduction after 120 days.
The coupling of anaerobic degradation and aerobic oxidation can be divided into two-stage (or sequential) anaerobic-aerobic degradation and concurrent (or alternating) anaerobic-aerobic degradation. Research showed that the latter method possesses better degradation ability. According to the results of experiment using anaerobic and aerobic degradation bacteria, and three facultative anaerobic bacteria, respectively, the degradation effect of the concurrent (or alternating) treatment mode was found to be better than a sequential process (Payne et al. 2013; Pathiraja et al. 2019b).
Bioremediation of PCBs-contaminated environments
Microorganisms play significant roles in the bioremediation of PCBs-contaminated environments (Passatore et al. 2014). Pure cultures, microbial consortia, and plant-microbe associations are discussed below along with their applications.
Due to the massive use of PCB congeners and the residues in the environment, researchers tried to use pure cultures to remove PCBs. Multiple PCBs-degrading bacteria have been isolated and researched as mentioned before. Many studies described the application of a single microorganism for the bioremediation of PCBs-contaminated soil. The strain QL could remove 45% of PCB 209 in soil within only 20 days (Qiu et al. 2016). Rhodococcus sp. IN306, closely related to the Rhodococcus jostii RHA1, degraded more than 54% of PCBs in commercially available PCBs‐polluted soil SQC068 after a 30-day test (Steliga et al. 2020). However, the degradation capability of a single strain to degrade PCBs is relatively low because of its genetic properties and the environmental factors, such as the pH, temperature, concentration of PCB congeners, etc (Horváthová et al. 2018). Moreover, pure cultures probably produce toxic metabolites in the process of aerobic degradation (Xing et al. 2020).
Microbial consortia have a more dominant impact on the bioremediation of pollutants contaminated soils compared with a single strain because different kinds of microorganisms have distinct substrate specificities (Mikeskova et al. 2012). Horváthová et al. (2018) used Achromobacter xylosoxidans, Stenotrophomonas maltophilia, Ochrobactrum anthropi, and Rhodococcus ruber in different combinations to compare the bioremediation effect. The best results were obtained with two bacterial strains consisting of R. ruber and A. xylosoxidans, and the consortia of O. anthropi, R. ruber, and A. xylosoxidans also displayed a good biodegradation activity. Because of the presence of viable but non-culturable strains and many uncultured bacteria, the PCBs degradation capacity of an artificial mixed consortium comprising isolated strains was lower than that of a natural mixed consortium (Mikeskova et al. 2012). Researchers applied extracellular organic matter from Micrococcus luteus to stimulate the growth of the indigenous bacterial community responsible for metabolizing PCBs and enhance the bioremediation of long-term PCBs-contaminated sites (Su et al. 2015). Natural or artificial microbial consortia are of great significance for the degradation and removal of PCBs. However, the species interactions in microbial consortia, the effect of the microbial interactions on the biodegradation potential, and their bioremediation performance are still unclear. The microbial consortia need to be developed and optimized by using a proper strategy when they are put into use (Mikeskova et al. 2012).
Some plant species could enhance the PCBs removal efficiency in soils and sediments (Hayat et al. 2019). Plants remarkably encourage PCBs degradation by providing habitats and nutrients for PCBs-degrading microorganisms and releasing organic exudates as inducers, surfactants, and microbial growth factors (Sharma et al. 2017). These microorganisms could significantly improve plant growth and the ability of PCBs uptake by plants, thereby enhancing the remediation of PCBs-contaminated soil (Teng et al. 2016). Liang et al. (2014) found that switchgrass could improve PCBs removal in soil because of phytoextraction processes and enhanced microbial activity in the rhizosphere. After inoculation with Paraburkholderia xenovorans LB400, the amounts of PCBs in switchgrass-treated soils were reduced by 43%. Salimizadeh et al. (2018) assessed the dissipation of PCB congeners in a transformer oil-contaminated soil using Pseudomonas spp. S5 treated or not by the maize plantation. The results showed that the soil inoculated with Pseudomonas spp. S5 and cultivated with maize had the best degradation effect after 10 weeks. In pot experiments, the soil PCBs concentrations of the planting Astragalus sinicus inoculated with Mesorhizobium sp. ZY1 was decreased by 53%, but the single incubation of Mesorhizobium sp. ZY1 and the single planting A. sinicus decreased PCBs amount in soil by 20% and 23%, respectively (Teng et al. 2016). Therefore, plant-microbe associated bioremediation techniques are effective and cost-efficient methods of cleaning polluted sites (Jing et al. 2018). Bioremediation using the plant-microbe associations is a promising method and will be used widely to significantly remove PCBs from the environment.
Concluding remarks and future perspectives
Sustainable development is committed to meeting the needs of contemporary people and ensuring the environmental rights of future generations. The removal of environmental pollutants and the restoration of polluted sites are primary challenges for modern society. Biodegradation based on the catabolic activity of PCBs-degrading microorganisms has emerged as the economical, ecofriendly, and promising strategy to remediate PCBs contaminations. The research mainly focused on the screening of PCBs-degrading strains and relevant enzymes and the introduction of practical applications of degrading strains. It is important for us to better understand the transport and fate of PCBs in the environments, and the regulatory genes and enzymes involved in the degradation pathways of PCBs. As treatment for enhanced biodegradation of PCBs, advanced molecular approaches, enzyme engineering, and an integrated approach for microorganisms mixed with emerging novel materials will provide better tools for the remediation of PCBs pollutants.
References
Adebusoye SA, Ilori MO, Picardal FW, Amund OO (2008) Metabolism of chlorinated biphenyls: use of 3,3'- and 3,5-dichlorobiphenyl as sole sources of carbon by natural species of Ralstonia and Pseudomonas. Chemosphere 70:656–663. https://doi.org/10.1016/j.chemosphere.2007.06.079
Adebusoye SA, Picardal FW, Ilori MO, Amund OO (2008) Evidence of aerobic utilization of di-ortho-substituted trichlorobiphenyls as growth substrates by Pseudomonas sp. SA-6 and Ralstonia sp. SA-4. Environ Microbiol 10:1165–1174. https://doi.org/10.1111/j.1462-2920.2007.01533.x
Adrian L, Dudkova V, Demnerova K, Bedard DL (2009) Dehalococcoides sp. strain CBDB1 extensively dechlorinates the commercial polychlorinated biphenyl mixture aroclor 1260. Appl Environ Microbiol 75:4516–4524. https://doi.org/10.1128/AEM.00102-09
Bedard DL (2014) PCB dechlorinases revealed at last. Proc Natl Acad Sci USA 111:11919–11920. https://doi.org/10.1073/pnas.1412286111
Bedard DL, Ritalahti KM, Loffler FE (2007) The Dehalococcoides population in sediment-free mixed cultures metabolically dechlorinates the commercial polychlorinated biphenyl mixture aroclor 1260. Appl Environ Microbiol 73:2513–2521. https://doi.org/10.1128/AEM.02909-06
Cai M, Song G, Li Y, Du K (2018) A novel Aroclor 1242-degrading culturable endophytic bacterium isolated from tissue culture seedlings of Salix matsudana f. pendula Schneid. Phytochem Lett 23:66–72. https://doi.org/10.1016/j.phytol.2017.11.012
Chain PS, Denef VJ, Konstantinidis KT, Vergez LM, Agullo L, Reyes VL, Hauser L, Cordova L, Gomez L, Gonzalez M, Land M, Lao V, Larimer F, LiPuma JJ, Mahenthiralingam E, Malfatti SA, Marx CJ, Parnell JJ, Ramette A, Richardson P, Seeger M, Smith D, Spilker T, Sul WJ, Tsoi TV, Ulrich LE, Zhulin IG, Tiedje JM (2006) Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc Natl Acad Sci USA 103:15280–15287. https://doi.org/10.1073/pnas.0606924103
Chen C, Yu C, Shen C, Tang X, Qin Z, Yang K, Hashmi MZ, Huang R, Shi H (2014) Paddy field-a natural sequential anaerobic-aerobic bioreactor for polychlorinated biphenyls transformation. Environ Pollut 190:43–50. https://doi.org/10.1016/j.envpol.2014.03.018
Elangovan S, Pandian SBS, Geetha SJ, Joshi SJ (2019) Polychlorinated biphenyls (PCBs): environmental fate, challenges and bioremediation. In: Arora PK (ed) Microbial Metabolism of Xenobiotic Compounds. Springer, Singapore, pp 165-188. https://doi.org/10.1007/978-981-13-7462-3_8
Ewald JM, Humes SV, Martinez A, Schnoor JL, Mattes TE (2019) Growth of Dehalococcoides spp. and increased abundance of reductive dehalogenase genes in anaerobic PCB-contaminated sediment microcosms. Environ Sci Pollut Res Int. https://doi.org/10.1007/s11356-019-05571-7
Fennell DE, Nijenhuis I, Wilson SF, Zinder SH, Haggblom MM (2004) Dehalococcoides ethenogenes strain 195 reductively dechlorinates diverse chlorinated aromatic pollutants. Environ Sci Technol 38:2075–2081. https://doi.org/10.1021/es034989b
Field JA, Sierra-Alvarez R (2008) Microbial transformation and degradation of polychlorinated biphenyls. Environ Pollut 155:1–12. https://doi.org/10.1016/j.envpol.2007.10.016
Frederiksen M, Andersen HV, Haug LS, Thomsen C, Broadwell SL, Egsmose EL, Kolarik B, Gunnarsen L, Knudsen LE (2020) PCB in serum and hand wipes from exposed residents living in contaminated high-rise apartment buildings and a reference group. Int J Hyg Environ Health 224:113430. https://doi.org/10.1016/j.ijheh.2019.113430
Furukawa K (1994) Molecular genetics and evolutionary relationship of PCB-degrading bacteria. Biodegradation 5:289–300. https://doi.org/10.1007/bf00696466
Garrido-Sanz D, Sansegundo-Lobato P, Redondo-Nieto M, Suman J, Cajthaml T, Blanco-Romero E, Martin M, Uhlik O, Rivilla R (2020) Analysis of the biodegradative and adaptive potential of the novel polychlorinated biphenyl degrader Rhodococcus sp WAY2 revealed by its complete genome sequence. Microb Genom. https://doi.org/10.1099/mgen.0.000363
Haddock JD, Gibson DT (1995) Purification and characterization of the oxygenase component of biphenyl 2,3-dioxygenase from Pseudomonas sp. strain LB400. J Bacteriol 177:5834–5839. https://doi.org/10.1128/jb.177.20.5834-5839.1995
Harayama S, Kok M, Neidle EL (1992) Functional and evolutionary relationships among diverse oxygenases. Annu Rev Microbiol 46:565–601. https://doi.org/10.1146/annurev.mi.46.100192.003025
Hayat A, Hussain I, Soja G, Iqbal M, Shahid N, Syed JH, Yousaf S (2019) Organic and chemical amendments positively modulate the bacterial proliferation for effective rhizoremediation of PCBs-contaminated soil. Ecol Eng 138:412–419. https://doi.org/10.1016/j.ecoleng.2019.07.038
Hendrickson ER, Payne JA, Young RM, Starr MG, Perry MP, Fahnestock S, Ellis DE, Ebersole RC (2002) Molecular analysis of Dehalococcoides 16S ribosomal DNA from chloroethene-contaminated sites throughout North America and Europe. Appl Environ Microbiol 68:485–495. https://doi.org/10.1128/aem.68.2.485-495.2002
Hirose J, Fujihara H, Watanabe T, Kimura N, Suenaga H, Futagami T, Goto M, Suyama A, Furukawa K (2019) Biphenyl/PCB degrading bph genes of ten bacterial strains isolated from biphenyl-contaminated soil in Kitakyushu, Japan: comparative and dynamic features as integrative conjugative elements (ICEs). Genes 10:404. https://doi.org/10.3390/genes10050404
Hofer B, Backhaus S, Timmis KN (1994) The biphenyl/polychlorinated biphenyl-degradation locus (bph) of Pseudomonas sp. LB400 encodes four additional metabolic enzymes. Gene 144:9–16. https://doi.org/10.1016/0378-1119(94)90196-1
Horváthová H, Lászlová K, Dercová K (2018) Bioremediation of PCB-contaminated shallow river sediments: the efficacy of biodegradation using individual bacterial strains and their consortia. Chemosphere 193:270–277. https://doi.org/10.1016/j.chemosphere.2017.11.012
Hu J, Qian M, Zhang Q, Cui J, Yu C, Su X, Shen C, Hashm MZ, Shi J (2015) Sphingobium fuliginis HC3: a novel and robust isolated biphenyl- and polychlorinated biphenyls-degrading bacterium without dead-end intermediates accumulation. PLoS One 10:e0122740. https://doi.org/10.1371/journal.pone.0122740
Ilori MO, Robinson GK, Adebusoye SA (2008) Degradation and mineralization of 2-chloro-, 3-chloro- and 4-chlorobiphenyl by a newly characterized natural bacterial strain isolated from an electrical transformer fluid-contaminated soil. J Environ Sci (China) 20:1250–1257. https://doi.org/10.1016/s1001-0742(08)62217-2
Jahnke JC, Hornbuckle KC (2019) PCB emissions from paint colorants. Environ Sci Technol 53:5187–5194. https://doi.org/10.1021/acs.est.9b01087
Jing R, Fusi S, Kjellerup BV (2018) Remediation of polychlorinated biphenyls (PCBs) in contaminated soils and sediment: state of knowledge and perspectives. Front Environ Sci 6:79. https://doi.org/10.3389/fenvs.2018.00079
Kikuchi Y, Yasukochi Y, Nagata Y, Fukuda M, Takagi M (1994) Nucleotide sequence and functional analysis of the meta-cleavage pathway involved in biphenyl and polychlorinated biphenyl degradation in Pseudomonas sp. strain KKS102. J Bacteriol 176:4269–4276. https://doi.org/10.1128/jb.176.14.4269-4276.1994
Klocke C, Lein PJ (2020) Evidence implicating non-dioxin-like congeners as the key mediators of polychlorinated biphenyl (PCB) developmental neurotoxicity. Int J Mol Sci. https://doi.org/10.3390/ijms21031013
LaRoe SL, Fricker AD, Bedard DL (2014) Dehalococcoides mccartyi strain JNA in pure culture extensively dechlorinates Aroclor 1260 according to polychlorinated biphenyl (PCB) dechlorination process N. Environ Sci Technol 48:9187–9196. https://doi.org/10.1021/es500872t
Lehtinen T, Mikkonen A, Sigfusson B, Olafsdottir K, Ragnarsdottir KV, Guicharnaud R (2014) Bioremediation trial on aged PCB-polluted soils-a bench study in Iceland. Environ Sci Pollut Res Int 21:1759–1768. https://doi.org/10.1007/s11356-013-2069-z
Liang Y, Meggo R, Hu D, Schnoor JL, Mattes TE (2014) Enhanced polychlorinated biphenyl removal in a switchgrass rhizosphere by bioaugmentation with Burkholderia xenovorans LB400. Ecol Eng 71:215–222. https://doi.org/10.1016/j.ecoleng.2014.07.046
Masai E, Yamada A, Healy JM, Hatta T, Kimbara K, Fukuda M, Yano K (1995) Characterization of biphenyls catabolic genes of gram-positive polychlorinated biphenyls degrader Rhodococcus sp. strain RHA1. Appl Environ Microbiol 61:2079–2085
Matturro B, Di Lenola M, Ubaldi C, Rossetti S (2016) First evidence on the occurrence and dynamics of Dehalococcoides mccartyi PCB-dechlorinase genes in marine sediment during Aroclor1254 reductive dechlorination. Mar Pollut Bull 112:189–194. https://doi.org/10.1016/j.marpolbul.2016.08.021
Matturro B, Ubaldi C, Rossetti S (2016) Microbiome dynamics of a polychlorobiphenyl (PCB) historically contaminated marine sediment under conditions promoting reductive dechlorination. Front Microbiol 7:1502. https://doi.org/10.3389/fmicb.2016.01502
Meckenstock RU, Boll M, Mouttaki H, Koelschbach JS, Cunha Tarouco P, Weyrauch P, Dong X, Himmelberg AM (2016) Anaerobic degradation of benzene and polycyclic aromatic hydrocarbons. J Mol Microb Biotech 26:92–118. https://doi.org/10.1159/000441358
Mikeskova H, Novotny C, Svobodova K (2012) Interspecific interactions in mixed microbial cultures in a biodegradation perspective. Appl Microbiol Biotechnol 95:861–870. https://doi.org/10.1007/s00253-012-4234-6
Mizukami-Murata S, Sakakibara F, Fujita K, Fukuda M, Kuramata M, Takagi K (2016) Detoxification of hydroxylated polychlorobiphenyls by Sphingomonas sp. strain N-9 isolated from forest soil. Chemosphere 165:173–182. https://doi.org/10.1016/j.chemosphere.2016.08.127
Mortan SH, Martin-Gonzalez L, Vicent T, Caminal G, Nijenhuis I, Adrian L, Marco-Urrea E (2017) Detoxification of 1,1,2-trichloroethane to ethene in a bioreactor co-culture of Dehalogenimonas and Dehalococcoides mccartyi strains. J Hazard Mater 331:218–225. https://doi.org/10.1016/j.jhazmat.2017.02.043
Mukerjee-Dhar G, Shimura M, Miyazawa D, Kimbara K, Hatta T (2005) bph genes of the thermophilic PCB degrader, Bacillus sp. JF8: characterization of the divergent ring-hydroxylating dioxygenase and hydrolase genes upstream of the Mn-dependent BphC. Microbiology 151:4139–4151. https://doi.org/10.1099/mic.0.28437-0
Murínová S, Dercová K, Dudášová H (2014) Degradation of polychlorinated biphenyls (PCBs) by four bacterial isolates obtained from the PCB-contaminated soil and PCB-contaminated sediment. Int Biodeterior Biodegrad 91:52–59. https://doi.org/10.1016/j.ibiod.2014.03.011
Nam IH, Chon CM, Jung KY, Kim JG (2014) Biodegradation of biphenyl and 2-chlorobiphenyl by a Pseudomonas sp. KM-04 isolated from PCBs-contaminated coal mine soil. Bull Environ Contam Toxicol 93:89–94. https://doi.org/10.1007/s00128-014-1286-6
Passatore L, Rossetti S, Juwarkar AA, Massacci A (2014) Phytoremediation and bioremediation of polychlorinated biphenyls (PCBs): state of knowledge and research perspectives. J Hazard Mater 278:189–202. https://doi.org/10.1016/j.jhazmat.2014.05.051
Pathiraja G, Egodawatta P, Goonetilleke A, Te’o VSJ (2019) Solubilization and degradation of polychlorinated biphenyls (PCBs) by naturally occurring facultative anaerobic bacteria. Sci Total Environ 651:2197–2207. https://doi.org/10.1016/j.scitotenv.2018.10.127
Pathiraja G, Egodawatta P, Goonetilleke A, Te'o VSJ (2019) Effective degradation of polychlorinated biphenyls by a facultative anaerobic bacterial consortium using alternating anaerobic aerobic treatments. Sci Total Environ 659:507–514. https://doi.org/10.1016/j.scitotenv.2018.12.385
Payne RB, Fagervold SK, May HD, Sowers KR (2013) Remediation of polychlorinated biphenyl impacted sediment by concurrent bioaugmentation with anaerobic halorespiring and aerobic degrading bacteria. Environ Sci Technol 47:3807–3815. https://doi.org/10.1021/es304372t
Praveckova M, Brennerova MV, Holliger C, De Alencastro F, Rossi P (2016) Indirect evidence link PCB dehalogenation with Geobacteraceae in anaerobic sediment-free microcosms. Front Microbiol 7:933. https://doi.org/10.3389/fmicb.2016.00933
Qiao W, Luo F, Lomheim L, Mack EE, Ye S, Wu J, Edwards EA (2018) A Dehalogenimonas population respires 1,2,4-trichlorobenzene and dichlorobenzenes. Environ Sci Technol 52:13391–13398. https://doi.org/10.1021/acs.est.8b04239
Qiu L, Wang H, Wang X (2016) Isolation and characterization of a cold-resistant PCB209-degrading bacterial strain from river sediment and its application in bioremediation of contaminated soil. J Environ Sci Health A 51:204–212. https://doi.org/10.1080/10934529.2015.1094324
Rotaru AE, Woodard TL, Nevin KP, Lovley DR (2015) Link between capacity for current production and syntrophic growth in Geobacter species. Front Microbiol 6:744. https://doi.org/10.3389/fmicb.2015.00744
Salimizadeh M, Shirvani M, Shariatmadari H, Nikaeen M, Leili Mohebi Nozar S (2018) Coupling of bioaugmentation and phytoremediation to improve PCBs removal from a transformer oil-contaminated soil. Int J Phytoremediat 20:658–665. https://doi.org/10.1080/15226514.2017.1393388
Sandhu M, Jha P, Paul AT, Singh RP, Jha PN (2020) Evaluation of biphenyl- and polychlorinated-biphenyl (PCB) degrading Rhodococcus sp. MAPN-1 on growth of Morus alba by pot study. Int J Phytoremediat. https://doi.org/10.1080/15226514.2020.1784088
Sha’arani S, Hara H, Araie H, Suzuki I, Mohd Noor MJM, Akhir FNM, Othman N, Zakaria Z (2019) Whole gene transcriptomic analysis of PCB/biphenyl degrading Rhodococcus jostii RHA1. J Gen Appl Microbiol 65:173–179. https://doi.org/10.2323/jgam.2018.08.003
Sharma JK, Gautam RK, Nanekar SV, Weber R, Singh BK, Singh SK, Juwarkar AA (2017) Advances and perspective in bioremediation of polychlorinated biphenyl-contaminated soils. Environ Sci Pollut R 25:16355–16375. https://doi.org/10.1007/s11356-017-8995-4
Shuai J, Yu X, Zhang J, Xiong A, Xiong F (2016) Regional analysis of potential polychlorinated biphenyl degrading bacterial strains from China. Braz J Microbiol 47(3):536–541. https://doi.org/10.1016/j.bjm.2014.12.001
Steliga T, Wojtowicz K, Kapusta P, Brzeszcz J (2020) Assessment of biodegradation efficiency of polychlorinated biphenyls (PCBs) and petroleum hydrocarbons (TPH) in soil using three individual bacterial strains and their mixed culture. Molecules 25(3):709. https://doi.org/10.3390/molecules25030709
Su XM, Liu YD, Hashmi MZ, Ding LX, Shen CF (2015) Culture-dependent and culture-independent characterization of potentially functional biphenyl-degrading bacterial community in response to extracellular organic matter from Micrococcus luteus. Microb Biotechnol 8:569–578. https://doi.org/10.1111/1751-7915.12266
Suenaga H, Fujihara H, Kimura N, Hirose J, Watanabe T, Futagami T, Goto M, Shimodaira J, Furukawa K (2017) Insights into the genomic plasticity of Pseudomonas putida KF715, a strain with unique biphenyl-utilizing activity and genome instability properties. Environ Microbiol Rep 9:589–598. https://doi.org/10.1111/1758-2229.12561
Taguchi K, Motoyama M, Iida T, Kudo T (2007) Polychlorinated biphenyl/biphenyl degrading gene clusters in Rhodococcus sp. K37, HA99, and TA431 are different from well-known bph gene clusters of Rhodococci. Biosci Biotechnol Biochem 71:1136–1144. https://doi.org/10.1271/bbb.60551
Takeda H, Yamada A, Miyauchi K, Masai E, Fukuda M (2004) Characterization of transcriptional regulatory genes for biphenyl degradation in Rhodococcus sp. strain RHA1. J Bacteriol 186:2134–2146. https://doi.org/10.1128/jb.186.7.2134-2146.2004
Teng Y, Li X, Chen T, Zhang M, Wang X, Li Z, Luo Y (2016) Isolation of the PCB-degrading bacteria Mesorhizobium sp. ZY1 and its combined remediation with Astragalus sinicus L. for contaminated soil. Int J Phytoremediation 18:141–149. https://doi.org/10.1080/15226514.2015.1073667
Tomza-Marciniak A, Pilarczyk B, Witczak A, Rzad I, Pilarczyk R (2019) PCB residues in the tissues of sea ducks wintering on the south coast of the Baltic Sea, Poland. Environ Sci Pollut Res Int 26:11300–11313. https://doi.org/10.1007/s11356-019-04586-4
Voronina AO, Egorova DO, Korsakova ES, Plotnikova EG (2019) Diversity of the bphA1 genes in a microbial community from anthropogenically contaminated soil and isolation of new Pseudomonads degrading biphenyl/chlorinated biphenyls. Microbiology 88(4):433–443. https://doi.org/10.1134/S0026261719030172
Wagner DD, Hug LA, Hatt JK, Spitzmiller MR, Padilla-Crespo E, Ritalahti KM, Edwards EA, Konstantinidis KT, Loffler FE (2012) Genomic determinants of organohalide-respiration in Geobacter lovleyi, an unusual member of the Geobacteraceae. BMC Genomics 13:200. https://doi.org/10.1186/1471-2164-13-200
Wang H, Hu J, Xu K, Tang X, Xu X, Shen C (2018) Biodegradation and chemotaxis of polychlorinated biphenyls, biphenyls, and their metabolites by Rhodococcus spp. Biodegradation 29:1–10. https://doi.org/10.1007/s10532-017-9809-6
Wang S, Chng KR, Wilm A, Zhao S, Yang KL, Nagarajan N, He J (2014) Genomic characterization of three unique Dehalococcoides that respire on persistent polychlorinated biphenyls. P Natl Acad Sci USA 111:12103–12108. https://doi.org/10.1073/pnas.1404845111
Wang S, He J (2012) Two-step denaturing gradient gel electrophoresis (2S-DGGE), a gel-based strategy to capture full-length 16S rRNA gene sequences. Appl Microbiol Biotechnol 95:1305–1312. https://doi.org/10.1007/s00253-012-4251-5
Wang X, Teng Y, Luo Y, Dick RP (2016) Biodegradation of 3,3',4,4'-tetrachlorobiphenyl by Sinorhizobium meliloti NM. Bioresour Technol 201:261–268. https://doi.org/10.1016/j.biortech.2015.11.056
Warenik-Bany M, Maszewski S, Mikolajczyk S, Piskorska-Pliszczynska J (2019) Impact of environmental pollution on PCDD/F and PCB bioaccumulation in game animals. Environ Pollut 255:113159. https://doi.org/10.1016/j.envpol.2019.113159
Wójcik A, Perczyk P, Wydro P, Broniatowski M (2020) Dichlorobiphenyls and chlorinated benzoic acids-Emergent soil pollutants in model bacterial membranes. Langmuir monolayer and grazing incidence X-ray diffraction studies. J Mol Liq 307:112997. https://doi.org/10.1016/j.molliq.2020.112997
Wu Q, Milliken CE, Meier GP, Watts JE, Sowers KR, May HD (2002) Dechlorination of chlorobenzenes by a culture containing bacterium DF-1, a PCB dechlorinating microorganism. Environ Sci Technol 36:3290–3294. https://doi.org/10.1021/es0158612
Xing Z, Hu T, Xiang Y, Qi P, Huang X (2020) Degradation mechanism of 4-chlorobiphenyl by consortium of Pseudomonas sp. strain CB-3 and Comamonas sp. strain CD-2. Curr Microbiol 77:15–23. https://doi.org/10.1007/s00284-019-01791-9
Yang X, Liu X, Song L, Xie F, Zhang G, Qian S (2007) Characterization and functional analysis of a novel gene cluster involved in biphenyl degradation in Rhodococcus sp. strain R04. J Appl Microbiol 103:2214–2224. https://doi.org/10.1111/j.1365-2672.2007.03461.x
Yang Y, Higgins SA, Yan J, Simsir B, Chourey K, Iyer R, Hettich RL, Baldwin B, Ogles DM, Loffler FE (2017) Grape pomace compost harbors organohalide-respiring Dehalogenimonas species with novel reductive dehalogenase genes. ISME J 11:2767–2780. https://doi.org/10.1038/ismej.2017.127
Acknowledgements
This work was funded by the National Natural Science Fund of China (41671317), the Fundamental Research Funds for the Central Universities (KYYJ202002), and the Jiangsu Agriculture Science and Technology Innovation Fund (CX(18)1005).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Ethical statement
The authors declared that the work has been approved by Ethical Committee. And the work we submitted has not been published elsewhere, either completely, in part, or in another form. The manuscript has not been submitted to another journal and will not be published elsewhere.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Xiang, Y., Xing, Z., Liu, J. et al. Recent advances in the biodegradation of polychlorinated biphenyls. World J Microbiol Biotechnol 36, 145 (2020). https://doi.org/10.1007/s11274-020-02922-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-020-02922-2