Abstract
Acidithiobacillus ferrooxidans (A. ferrooxidans) ATCC 23270 is a model bacteria for bioleaching research. Because of the use of extractant in metal extraction industry, A. ferrooxidans needs to cope with the water-organic two-phase system. To get insight into the molecular response of A. ferrooxidans to organic solvent, global gene expression pattern was examined in A. ferrooxidans ATCC 23270 cells subjected to Lix984n (an organic extractant) using the method of whole-genome DNA microarray. The data suggested that the global response of A. ferrooxidans to Lix984n stress was characterized by the up-regulation of genes involved in pentose phosphate pathway, fatty acid and glutamate biosynthesis. In further study, compared to heterotrophic bacteria in dealing with short-time stress, A. ferrooxidans has a special strategy of continuously enhancing the expression of genes encoding proteins involved in electron transport, such as petI, petII, cyo and cyd. Besides, acrAB-tolC operon encoding organic solvent efflux pump and its positive regulator gene ostR were addressed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bioleaching aims to extract valuable metals, such as copper, nickel, and gold through microbial oxidation of minerals (Rawlings 2002). In this process, extractant is usually added to enrich metal ions, and then the solubilized metals ions are transformed into metal by electrowinning. It is considered to be an economical and an ecological friendly process commonly used by the miners as an alternative method to roasting or smelting, especially when there are lower concentrations of metal in the ore (Plessis et al. 2007).
There are two important steps that influence the operation of bioleaching industry, namely leaching of metal ions from ores and extracting metal ions from the resulting leaching solution using high efficient extractant. Microbes play a key role in mineral leaching stage. Although the molecular mechanism of bioleaching was still unclear, the leaching rate with the microbes is significantly higher than the one without microbes (Fowler and Crundwell 1999; Rohwerder et al. 2003). During the extraction process, physical and chemical properties of extractant exert significant effects on the extraction efficiency of metal ion. Among all extractants used in copper extraction industry, Lix984n, a mixture of aldoxime and ketoxime (Henkel Corporation), is of widespread availability (Sengupta et al. 2006). Because the raffinate containing organic solvents from extraction system usually returns to leaching solution, it is inevitable for bioleaching microbes to be immersed in the water-organic two-phase system. Torma and Itzkovitch (1976) had studied the effects of nineteen extractant reagents on A. ferrooxidans, a typical bacterium in bioleaching environments. Their study showed that the oxidation ability of microorganism was impaired to varying degrees by different reagents, and LIX reagents demonstrated a moderate damage to A. ferrooxidans. Alexander et al. (1987) reported that organic acid will result in acidification of cytoplasm and impair bacteria respiration by influencing their oxygen reduction process. In recent years, real-time PCR has been used to study the response of A. ferrooxidans to organic solvent, revealing that the expression of cbb genes were substantially changed in A. ferrooxidans under Lix984n stress (Wang et al. 2008). Few studies, especially on a whole-genome scale, are reported on the response of bioleaching bacteria to organic solvent stress (Wang et al. 2008; Torma and Itzkovitch 1976; Alexander et al. 1987).
Transcriptional level analyses have been carried out concerning gram-negative heterotrophic bacteria under stress responses such as heat shock, cold shock and organic shock using whole-genome DNA microarray (Cuevas et al. 2006; Gao et al. 2004, 2006), but relatively few studies have been conducted on autotrophic bacteria. In 2008, a whole-genomic 50-mer oligonucleotide microarray was developed based on the 3,131 open read frames (ORFs) of A. ferrooxidans ATCC 23270 genome in our laboratory (Li et al. 2008). It offers a new way to explore the stress responses of bioleaching bacteria at transcriptome level.
In this work, the whole-genome array was employed to conduct the transcriptome analysis of A. ferrooxidans ATCC 23270 in response to 1% (v/v) Lix984n stress. According to the result, there was significant difference of gene expression pattern between treated and untreated samples. Meanwhile, real-time quantitative reverse transcription-PCR was used to measure the mRNA expression level of some important genes. It is anticipated that knowledge of global transcriptional responses to Lix984n stress of A. ferrooxidans will be useful in elucidating the specific mechanisms that function in cellular survival and adaptation to organic stress.
Materials and methods
Bacterial strains, culture, and growth conditions
Acidithiobacillus ferrooxidans ATCC 23270 obtained from American Type Culture Collection (ATCC) was cultivated at 30°C in medium containing 0.5 g/l MgSO4·7H2O, 3.0 g/l (NH4)2SO4, 0.5 g/l K2HPO4, 0.1 g/l KCl and 0.01 g/l Ca(NO3)2. The medium (pH = 2.0) was sterilized at 120°C for 20 min and added elemental sulfur to 10.0 g/l.
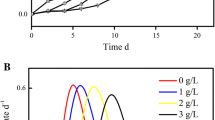
To understand the impact of Lix984n on the growth of A. ferrooxidans ATCC 23270, A. ferrooxidans cells (density is 7.33 × 107 cells/ml) were exposed to medium containing different amounts of Lix984n [ranging from 0 to 10% (v/v)] when the cells reached exponential phase (~95 h) (Fig. 1). Growth yield was measured every 4 h for each condition. In the present study, 1% Lix984n was chosen to investigate the endurance mechanism of A. ferrooxidans ATCC 23270.
For microarray analysis, when the cells were in mid-log phase (~95 h), eight volumes (400 ml) of bacterial culture were condensed to one volume (50 ml) by centrifugation (10,000×g) for 10 min at 4°C in a 5,804 R centrifuge (Eppendorf, Wesbury, NY) and re-suspended with sterilized fresh medium when the cells were in mid-log phase. Samples (cellular density is about 5.86 × 108 cells/ml) were collected from three independent experiments grown under 0% (non stress condition) and 1% (stress condition) Lix984n for 0 (control), 5, 20, 40, 80 min, respectively, and centrifuged at 10,000×g for 10 min at 4°C in a 5804R centrifuge (Eppendorf, Wesbury, NY). The resulting 27 pellets were frozen immediately in liquid nitrogen and stored at −80°C before RNA extraction.
Whole genome array of A. ferrooxidans ATCC 23270
The whole genome array used in this work has been described in detail previously (Li et al. 2008). The 50-mer oligonucleotide whole-genome array was developed based on the 3,131 ORFs annotated in A. ferrooxidans ATCC 23270 genome (http://www.ncbi.nlm.nih.gov/). Twenty categories are included and described in the legend of Fig. 2. Each probe was printed in triplicate onto silicon chip.
Differential expression of genes grouped by functional classification according to the NCBI A. ferrooxidans ATCC 23270 genome databases and COGs classification (Tatusov et al. 1997). Columns: 1 biosynthesis of cofactors, prosthetic groups, and carriers; 2 central intermediary metabolism; 3 mobile and extrachromosomal element functions; 4 amino acid biosynthesis; 5 cell envelope; 6 cellular processes; 7 disrupted reading frame; 8 DNA metabolism; 9 energy metabolism; 10 fatty acid and phospholipid metabolism; 11 hypothetical proteins-conserved; 12 hypothetical proteins; 13 protein fate; 14 protein synthesis; 15 purines, pyrimidines, nucleosides, and nucleotides; 16 regulatory functions; 17 signal transduction; 18 transcription; 19 transport and binding proteins; 20 unknown function
Total RNA extraction and purification
Total cellular RNA from A. ferrooxidans grown in the presence and absence of Lix984n was isolated using the TRIzol Reagent (Invitrogen, Carlsbad, USA) following the manufacturer’s instructions. RNA samples were treated with RNase-free DNaseI (QIAGEN, Carlsbad, USA) to digest residual chromosomal DNA and then purified with an RNeasy kit (Qiagen, Valencia, USA). Total cellular RNA was quantified at OD260 and OD280 with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). Three replicates of purified RNA for each sample mixed and the mixed RNA served as the template for labeling.
Labeling and hybridization conditions
RNA samples from A. ferrooxidans in non stress and stress conditions were labeled with Cy3-labeled dUTP during cDNA production. 10 μg of total cellular RNA was mixed with 10 μg of random primer at 70°C for 10 min, then immediately chilled by ice for 2 min. The labeling reaction mixture, which contains 5× reverse transcription buffer; 5 mM MgCl2; 5 mM each of dATP, dTTP, and dGTP; 2.5 mM dCTP (New England Biolabs, Beverly, Mass); 1 mM Cy3 dUTP (Amersham Pharmacia Biotech, Piscataway, USA); and 40 units of RNAase OUT (Invitrogen), was added to the RNA primer solution and then incubated at 25°C for 5 min. Superscript II reverse transcriptase (200 units, Invitrogen) was added to the solution and the mixture was incubated at 25°C for 10 min and then 42°C for 2 h. The labeled cDNA probes was treated with 1 mM NaOH to remove residual RNA, and purified with a Qiagen PCR purification column, then concentrated in speedvac at 45°C for 30 min.
The purified and fluorescently labeled probes were dissolved in 30 μl of hybridization buffer containing 50% (v/v) formamide, 3× SSC (containing 0.45 mM NaCl and 0.045 mM sodium citrate), 20 μg of unlabeled herring sperm DNA (Promega, USA), and 0.3% (v/v) sodium dodecyl sulfate (SDS). The hybridization mixture was heated at 98°C for 2 min, applied directly onto the microarray slide, then covered with a glass coverslip. After that the microarray was placed into a hybridization chamber (Corning, USA), and immersed into the preheated water bath (45°C) immediately for 10–12 h. After hybridization, each microarray slide was taken out, and washed sequentially in 1× SSC-0.2% SDS for 5 min, 0.1× SSC-0.2% SDS for 10 min and 0.1× SSC for 30 s. Drying of the microarray slide was necessary before data scanning.
Data analysis
After hybridization, microarrays were scanned with a ScanArray 4100 Microarray Analysis System (AXON instruments, Inc, USA).The emitted fluorescent signal was detected by a photomultiplier tube (PMT) at 532 nm. After background subtraction, signal intensities for each spot were quantified using GenePix Pro version 6.0 (AXON instruments, Inc, USA). The SNRs from three replicate data sets were then averaged to represent the SNR for a particular probe. A commonly accepted criterion for the minimum signal (threshold) that can be accurately quantified is Signal to Noise Ratios (SNRs) ≥2. Spots that appeared to be lower than the threshold value were removed from the data set. The data sheet from GenePix Pro was then exported to Excel for further processing. Spot intensities and median values for each spot were normalized by average intensity of each slide to account for any difference in total intensity between the scanned images. The processed stress and non stress signal intensities for each spot were used for calculating the Log transformed expression ratio (log2 stress/non stress). Genes with a |log2 ratio| larger than 1.5 are considered differentially expressed and genes P value <0.05 are considered significant. Three replicate data sets in each slide were used for further statistical analysis of the data. The raw data, as well as the processed data, for all hybridizations were submitted to the Gene Expression Omnibus database and are available under the accession code GSE28538. The time-course changes of genes during Lix984n shock were determined using hierarchical cluster analysis (CLUSTER) and visualized with TREEVIEW (Eisen et al. 1998).
Reverse transcription-quantitative PCR analysis
To validate the microarray results, four genes were selected for real-time quantitative reverse transcription-PCR (RT-PCR) analysis with the same RNA samples used in the array hybridization. Primer pairs were designed using Primer Premier 5.0 software and synthesized by Sagon Biotech, Shanghai, China. All primers were listed in Table 1. The results were normalized against a control gene (16S rRNA) to correct sample-to-sample variation. The standard curve was derived from PCR product representing each gene with genomic DNA as the template and serially diluted to 5–6 orders of magnitude. The reactions were performed with 40 cycles of 30 s at 94°C, 30 s at 58°C, and 1 min at 72°C, and monitored in an iCycler iO Real-time PCR detection system (Bio-Rad, Hercules, Calif.). The expression of each gene was determined from three replicates on a single RT-PCR experiment. The ratios of the gene expression data under different treatments were calculated by the ratios of signal intensities between stress and non stress, and normalized by 16S rRNA gene expression and diluted folds.
Results and discussion
Growth response
Growth curves of A. ferrooxidans grown in five different concentrations [0% as control, 1, 3, 6, 10% (v/v)] of Lix984n were showed in Fig. 1. After about 32 h in the exponential phase, A. ferrooxidans cells under non stress condition reached the stationary phase with the highest cell density (1.1 × 108 cells/ml) at 100 h. Results showed that the growth of A. ferrooxidans was inhibited immediately and resumed after a lag period (~20 h) when logarithmically growing A. ferrooxidans cells were exposed to 1% Lix984n. This phenomenon indicated that 1% Lix984n concentration is permissive for growth. Cells growing in the medium containing 1% Lix984n entered stationary-phase with almost equal cell density (1.07 × 108 cells/ml) to the untreated samples. The growth rate decreased significantly under 3% Lix984n stress, and cell growth was drastically reduced under 6 and 10% Lix984n stress, demonstrating that those toxic concentrations are lethal. In the industrial operations, the concentration of Lix984n is usually 1–10% (Readett et al. 2003; Liu et al. 2002; Lan et al. 2005). When the concentration of Lix984 is 1%, the copper extraction rate is 96.94% after an equilibration time of 5 min (Lan et al. 2005). Therefore, 1% Lix984n was chosen to be the stress concentration for the following transcriptional analysis.
General overview of transcriptome response
Comprehensive investigation on molecular responses of A. ferrooxidans to Lix984n stress was performed through hybridization of whole genome array with total cellular cDNA acquired from samples taken at different time points during cultivation. 401 (248/153, induced/repressed), 873 (860/13), 512 (323/189), and 719(58/661) genes exhibited significant expression differences at 5, 20, 40 and 80 min, respectively (Supplementary Tables). These genes correspondingly represented 13% (5 min), 28% (20 min), 16% (40 min), and 23% (80 min) of the 3,131 ORFs spotted on the array. Figure 2 summarized differently expressed genes grouped by functional classification at each of the four time points. Genes showed significant change in expression level involved in the process of central intermediary metabolism, cell envelope, cellular processes, fatty acid and phospholipid metabolism, amino acid biosynthesis, DNA metabolism, energy metabolism and protein fate and the classification procedure used was based on COGs classification (Tatusov et al. 1997). The number of up-regulated genes increased after Lix984n stress, peaked at 20 min, and then declined over time. The number of down-regulated genes had a temporary decline, obtaining the minimum at 20 min, and then increased to the maximum at 80 min. The time point of 20 min is considered to be a boundary for the gene expression pattern. Hence, the phases were divided into short-time treatment phases including 5 and 20 min and long-time treatment phases including 40 and 80 min.
Membrane level regulation
Cell envelope is the first barrier to defense against Lix984n toxicity. In this study, four aspects were proposed to explicate the antitoxic mechanism of cells in membrane level. Firstly, membrane components were changed to enhance membrane rigidity. Three genes involved in fatty acid and phospholipid anabolism were significantly induced in short-time phase (AFE_2067, AFE_1905, AFE_2124) and AFE_1905 was repressed in long-time phase, yet AFE_2124 was constantly up-regulated according to genome-wide analysis (Table 2). Cell surface hydrophobicity and altered membrane components can play a vital role in preventing the penetration of organic solvent molecules from entering the cell (Abe et al. 2003; Aono and Kobayashi 1997). In Pseudomonas putida, two important phospholipid alterations were induced under short-term organic solvent stress, such as (1) rapid transformation of the cis fatty acids C16: 1, 9 and C18: 1, 9 into their trans isomers and (2) changes of the phospholipid head-group composition (Junker and Ramos 1999; Ramos et al. 1997). A similar strategy might be employed by A. ferrooxidans during organic stress response. Unfortunately, there have been few studies on the response change under organic solvent stress of A. ferrooxidans to date.
Secondly, a series of genes with potential functions as membrane transportation and binding proteins (unknown substrate) exhibited obvious co-regulation, which up-regulated during short-time stress and returned to a normal level during long-time stress (Table 2; Fig. 3). The genes include an outer membrane efflux protein gene tolC, two efflux transporter genes acrAB, a conserved hypothetical protein gene hyp, and a rhodanese-like domain protein gene rhd. These results suggested that it is possible for these genes acrAB-tolC, hyp and rhd to constitute an operon that can encode an efflux pump closely related to organic tolerance. AcrAB-tolC was extensively reported in Escherichia coli and other gram-negative bacterium (Nishino et al. 2003; Poole 2004). It belongs to resistance-nodulation-division (RND) family and functions as an efflux pump associated with antibiotics and organic solvents transportation. RND efflux pumps (AcrAB-TolC) are reported to transport two kinds of substrates: (1) some drugs such as tetracycline, chloramphenicol and fluoroquinolones (2) some harmful solvent such as octane, heptane, and n-hexane (Nikaido 1996; Tsukagoshi and Aono 2000). The substrates are usual small molecule. Lix984n, with a molecular weight of no more than three hundred, is composed of Lix84 (2-hydroxy-5-nonylacetophenoneoxime) and Lix860N (5-nonylsalicylaldoxime), and it is a potential substrate for RND efflux pumps (Liu et al. 2002). More work should be done in this supposition. An attempt is taken by Takatsuka et al. (2010) to discover the recognition mechanism of compounds for AcrB via computer simulation of protein structure. They found that many compounds are predicted to bind a pocket in the periplasmic domain of the binding protomer thought a narrow groove or a wide cave. Another gene, ostR, encoding a transcriptional regulator located in the upstream of acrAB-tolC, had similar expression change with acrAB-tolC. OstR was presumed to be a positive regulator of acrAB-tolC since it shared a similar expression pattern with acrAB-tolC., The rhd gene encoding a rhodanese-like protein was reported to have 3-mercaptopyruvate/cyanide sulfurtransferase (MST) activities (Acosta et al. 2005). More work remains to be done with regard to the operon.
Thirdly, A. ferrooxidans genes involved in sulfur energy metabolism were up-regulated during short-time Lix984n stress. For these chemoautotrophic bacteria, A. ferrooxidans, the sulfur oxidation is the main energy source. During the process of sulfur metabolism, proton gradient generates between the two sides of intercellular membrane, and the oxygen is the terminal electron acceptor (Quatrini et al. 2006; Valdes et al. 2008). Although the identification of some enzymes in this process such as thiosulfate quinol reductase (TQR) are controversial, some operons encoding protein involved in electron transport anchoring on the intracellular membrane are acknowledged (Fig. 4b). The gene expression of electron transport genes could reflect energy metabolism situation. In this study, in general, energy metabolism enhanced during short-time treatment, and decreased thereafter. Genes encoding electron transport proteins such as operon cyd, operon cyo, operon petI and operon petII were significantly induced during short-time treatment and repressed afterwards (Table 2; Fig. 4a). (1) For cyd operon (including genes cydA and cydB), which encodes bd ubiquinol oxidase, expressed in a typical pattern (increasing at short-time phase and decreasing at long-time phase). (2) For cyo operon (including genes cyoA, cyoB, cyoC and cyoD), which encodes bo 3 ubiquinol oxidase, only cyoC showed down-regulation in long-time phase and other three genes unchanged significantly. (3) In petI operon (petC-1, petB-1, petA-1 and cycA-1), the expression of the first three genes markedly changed. (4) Compared to petI operon, petII operon (including genes petC-2, petB-2, petA-2, cycA-2 and iro) showed a hysteretic pattern with petC-2 and cycA-2 down-regulated in long-time phase.
a Expression profile for genes encoding electron transport protein. b A. ferrooxidans sulfur energy metabolism model derived from Quatrini et al. (2006) and Zhang et al. (2007). The transcriptional units and the corresponding redox proteins are presented in the same color: operon petI and operon petII (bc 1 complex, CycA and iro); operon cyo (bo 3 ubiquinol oxidase); operon cyd (bd ubiquinol oxidase). TQR, thiosulfate quinol reductase; SQR, sulfide quinone reductase
Few investigations of stress response in chemolithoautotrophic bacteria have been carried out under organic stress condition. Interestingly, energy metabolism responding to Lix984n in A. ferrooxidans was distinct in comparison with other works on heterotrophic bacteria. The energy metabolism in Pseudomonas putida was down-regulated when it suffered from toluene, o-xylene or 3 MB in the short-time treatment (15 min) (Cuevas et al. 2006), but A. ferrooxidans had a short-time increase of energy metabolism before a long-time decrease. Several hypotheses were proposed to explain this phenomenon. (1) Trophic type (autotrophic or heterotrophic) is a key factor for different energy consumption. Acidithiobacillus ferrooxidans lack curial enzymes in Embden-Meyerhof-Parnas (EMP), Entner-Doudoroff (ED) and Tricarboxylic Acid cycle (TCA), which results in the fact that A. ferrooxidans suffer from energy deficiency from substrate-level phosphorylation (Valdes et al. 2008). Therefore, A. ferrooxidans need to utilize more sulfur to subsist. Another proof for this is that operon petII plays a major role in sulfur oxidation compared to operon petI (Quatrini et al. 2006). But we have identified that genes encoding operon petI presented longer induction time than operon petII. (2) Under Lix984n stress, different organic toxicity may trigger different response. In our work, less intense organic toxicity may make the A. ferrooxidans adjust to organic environment more easily. Growth restoration under 1% Lix984n can be another evidence for this hypothesis (Fig. 1).
Cytoplasm level regulation
Carbon metabolism associated genes were analyzed in A. ferrooxidans ATCC 23270 in order to get insights about some special pathway under Lix984n stress. Central carbon metabolism can be divided into five parts: fixation of carbon, pentose phosphate pathway, glycolysis, TCA cycle and glycogen metabolism (Table 2; Fig. 5). Main approaches and the corresponding functional genes were displayed. Figure 5 illuminated that the expression of genes involved in carbon metabolism comprehensively changed, especially in fatty acids, phospholipids and pentose phosphate pathway. Additionally, the expression of genes associated with incomplete TCA cycle might be concernful when A. ferrooxidan suffered form Lix984n stress.
Central carbon metabolism derived from Valdes et al. (2008). (1) Up-regulated, (2) down-regulated, (S) short-time phase, (L) long-time phase
In this study, the first potential strategy for A. ferrooxidans to accommodate Lix984n stress in cytoplasm level is associated with enhancing anabolism, especially for fatty acids, phospholipids and glycogens. It can be explained that fatty acids, phospholipids and glycogens are essential materials for cell envelope buildup (polysaccharides and lipopolysaccharides). Besides, fatty acid biosynthesis was reported to be involved in solvent tolerance in Pseudomonas putida (Segura et al. 2004). Although over-expression of efflux pump operons was detected in the solvent-sensitive mutant of Pseudomonas putida, this mutant showed a less efficient in solvent extrusion than the wild-type cells. By the approach of isotopic tracing, negligible synthesis of fatty acids from 13CH3–13COOH was obtained in the presence of sublethal concentrations of toluene (Segura et al. 2004).
Active amino acid metabolism, such as glutamate, tryptophan, serine and cysteine metabolism can reflect active protein metabolism. We inferred that it may be another protection method to resist Lix984n stress. Impressively, during short-time shock, the biosynthesis of glutamate family amino acid (glutamine, arginine, and proline) was induced. Glutamine synthetase gene AFE_0466/glnA, N-acetylglutamate synthase gene AFE_0664/argA, acetylglutamate kinase gene AFE_0256/argB, argininosuccinate synthase gene AFE_0404/argG, and glutamate 5-kinase gene AFE_1783/proB-2 were 9.99, 9.72, 9.69, 10.17, and 9.90-fold induced, respectively. Moreover, genes involved in tryptophan synthesis were decreasingly regulated, such as AFE_2068 (trpA, tryptophan synthase, alpha subunit: −10.15-fold) AFE_2069 (trpB, tryptophan synthase, beta subunit: −11.71-fold). Besides, cysteine biosynthesis genes (e.g. cysM, cysteine synthase B: 10.42-fold) was induced in early response. Oxidative stress caused by invasion of Lix984n through impaired membrane probably resulted in inaccurate synthesis of protein and nucleic acid, which will lead to low efficiency of cell function. Under Lix984n stress condition, A. ferrooxidans showed enhanced metabolism regarding to amino acid and inductive synthesis of proteins associated with protein folding and stabilization, DNA recombination and repair. The RecA protein was reported to play an essential role in homologous genetic recombination, DNA repair, induction of the SOS response, and initiation of stable DNA replication in E. coli (Roca and Cox 1997). Interestingly, recX (1.11-fold in short-time phase and 1.09-fold in long-time phase) that encodes regulatory protein for recA was continuously induced in the course of the treatment, but expression of recA gene in A. ferrooxidans remained unchanged. This suggested that RecA might play a minor role in DNA repair in A. ferrooxidans, and RecX might launch other RecA-like protein to perform the emergency responses. A recA mutant reported in A. ferrooxidans could be an evidence to our viewpoint, indicating that the A. ferrooxidans recA mutants were less sensitive to these physical DNA-damaging agents than the recA mutants described in other bacterial species (e.g. E. coli) (Liu et al. 2000).
Pentose is continuously synthesized during the pentose phosphate pathway (Table 2; Fig. 5). Three genes (fba-2, tkt-1, tkt-2) out of four encoding important enzymes during erythrose-4-phosphate changed into ribose-5-phosphate were induced in short-time phase. Intriguingly, gene AFE_2679 encoding metallo-beta-lactamase family protein, which catalyzes the reaction of the transformation from ribose-5-phosphate to ribulose-5-phosphate, was highly induced in short-time phase (9.68-fold in short-time phase). For one thing, active DNA metabolism is a reasonable explanation for pentose biosynthesis; for another, pentose phosphate pathway is a main source of NADPH, which has vital effect on maintaining the redox condition against oxidative stress. Besides, derivatives of pentose metabolism such as NAD+, FAD and Coenzyme A are important coenzymes for many metabolic processes (e. g. carbon and nitrogen fixation) (McKee and McKee 1999). The above analyses suggested that pentose phosphate pathway is a pivotal pathway responding to Lix984n stress.
Acidithiobacillus ferrooxidans have an incomplete TCA cycle since α-ketoglutarate dehydrogenase has not been discovered (Valdes et al. 2008). Genes involved in glutamine biosynthesis from α-ketoglutarate were up-regulated in A. ferrooxidans under Lix984n stress, which was also found in other gram-negative bacterium under adverse environment stress. Cuevas et al. (2006) found that glutamine biosynthesis increased in Pseudomonas putida after short-time toluene stress, and deduced that it may relate to signal transfer. Genes putatively involved in glutamine biosynthesis were up-regulated in Shewanella oneidensis MR-1 under high salinity stress (Liu et al. 2005). We speculated that it may involve in glutathione biosynthesis against oxidative stress since cysteine biosynthesis genes were also induced. Furthermore, carbon and sulfur metabolism under Lix984n stress had similar regulation pattern: enhancement in short-time phase and inhibition in long-time phase. Glutamine works as a potential material of signal transmission in A. ferrooxidans.
Reverse transcription-quantitative PCR validation
Reverse transcription-quantitative PCR (RT-PCR) was utilized to measure the mRNA expression levels of four genes in order to examine the validity of microarray data. The microarray and RT-PCR analysis was consistent (gene name: RT-PCR results/gene microarrays result; AFE_2347: 1.08/1.75-fold; omp40, major outer membrane protein 40: 1.49/1.20-fold; sdhC, succinate dehydrogenase/fumarate reductase, C subunit: −1.91/−2.23-fold; cbbR, rubisco operon transcriptional regulator: −0.66/−0.25-fold), with the correlation coefficient of R 2 = 0.9208, providing cogent evidence to support the microarray data.
Conclusions
This study provides the first as well as comprehensive profile to elucidate the Lix984n stress upon A. ferrooxidans ATCC 23270. The tolerance mechanism can be divided into membrane level and cytoplasm level. These two levels are intimately connected and together determine the tolerance mechanism of bacteria. Membrane level mechanism includes: (1) enhanced membrane mechanical rigidity to prevent Lix984n from being penetrated by changing surface shape and its chemical components; (2) effective organic solvent efflux pump was induced to reduce the concentration of Lix984n in periplasm space; (3) supplied energy for increasing bacteria activities through significantly activating electron transport genes during early response; cytoplasm level mechanism includes: (1) up-regulation of fatty acid, phospholipid, and glycogen metabolism genes to provide materials for maintaining membrane mechanical rigidity; (2) alleviation and repairment of the damage on protein and nucleic acid caused by Lix984n stress by activating the metabolism of multiple amino acids and other substrates and inducing genes associated with protein folding and stabilization, DNA recombination and repairing; (3) continuously up-regulation of pentose phosphate pathway could not only supply NADPH to resist oxidative stress caused by membrane impairment, but also offer pentose for DNA metabolism; (4) enhanced glutamine biosynthesis could not only supply original material for antioxidant glutathione but also play a key role in signal transduction for stress tolerance.
References
Abe S, Okutsu T, Nakajima H, Kakuda N, Ohtsu I, Aono R (2003) n-Hexane sensitivity of Escherichia coli due to low expression of imp/ostA encoding an 87 kDa minor protein associated with the outer membrane. Microbiology 149:1265–1273
Acosta M, Beard S, Ponce J, Vera M, Mobarec JC, Jerez CA (2005) Identification of putative sulfurtransferase genes in the extremophilic Acidithiobacillus ferrooxidans ATCC 23270 genome: structural and functional characterization of the proteins. OMICS 9:13–29
Alexander B, Leach S, Ingledew WJ (1987) The relationship between chemiosmotic parameters and sensitivity to anions and organic acids in the acidophile Thiobacillus ferrooxidans. Microbiology 133:1171–1179
Aono R, Kobayashi H (1997) Cell surface properties of organic solvent-tolerant mutants of Escherichia coli K-12. Appl Environ Microbiol 63:3637–3642
Cuevas PD, Pastor JE, Marques S, Ramos JL, Lorenzo V (2006) Transcriptional tradeoff between metabolic and stress-response programs in Pseudomonas putida KT2440 cells exposed to toluene. J Biol Chem 281:11981–11991
Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95:14863–14868
Fowler TA, Crundwell FK (1999) Leaching of Zinc sulfide by Thiobacillus ferrooxidans: bacterial oxidation of the sulfur product layer increases the rate of Zinc sulfide dissolution at high concentrations of ferrous ions. Appl Environ Microbiol 65:5285–5292
Gao HC, Wang Y, Liu XD, Yan TF, Wu LT, Alm E, Arkin A, Thompson DK, Zhou JZ (2004) Global transcriptome analysis of the heat shock response of Shewanella oneidensis. J Bacteriol 186:7796–7803
Gao HC, Yang ZK, Wu LY, Thompson DK, Zhou JZ (2006) Global transcriptome analysis of the cold shock response of Shewanella oneidensis MR-1 and mutational analysis of its classical cold shock proteins. J Bacteriol 188:4560–4569
Junker F, Ramos JL (1999) Involvement of the cis/trans isomerase Cti in solvent resistance of Pseudomonas putida DOT-T1E. J Bacteriol 181:5693–5700
Lan ZY, Hu YH, Liu JS, Wang J (2005) Solvent extraction of copper and zinc from bioleaching solutions with LIX984 and D2EHPA. J Cent S Univ Technol 12:45–49
Li Q, Shen L, Luo HL, Yin HQ, Liao LQ, Qiu GZ, Liu XD (2008) Development and evaluation of whole-genome oligonucleotide array for Acidithiobacillus ferrooxidans ATCC 23270. Trans Nonferrous Met Soc China 18:1343–1351
Liu ZY, Guiliani N, Ayme CA, Borne F, Ratouchniak J, Bonnefoy V (2000) Construction and characterization of a recA mutant of Thiobacillus ferrooxidans by marker exchange mutagenesis. J Bacteriol 182:2269–2276
Liu JS, Qiu GZ, Ge YQ, Xu J (2002) Extraction of copper from bacterial leach solution using Lix984. Trans Nonferrous Met Soc China 12:313–316
Liu YQ, Gao WM, Wang Y, Wu LY, Liu XD, Yan TF, Alm E, Arkin A, Thompson DK, Fields MW, Zhou JZ (2005) Transcriptome analysis of Shewanella oneidensis MR-1 in response to elevated salt conditions. J Bacteriol 187:2501–2507
McKee T, McKee JR (1999) Biochemistry: an introduction. McGraw-Hill, New York
Nikaido H (1996) Multidrug efflux pumps of gram-negative bacteria. J Bacteriol 178:5853–5859
Nishino K, Yamada J, Hirakawa H, Hirata T, Yamaguchi A (2003) Roles of TolC-dependent multidrug transporters of Escherichia coli in resistance to ß-Lactams. Antimicrob Agents Ch 47:3030–3033
Plessis CA, Batty JD, Dew DW (2007) Commercial applications of thermophile bioleaching biomining. In: Rawlings DE, Johnson DB (eds) Biomining. Springer, Berlin, pp 61–64
Poole K (2004) Efflux-mediated multiresistance in gram-negative bacteria. Clin Microbiol Infect 10:12–26
Quatrini R, Ayme CA, Denis Y, Ratouchniak J, Veloso F, Valdes J, Lefimil C, Silver S, Roberto F, Orellana O, Denizot F, Jedlicki E, Holmes D, Bonnefoy V (2006) Insights into the iron and sulfur energetic metabolism of Acidithiobacillus ferrooxidans by microarray transcriptome profiling. Hydrometallurgy 83:263–272
Ramos JL, Duque E, Herva JR, Godoy P, Haidour A, Reyes F, Barrero AF (1997) Mechanisms for solvent tolerance in bacteria. J Biol Chem 272:3887–3890
Rawlings DE (2002) Heavy metal mining using microbes. Annu Rev Microbiol 56:65–91
Readett DJ, Hanlon M, Dudley KA, Crane PA (2003) Evolution of a copper solvent extraction circuit: how operating parameters were managed to counter varying leach solution conditions throughout the life of Girilambone copper company’s solvent extraction circuit. In: Proceedings of the Alta Copper conference. Perth, Jo-Al Enterprises, Melbourne
Roca AL, Cox MM (1997) RecA protein: structure, function, and role in recombinational DNA repair. In: Cohn WE (ed) Progress in nucleic acid research and molecular biology. Academic Press, California, pp 130–138
Rohwerder T, Gehrke T, Kinzler K, Sand W (2003) Bioleaching review part A: progress in bioleaching: fundamentals and mechanisms of bacterial metal sulfide oxidation. Appl Microbiol Biotechnol 63:239–248
Segura A, Duque E, Rojas A, Godoy P, Delgado A, Hurtado A, Cronan JE, Ramos JL (2004) Fatty acid biosynthesis is involved in solvent tolerance in Pseudomonas putida DOT-T1E. Environ Microbiol 6:416–423
Sengupta B, Sengupta R, Subrahmanyam N (2006) Copper extraction into emulsion liquid membranes using LIX 984 N-C®. Hydrometallurgy 81:67–73
Takatsuka Y, Chen C, Nikaido H (2010) Mechanism of recognition of compounds of diverse structures by the multidrug efflux pump AcrB of Escherichia coli. Proc Natl Acad Sci USA 107:6559–6565
Tatusov RL, Koonin EV, Lipman DJ (1997) A genomic perspective on protein families. Science 278:631–637
Torma RE, Itzkovitch RJ (1976) Influence of organic solvents on chalcopyrite oxidation ability of Thiobacillus ferrooxidans. Appl Environ Microbiol 32:102–107
Tsukagoshi N, Aono R (2000) Entry into and release of solvents by Escherichia coli in an organic-aqueous two-liquid-phase system and substrate specificity of the AcrAB-TolC solvent-extruding pump. J Bacteriol 182:4803–4810
Valdes J, Pedroso I, Quatrini R, Dodson RJ, Tettelin H, Blake R, Eisen JA, Holmes DS (2008) Acidithiobacillus ferrooxidans metabolism: from genome sequence to industrial applications. BMC Genom 9:597–620
Wang W, Xiao SM, Chao J, Chen QJ, Qiu GZ, Liu XD (2008) Regulation of CO2 fixation gene expression in Acidithiobacillus ferrooxidans ATCC 23270 by Lix984n shock. J Microbiol Biotechnol 18:1747–1754
Zhang CG, Xia JL, Wang J, Qiu GZ (2007) Progress on researches of sulfur oxidation system of Acidithiobacillus spp. Biotechnol Bull China 1:59–65
Acknowledgments
We thank Ming Xie, Xian Fu and Xiangru Fan for participating in revising the manuscript. This research was supported by the National Natural Science Foundation of China (No. 50621063, No. 31070104, No. 30900203), Doctoral Fund of Ministry of Education of China No. 200805330035), New Teacher Fund for Doctor Station, the Ministry of Education (No. 200805331078) and the National Basic Research Program (No. 2010CB630901).
Author information
Authors and Affiliations
Corresponding authors
Additional information
These authors Zhijun Zhou and Yun Fang contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhou, Z., Fang, Y., Li, Q. et al. Global transcriptional analysis of stress-response strategies in Acidithiobacillus ferrooxidans ATCC 23270 exposed to organic extractant—Lix984n. World J Microbiol Biotechnol 28, 1045–1055 (2012). https://doi.org/10.1007/s11274-011-0903-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-011-0903-3