Abstract
Recently, the importance of the Geminiviruses infecting cereal crops has been appreciated, and they are now being studied in detail. Barley and wheat strains of Wheat dwarf virus are recorded in most European countries. Information on complete sequences of isolates from the United Kingdom, Spain, and Austria are reported here for the first time. Analysis revealed that their sequences are very stable. Recombination between strains was recorded only for the barley strain. We identified several defective forms of the barley strain from barley and wheat, which do not influence symptom expression. Sequences of barley isolates infecting wheat were obtained that did not differ from the isolates from barley. Based on specific features of the SIR of the barley strains, it is suggested that they are assigned to one of the two proposed new clusters, A1 or A2.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cereal-infecting DNA viruses, such as Wheat dwarf virus (WDV), are members of the family Geminiviridae [1]. Their circular single-stranded (ss) DNA is packed in a geminate (twinned) virion [2]. Mastreviruses, of which Maize streak virus is the type member, are transmitted by leafhoppers, have a monopartite genome and mostly infect monocotyledonous plants belonging to the family of Poaceae. Recently, a phylogenetic analysis of the wheat (WDV-W) and barley (WDV-B) strains of WDV and Oat dwarf virus (ODV) indicates that they are separate species [3]. In addition, both forms can be further categorized as strains A to E [3].
The broad application of the Rolling circle amplification (RCA) of circular DNAs (circomics, [4]) resulted in the discovery of numerous new viruses belonging to this genus [5–9].
Wheat dwarf virus was detected first in Triticum aestivum L. in Czechoslovakia [10] and subsequently in several regions of Europe, Asia, and Africa [11–20]. It may cause yellowing, streaking of leaves, and severe stunting or even death of an infected plant. In 2007, Oat dwarf virus, which is similar to WDV, was recorded in Germany [5]. WDV and ODV are transmitted by the leafhopper Psammotettix alienus (Dahlb.) and P. provincialis may also be a vector [19].
There are contradictory reports on whether under natural conditions the barley strain can infect wheat and the wheat strain barley [21] or not [22]. Recently, Köklü et al. [23] reported the occurrence of WDV-B in wheat and Kundu et al. [18] of WDV-W in barley. A survey carried out in Germany (Drechsler et al., unpublished) indicates that in rare cases, WDV-W occurs in barley and WDV-B in wheat, which supports our previous findings [5]. Tobias et al. [20] record the wheat strain in barley and wheat, but only the barley strain in barley. Using agroinfection, it is possible to infect both barley and wheat with the barley strain [24].
It is known that different strains of geminiviruses infecting mono- and dicotyledonous plants may recombine and produce new variants of this virus [8, 25–29]. For WDV-B, there is only one case of recombination suspected [24], although there are more than 100 complete sequences of the different isolates published. In this case, a RDP-analysis indicates that recombination with an unknown virus was expected in the SIR. To identify possible recombinants, which might be a threat to cereal production, we regularly do circomics of samples, which are suspected of being infected with WDV/ODV. Some of the data are included in this report.
In 2012, a severe outbreak of WDV occurred in Austria [30, 31] and it is unknown whether this was attributable to some special genotype. There is no information on the presence of WDV in the UK. There is a report of its presence in Spain by Achon et al. [32] but no sequence data for its complete genomes. A complete sequence of a WDV isolate from France was the first to be obtained [33]. Sequencing of a more recently collected isolate from France would provide an indication of the stability of its genome.
Materials and methods
Plant material
The origin of the plant material analyzed is given in Table 1. In the first step, the material was analyzed for the presence of WDV by means of DAS-ELISA using polyclonal antibodies prepared in cooperation with Frank Rabenstein (JKI Quedlinburg).
Nucleic acid preparation
DNA was isolated from air dried 20 mg samples using a DNA isolation kit (NucleoSpin Plant, Machery-Nagel) following the manufacturer’s instructions. In some cases, the dried samples were stored at 4 °C for several months. DNA was eluted from the spin columns using 50 μl of elution buffer.
Rolling circle amplification, cloning, and sequencing
RCA using the Phi29 DNA polymerase in the TempliPhi kit (General Electric Healthcare) was done using 2 μl of eluted DNA. DNA was denatured by adding 5 μl of the sample buffer provided at 95 °C for 3 min. The RCA was carried out at 30 °C for 16 h after the addition of 5 μl of the reaction buffer and 0.2 μl of the enzyme mix. 2 μl of the amplified DNA were cleaved using 1 μl of a restriction enzyme (Fast Digest, Thermo Scientific) in 30 μl of Fast Digest buffer over a period of 15 min at 37 °C. DNA fragments were separated on 1 % agarose gels in TAE buffer and visualized by staining with ethidium bromide. Restricted DNA was gel-purified (Macherey–Nagel, DNA gel extraction kit) and inserted into pBluescript-AB (Stratagene) restricted by an appropriate endonuclease. Transformation was carried out into chemically competent cells of Escherichia coli XL-1 (Stratagene). Two independent clones of each product were chosen for sequencing using reverse, universal, and sequence-specific primers using a CEQ-DTCS Quick starter sequencing kit (Beckman-Coulter) on a Beckman-Coulter CEQ 8800 sequencer. Base calling was performed using Chromas (Technelysium) and SeqMan (DNAStar) software. Complete sequences were deposited in the EMBL database under the accession numbers listed in Table 1.
Sequence analysis and comparison
As suggested by Muhire et al. [3], the phylogenetic analysis was done using MUSCLE software implemented in Lasergene 11 (Megalign Pro, DNAStar). Sequence homology [%] was calculated using DNAMAN7 software (Lynnon Corp.).
Recombination analysis was done using Recombination Detection Program v. 4.22 (beta version, [34]).
The sequences obtained earlier using PCR amplification [35], but not submitted at that time, are also now included in the public databases (Table 1).
Results
In order to reveal variations in the sequences and differentiate between the barley and wheat strains, the DNA was first digested using restriction endonucleases HindIII, BamHI, and EcoRI [5]. If possible, complete fragments of the genome were cloned. The type of fragments cloned is given in Table 1.
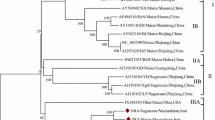
Kundu et al. [18] define typical clades for complete sequences of WDV, two for WDV-W and two for WDV-B. Muhire et al. [3] suggest that ODV should be regarded as a separate virus whereas the isolates from wheat, barley, and other cereals should be designated as strains A to E of WDV (see Fig. 1). Consequently, for the first phylogenetic analysis of the new sequence data typical members of these clades/strains were included (results not shown). The sequences of the first completely sequenced isolates of the wheat ([33]—sequence data not in databases, designated always as Matzeit; [29]—X02869) and barley strains [35] were also included. As a rule, sequences of parallel clones of the isolates investigated were nearly identical and thus not included in subsequent analyses. Of the primary sequences, only typical samples were retained for the subsequent analyses. The result of the phylogenetic analysis of the remaining 30 typical complete sequences is shown in Fig. 1.
There are two main clusters, one each for the wheat and barley isolates. The new wheat isolate from France forms a separate minor cluster together with the UK wheat isolate, which is distinct from the older French sequence. The new German wheat isolate is similar to the older German isolates. The new barley isolates from Spain form a separate cluster. The new barley isolate from Austria is very similar to some Czech isolates. The German barley isolates form a separate cluster together with the Bulgarian isolate. The clusters suggested by Muhire et al. [3] apply to all the newly sequenced isolates (Fig. 1). The subdivision of strain A into two substrains A1 and A2 seems reasonable. It is based mainly on differences in the SIR.
Recombination analysis was done using RDP and the reduced set of 30 sequences (Fig. 1) or a set of 26 sequences in which that of ODV and the non-typical isolates of strains B, C, and D were not included. For these analyses, the algorithms included in RDP were GeneConv, Chimaera, MaxChi, BootScan, SiScan and 3Seq, with default values, but with a window size of 20 and step interval of 10. Only those recombination events were displayed that were detected by more than four methods. In both cases, the programs detected only recombinants between isolates of the barley strains (results not shown). In contrast to this, a visual inspection of aligned sequences revealed several possible regions of recombination in which fragments of the wheat strain might have been integrated into the genome of the barley strain. They are shown in Fig. 2 for sites with an exchange of at least four consecutive nucleotides.
Recombination has occurred in the central part of the CP (position ~720), the 3′-terminal part of Rep (position ~1,460), the 5′-terminal part of Rep/RepA (position ~2,255), and in the SIR (position ~1.280 to ~1.340). The alignment also supports the phylogenetic analysis in indicating that cluster A consists of two sub-clusters because sequences of large parts of the SIR’s differ. Figure 3 gives the level of sequence homology for the SIR of a reduced number of isolates. For the members of sub-cluster A2 investigated recombination occurred mainly in the SIR and in A1 in the coding regions. Thus, it is likely that the recombination of WDV-B suggested by Ramsell et al. [24] for the Turkish isolate of WDV-B is not an exception but a typical feature of subcluster A1.
Circomics of 3 of 13 wheat samples investigated revealed the typical restriction pattern of a barley isolate. From these samples, several full-length sequences were obtained: AM942045, AM921991, AM922261, AM922260, AM921993, and AM942044 (Table 1). The phylogenetic analysis revealed that they are typical barley strains (supplementary data, Fig. S1). This analysis also demonstrated that AM942044 is slightly different from the other sequences of the virus from the same host plant. This indicates that in one host, there can be a population of WDV with different sequences.
In several cases, circomics also revealed restriction fragments of isolates from wheat and barley that were of an unexpected size. From two isolates, one from barley (isolate 43) and one from wheat (isolate 57), several bands of unexpected size were isolated, cloned, and sequenced. From the same samples also full-length sequences of the virus genome were investigated. The data are presented in Table 1.
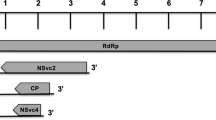
The fragments of unexpected size are defective forms of the WDV-B. The structure of the defective forms is given in Fig. 4. The defective sequences AM922264 and AM921995 originated from parental sequences AM921993 or AM922260. The defective sequences AM980883, AM980882, AM922261, AM922263, AM921649, AM932875, and AM932876 originated from the parental sequence AM922261.
Schematic drawing of the different defective forms of WDV-B. The structure of the defective form of WDV-W described by McDonald et al. [35] is included. Hollow lines represent deleted regions of the genome. Clones are labeled with corresponding accession numbers
It is characteristic of the defective forms that larger parts of the LIR and SIR are retained. AM980882 shows a partial genome duplication. The non-viral DNA integrated into clone AM980882 might have originated from wheat chromosome 5D (accession number CT009735, identities 199/305 [65 %], a truncated Class I, non-LTR retrotransposon) or 3B (accession number CR626934, identities 199/305[65 %], also a Class I, non-LTR retrotransposon). Shorter parts of this sequence (approx. 130–200 nt) are very similar to sequences from unrelated plant species like Lotus japonicus, Vitis vinifera or Solanum lycopersicum.
Discussion
Comparison of the sequences of WDV isolates from different geographical regions of Europe confirmed that their genomes are very stable. After nearly 30 years, there are no striking differences between the newly sequenced isolate of WDV-W from France and the isolates obtained in the 1980s (Matzeit [33] from France and X02869 from Sweden [11]). The new French isolate shows a sequence homology of 98 % with both of these sequences.
The WDV epidemic in Austria was not caused by a new variant of this virus. The sequence of the isolate investigated was similar to those of isolates collected in the neighbouring Czech Republic.
The conservation of the molecular structure of WDV previously recorded by Kvarnheden et al. [13], contrasts with what is recorded for the related Maize streak virus, which is highly diverse as its strains have undergone intensive recombination [25, 26]. Although there are approximately 100 sequences of WDV, there is no evidence of a WDV::BDV recombination. Only in the case of AJ783960 originating from Turkey [23, 24], there is a record of a recombination with an unknown relative. Our results demonstrated that this recombinant type is widespread. Recombination analysis of the isolates investigated revealed an obvious recombination in WDV-B in which small fragments of WDV-W are integrated into its genome.
The new geographic variants of WDV fit in one of the strains designated by Muhire et al. [3]. We suggest that cluster A is divided into two further clusters or strains as sequences of the SIRs of the members of this cluster are highly diverse.
The sequences of the barley strain that originated from wheat did not reveal characteristics that might account for why they infected this host. Thus, it is unclear why WDV-B is rarely reported infecting wheat. For practical purposes, these results mean that a spread of WDV from wheat to barley and vice versa cannot be excluded.
Defective forms of the genomes of several DNA viruses are known [36, 37] and they may influence symptom expression [38, 39]. They have previously been recorded and characterized [11, 40] for the wheat strain. Defective forms are previously reported for MSV, another mastrevirus [36]. As recorded for other Geminiviruses, they always contain the origin of replication and may contain duplications of viral sequences as well as integrated parts of the host genome. In case of the barley strain of WDV, we identified a partial genome duplication as well as an insertion of a host DNA. The plants containing defective forms did not show symptom attenuation. The presence of SIR/LIR seems to be essential for their replication because all investigated defective forms contained at least parts of these regions. As mentioned by MacDonald et al. [40], they always contain the TAATATT/AC-motif (LIR) and a primer binding site (SIR). The sequence for the primer binding site suggested by Hayes et al. [41] for the wheat strain is missing in the barley strain. The presence of both regions seems not to be necessary for MSV. Some characteristic sequence motifs were not observed at the cleavage sites (supplementary data, Table S1). Casado et al. [36] identified several encapsidated defective forms of MSV lacking either LIR or SIR regions or both. Defective forms might play a role in genome evolution of their host plants transferring DNA fragments from one species to another.
References
Stanley, D.M. Bisaro, R.W. Briddon, J.K. Brown, C.M. Fauquet, B.D. Harrison, E.P. Rybicki, D.C. Stenger, Virus Taxonomy, in VIII Report of the International Committee on Taxonomy of Viruses, ed. by L.A. Ball (Elsevier/Academic Press, London, 2005), pp. 301–326
W. Zhang, N.H. Olson, T.S. Baker, L. Faulkner, M. Agbandje-McKenna, M. Boulton, J.W. Davies, R. McKenna, Virology 279, 471–477 (2001)
B. Muhire, D.P. Martin, J.K. Brown, J. Navas-Castillo, E. Moriones, F.M. Zerbini, R. Rivera-Bustamante, V.G. Malathi, R.W. Briddon, A. Varsani, Arch. Virol. 158, 1411–1424 (2013)
P.S. Wyant, S. Strohmeier, B. Schäfer, B. Krenz, I.P. Assunção, G.S. de Andrade Lima, H. Jeske, Virology 427, 151–157 (2012)
J. Schubert, A. Habekuß, K. Kazmaier, H. Jeske, Virus Res. 127, 61–70 (2007)
A. Varsani, A.L. Monjane, L. Donaldson, S. Oluwafemi, I. Zinga, E.K. Komba, D. Plakoutene, N. Mandakombo, J. Mboukoulida, S. Semballa, R.W. Briddon, P.G. Markham, J.-M. Lett, P. Lefeuvre, E.D. Rybicki, D.P. Martin, Virology J. 6, 194 (2009). doi:10.1186/1743-422X-6-194
D.P. Martin, D. Linderme, P. Lefeuvre, D.N. Shepherd, A. Varsani, Arch. Virol. 156, 1299–1303 (2011)
S. Kraberger, J.E. Thomas, A.D.W. Geering, A. Dayarama, D. Staintona, J. Hadfield, M. Walters, K.S. Parmenter, S. van Brunschot, D.A. Collingsa, D.P. Martin, A. Varsani, Virus Res. 169, 127–136 (2012)
J. Kumar, P. Sudhir, S.P. Singh, J. Kumar, A. Tuli, Arch. Virol. 157, 2031–2034 (2012)
J. Vacke, Biol Plant Praha 3, 228–233 (1961)
S.W. MacDowell, H. MacDonald, W.D. Hamilton, R.H. Coutts, K.W. Buck, EMBO J. 4, 2173–2180 (1985)
W. Huth, J. Plant Dis, Protect 107, 406–414 (2000)
A. Kvarnheden, M. Lindblad, K. Lindsten, J.P.T. Valkonen, Arch. Virol. 147, 205–216 (2002)
S. Mehner, B. Manurung, M. Grüntzig, A. Habekuß, W. Witsack, E. Fuchs, J. Plant Dis, Protect 110, 313–323 (2003)
R.G. Kapooria, J. Ndunguru, OEPP/EPPO Bulletin 34, 413–419 (2004)
G. Köklü, Cereal Res Commun 32, 61–68 (2004)
J. Xie, X. Wang, Y. Liu, Y. Peng, G. Zhou, Plant Dis. 91, 111 (2007)
J.K. Kundu, S. Gadiou, G. Cervena, Virus Genes 38, 468–474 (2009)
A.M. Ekzayez, S.G. Kumari, Plant Dis. 95, 76 (2011)
I. Tobias, O. Shevchenko, B. Kiss, A. Bysov, H. Snihur, V. Polischuk, K. Salank, L. Palkovics, Pol. J. Microbiol. 60, 125–131 (2011)
U. Commandeur, W. Huth, Z. Pflanzenkr, Pflanzensch 106, 550–552 (1999)
K. Lindsten, J. Vacke, Acta Phytopath. Entomol. Hungarica 26, 175–180 (1991)
G. Köklü, J.N.E. Ramsell, A. Kvarnheden, Virus Genes 34, 359–366 (2007)
J.N.E. Ramsell, M.I. Boulton, D.P. Martin, J.P.T. Valkonen, A. Kvarnheden, Plant Pathol. 58, 1161–1169 (2009)
W.H. Schnippenkoetter, D.P. Martin, J.A. Willment, E.P. Rybicki, J. Gen. Virol. 82, 3081–3090 (2001)
B.E. Owor, D.P. Martin, D.N. Shepherd, R. Edema, A.L. Monjane, E.P. Rybicki, J.A. Thomson, A. Varsani, J. Gen. Virol. 88, 3154–3165 (2007)
A. Varsani, D.N. Shepherd, D. Dent, A.L. Monjane, E.P. Rybicki, D.P. Martin, Virology J. 6, 36 (2009)
E. van der Walt, E.P. Rybicki, A. Varsani, J.E. Polston, R. Billharz, L. Donaldson, A.L. Monjane, D.P. Martin, J. Gen. Virol. 90, 734–746 (2009)
J. Hadfield, D.P. Martin, D. Stainton, S. Kraberger, B.E. Owor, D.N. Shepherd, F. Lakay, P.G. Markham, R.S. Greber, R.W. Briddon, A. Varsani, Arch. Virol. 156, 335–341 (2011)
A. Habekuß, M. Oberforster, Inform 2, 7–10 (2012)
H. Huss, N.A. Gund, L. Seigner, A.M. Manschadi, Der Pflanzenarzt 1–2, 22–26 (2013)
M.A. Achon, L. Serrano, C. Ratti, C. Rubies-Autonell, Plant Dis. 90, 970 (2006)
V. Matzeit, Universität Köln (1988) Diss. 139 S
D.P. Martin, P. Lemey, M. Lott, V. Moulton, D. Posada, P. Lefeuvre, Bioinformatics 26, 2462–2463 (2010)
J. Schubert, A. Habekuß, F. Rabenstein, Plant Protect. Sci. 38, 43–48 (2002)
C.G. Casado, G.J. Ortiz, E. Padron, S.J. Bean, R. McKenna, M. Agbandje-McKenna, M.I. Boulton, Virology 323, 164–171 (2004)
B.L. Patil, N. Dutt, R.W. Briddon, S.E. Bull, D. Rothenstein, B.K. Borah, I. Dasgupta, J. Stanley, H. Jeske, Virus Res. 124, 59–67 (2007)
B.L. Patil, I. Dasgupta, Crit. Rev. Plant Sci. 25, 47–64 (2006)
S.A.A. Behjatnia, I.B. Dry, M.A. Rezaian, Arch. Virol. 152, 1127–1138 (2007)
H. MacDonald, R.H.A. Coutts, K.W. Buck, J. Gen. Virol. 69, 1339–1344 (1988)
R.J. Hayes, H. MacDonald, R.H.A. Coutts, K.W. Buck, J. Gen. Virol. 69, 1345–1350 (1986)
Acknowledgments
Authors wish to thank Toni Dixon for correcting English, M. Nielitz for technical assistance, and F. Rabenstein for providing isolates and antibodies for virus detection.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Schubert, J., Habekuß, A., Wu, B. et al. Analysis of complete genomes of isolates of the Wheat dwarf virus from new geographical locations and descriptions of their defective forms. Virus Genes 48, 133–139 (2014). https://doi.org/10.1007/s11262-013-0989-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-013-0989-9