Abstract
The genomic RNA of a novel chicken astrovirus was determined. The full length sequence is 7520 nucleotides and encodes three open reading frames (1a, 1b, 2) for three proteins. The genomic organization was similar to other astroviruses with two exceptions. The open reading frame of the RNA-dependent RNA polymerase contains its own start codon which is different from other astroviruses described to date, providing evidence for a replication mechanism different than what has previously been described for astroviruses. Furthermore, the stem-loop structure located at the potential ribosomal frameshift signal described for other astroviruses has been shown to be a hairpin structure for the novel chicken astrovirus. Phylogenic analysis of the full length sequence revealed that this chicken astrovirus formed a branch independent from other astroviruses, indicating that this astrovirus is significantly different from astroviruses described to date.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The determination of the full length sequences obtained from environmental samples will enhance the understanding of the biologic diversity of our world. To understand the steps taken during viral evolution, determination of full length sequences are valuable since only with their availability can mechanisms, such as recombination between viral genomes, be fully understood. Viruses belonging to the family Astroviridae have a non-enveloped capsid which contains a positive sense, ssRNA genome [1]. The viruses belong to a large group of small viruses with a diameter of approximately 28–30 nm. The genome length varies between 6.8 and 7.9 kb irrespective of the species of isolation. The genome encodes for three proteins, the nonstructural polyprotein (NS polyprotein), the RNA-dependent RNA polymerase (RdRp), and the capsid protein [2]. The NS polyprotein and the capsid protein are each encoded by an individual open reading frame (ORF), ORF1a and ORF2, while the RdRP (ORF1b) has been reported to be expressed via a ribosome shift mechanism [2] as a fusion protein to the NS protein [3]. Astroviruses have been isolated worldwide from several mammals (humans, cats, pigs, sheep, bat) as well as birds (ducks, chickens, turkeys) and are associated in general with gastroenteric diseases. While in mammals astroviruses mainly cause diarrhea, in birds astroviruses are associated with wider spectrum of diseases, including diarrhea, hepatitis, and nephritis. One of the diseases in poultry associated with astrovirus is the runting and stunting syndrome (RSS) in chickens. RSS is a transmissible disease of uncertain etiology. RSS affects chickens early in life and is characterized by growth retardation, ruffled feathers, and diarrhea resulting in considerable economic losses especially in commercial broiler production. The syndrome is also known as malabsorption syndrome, infectious stunting syndrome, broiler runting syndrome, and helicopter syndrome [4]. Currently, there is no effective licensed vaccine against the disease, mainly because of the absence of known etiologic agent/agents. One experimental vaccine, based on a recombinant baculovirus encoding for a novel astrovirus capsid protein, was recently described [5]. Clinical and pathological signs of RSS have been experimentally reproduced using oral inoculation of filtered and non-filtered intestinal homogenates from RSS affected chickens [5–8]. Based on preliminary sequence data of the capsid protein, obtained from gut samples collected from chickens experimentally exposed to RSS-contaminated litter, the full length sequence of astrovirus was determined. Comparisons to published astrovirus sequences indicated that the virus which harbors this genome belongs to a chicken astrovirus not previously described.
Materials and methods
Generation of material for sequence determination
One-day-old commercial broiler chickens were exposed to chicken litter transported from a commercial farm with chickens exhibiting RSS to a research isolation house, as previously described [5]. Chickens from this study were euthanatized with CO2 and the small intestine was harvested and homogenized with sterile phosphate buffered saline (PBS) at a 1:3 ratio (w/v) in a blender. The resulting homogenate was centrifuged at 3500×g for 20 min at 4°C. The supernatant obtained was centrifuged a second time at 16000×g for 20 min at 4°C, followed by a sequential filtration through a 0.45 μm and subsequently through a 0.22 μm filter (Whatman, Florham Park, NJ, USA). The filtrate was treated with chloroform and used for RNA purification using the High Pure RNA Isolation-Kit (Roche, Diagnostics GmbH, Mannheim, Germany). RNA was stored at −80°C until use.
Determination of the sequence of a novel chicken astrovirus from gut samples
RNA isolated and purified from the gut homogenate described above was used for 5′-rapid amplification of cDNA using the 5′ RACE System, Version 2.0 (Invitrogen, Carlsbad, CA, USA). The first primer used for the initial cDNA synthesis was located inside the open reading frame (ORF) of the capsid protein from a previously reported chicken astrovirus [5]. The subsequent PCR was performed with a nested astrovirus-specific primer and the anchor primer from the 5′ RACE System. The RT-PCR fragment obtained was gel eluted and purified using the QIAquick Gel Extraction Kit (Qiagen Sciences, Md, USA) and cloned into the pCR2.1 plasmid using the TOPO TA cloning kit (Invitrogen) and transformed into competent E. coli. The recombinant plasmids obtained were sequenced using the BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, CA, USA). Based on the novel sequence obtained, two novel astrovirus-specific oligonucleotides were delineated and used for the next 5′-RACE amplification. One was used for the initial cDNA synthesis, while the second oligonucleotide was used as nested primer for the subsequent PCR. Using the primer walking approach, the full length sequence was determined. To determine the extreme 5′ end of the viral genome, different 5′ RACE reactions were performed as described by Mundt and Müller [9]. In brief, for the determination of the first 5′-end nucleotide, the deoxynucleotide tailing reaction was performed using either dCTP or dGTP. The subsequent PCR was appropriately performed with either the anchor primer (dCTP-tailing) or a poly-C primer (dGTP tailing). The information regarding the primer sequences is available upon request from the corresponding author. Since the primer walking procedure was performed on a non-defined mixture present in the gut, the full length sequence was confirmed by amplification of overlapping 1 kb fragments using oligonucleotides delineated from the previously determined sequence. The RT-PCR fragments obtained were cloned and at least 3 plasmids were sequenced in both directions, obtaining a sixfold coverage of the sequence.
Multiple alignment and sequence analysis
Sequence data was analyzed using the DNAStar Lasergene 8 software package (DNASTAR Inc, Madison, WI, USA) for sequence alignments and in silico translation to amino acid sequences. Phylogenic analysis was performed using the MEGA-4.1 software (10), available as freeware online (http://www.megasoftware.net/mega4/mega41.html). The RNA secondary structure was determined using the RNA secondary structure prediction software available online (http://www.genebee.msu.su/services/rna2_reduced.html).
Results and discussion
Determination of the full length sequence of a novel chicken astrovirus
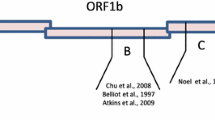
The determination of the full length sequence, using the method of primer walking, resulted in RT-PCR fragments between 400 and 1200 bp in length. The determination of the extreme 5′-end was performed as previously described [9]. The full length sequence of the virus genome is 7520 nucleotides (Genbank accession number JF414802), not including the poly-A tail sequence. A schematic of the viral genome is shown in Fig. 2. The 5′- and 3′-noncoding regions were determined with 21 and 282 nucleotides, respectively. The data correlated with full length sequences from other avian astroviruses where the 5′ NCR was also a short sequence from 10 nt (Turkey astrovirus 1, 11) to 23 nt (Duck astrovirus, 12). The 3′ NCR was also comparable in length to other bird astroviruses, with a range between 192 nt for turkey astrovirus 2 (13) and 305 nt (ANV1, 14). The alignment of the nucleotide sequences showed that the first five nucleotides (CCGAA), located at the 5′ end, were highly conserved between all bird astroviruses (Fig. 1). In addition, this sequence motif was also observed in close proximity to the start codon for the ORF2, likely encoding the viral capsid protein. This feature has also been described for the duck astrovirus [12] and turkey astrovirus 1 [11] and 2 [13]. In contrast, this motif was absent upstream of the proposed start codon for the capsid protein of ANV1 (14). Furthermore, turkey astroviruses 2, duck astrovirus, and the chicken astrovirus described in this article shared six homologous nucleotides at the very 3′ end (Fig. 1). Interestingly, when the ANV 1 sequence was also taken into consideration in the comparison of the 3′ end, the last three nucleotides were highly conserved between all astroviruses analyzed. The genome of the novel chicken astrovirus encodes three open reading frames (ORF), one protein each (Fig. 2) and follows the principal genomic structure for an astrovirus [2]. The first ORF (ORF1a) encodes for a protein of 1139 amino acids (aa), while the second ORF (ORF1b) encodes for 519 aa. ORF1a encodes for the NS polyprotein and ORF1b encodes for the viral RNA depended RNA-polymerase (RdRp) as previously proposed [2]. The third ORF (ORF2) encodes, with 743 aa, the viral capsid protein (see also 5). ORF1a and ORF1b are located in an overlapping position, while ORF2 is downstream from the ORF1b. Despite genomic similarities to other astroviruses described to date, slight differences were identified within the novel chicken astrovirus. Although there is a potential ribosomal frameshift signal, consisting of a heptanucleotide (5′-AAAAAAC-3′), previously described [2], the ORF1b contains its own start codon which makes this, by definition, a true ORF (Fig. 2). In addition, the proposed typical stem-loop structure was not present in the sequence determined, but rather a sequence was present in the proposed region which may form a strong hairpin structure with no possibility of forming a pseudo knot structure as proposed earlier [2]. The importance of this stem-loop structure is not clearly understood since changes in a model system in the structure did not abolish the expression of a pseudo ORF1a–ORF1b fusion protein but decreased the efficacy [3]. On the other hand, deletion of the ribosomal frameshift signal sequence and also only a point mutation within the ribosomal frameshift signal sequence abolished the translation of the fusion protein in this model system [3], thus this sequence likely plays a central role for the translation of the fusion protein. Based on the data described in this article, it is possible that the ORF1b encodes the RdRp in the classical mode containing a start and stop codon. The possibility exists that in case of the nucleotide sequence here described for the chicken astrovirus, either a ribosomal scanning at the viral RNA with initiation at the start codon of ORF1b, or a that the ribosomal unit dissociates at the ribosomal frameshift signal sequence and reinitiates at the methionine of ORF1b or there is a ORF1b mRNA transcribed by an unknown mechanism. The latter possibility is rather unlikely due to the nature of the ORF1b encoding protein, the RdRp.
Conserved nucleotides in the noncoding regions of bird astroviruses. The 5′- and 3′-noncoding regions (NCR) of the chicken astrovirus (described in this article, JF414802), turkey astrovirus 1 [11] and 2 [13], duck astrovirus [12], and avian nephritis virus 1 (ANV1, 14) were aligned. Highly conserved nucleotides in the 5′-NCR were marked by an asterisk. The highly conserved nucleotides in the 3′-NCR between chicken astrovirus, both turkey astroviruses, and the duck astrovirus were marked by an asterisk, while the highly conserved nucleotides between all analyzed sequences of the 3′-NCR were marked by a plus sign. The poly-A sequence at the 3′-NCR was labeled as (A)n
Schematic of the genomic organization of the chicken astrovirus. a The position of the open reading frames encoding for the nonstructural (NS) polyprotein, RNA-dependent RNA polymerase (RdRp), and the capsid protein (Capsid) are shown. The viral RNA associated poly-A tail [(A)n] is shown. b The heptanucleotide sequences (chicken and duck astrovirus) and octanucleotide sequences (turkey astrovirus 1 and 2) serving as the proposed “shifty” sequence as part of the potential ribosomal frameshift signal [2] were highlighted by bold type letters. The noncoding region for the NS protein is marked by an asterisk. The location of the methionine marked in single letter code, likely serving as start amino acid for the RdRP of the novel chicken astrovirus, is highlighted bold typed. c The secondary structure for the proposed region of the potential ribosomal frameshift signal is shown for the chicken astrovirus (JF414802) and human astrovirus 2 (L13745). The heptanucleotide sequence was highlighted by asterisks and the proposed hairpin structure (Chicken Astrovirus) and stem-loop structure (Human Astrovirus 2) was marked by a bracket. The nucleotide numbers shown in the structures is in accordance to the numbering in the sequence published in the Genbank
Analysis of the full length sequence with other Astroviruses
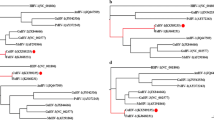
The nucleotide sequence obtained from the chicken astrovirus was compared in a phylogenetic analysis with other astroviruses using full length sequences. To this end, full length astrovirus sequences from several species [turkey astrovirus 1 (Y15936), turkey astrovirus 2 (EU143843), duck astrovirus 1(NC012437), avian nephritis virus 1 (NC003790), bat astrovirus (EU847155), human astrovirus VA1 (FJ973620), mink astrovirus (GU985458), ovine astrovirus (NC002469)] were included in this analysis (Fig. 3). The sequences were aligned using the ClustalW program (http://www.ebi.ac.uk/Tools/msa/clustalw2) and the multiple alignment obtained was analyzed using the program MEGA4.1. The neighbor-joining method and the minimum-evolution method were applied using 1000 replicates. The results of the neighbor-joining method clearly show that the novel sequence was significantly different (bootstrap value of 100) from other astrovirus sequences, including those described for ducks, turkeys, and chicken (Fig. 3) regardless of the algorithm used for the phylogenetic analysis. To further analyze the relatedness of this virus to other astroviruses, the amino acid sequences of all three in vitro translated ORFs were compared with published sequences (Table 1) using the pBlast search option in the NCBI database with one exception—the partial amino acid sequence for the ORF1a protein for a previously published chicken astrovirus [15] was taken from the publication and compared using the DNASTAR program package since this sequence is not available in the NCBI database. A 100% identity with a 99 aa partial ORF1a sequence was observed which has been described for a chicken astrovirus isolated in Europe [15]. A high similarity (99–89%) was observed with partial sequences of the capsid protein of previously described chicken astrovirus sequences obtained from US field samples [16]. Interestingly, the overall amino acid sequences indicated that the most similar relative to the novel chicken astrovirus was a recently described duck astrovirus which caused a fatal hepatitis in ducklings [12], followed by turkey astrovirus 2 [13, 17] and 1 [11]. A turkey astrovirus 3 capsid protein sequence showed a 38% identity [17]. Surprisingly, the deduced amino acid sequences of all three proteins of the novel chicken astrovirus similarly showed a low similarity to the corresponding sequences of ANV1 [14] and to the capsid protein sequence of ANV2 in addition to the expected lack of similarity observed with the mammalian astroviruses, such as ovine, mink, human, and bat astrovirus (see Table 1). This data indicates the high degree of variability between astroviruses isolated from the same species. In addition, the similarity to the RdRp amino acid sequence was always higher likely due its nature as a functional enzyme responsible for the replication of the virus genome. RdRp sequences appear over-represented in the NCBI database, likely due to their highly conserved nature, compared to the few sequences available for the remaining regions of the genome. This region also serves as a target for the development of diagnostic tools [18–20]. Determination of the full length sequences of viral genomes will certainly provide the basis for a better understanding of the biology of particular virus. Data obtained in this study support the need to determine the full length sequence of novel viruses since due only to the availability of the full length sequence was evidence found to suggest that this particular astrovirus may employ a different replication strategy than what has been described for other astroviruses. Based on the findings, experiments to isolate the virus in cell culture and to generate antisera against the proteins encoded by ORF1a and ORF1b are under way to determine the proposed alternate mechanism for astrovirus replication.
The sequence of the novel chicken astrovirus forms a new branch. The full length sequences of published full length sequences of turkey astrovirus (TkAstV) 1 and 2, duck astrovirus (DkAstV), chicken astrovirus (CkAstV), avian nephritis virus 1(ANV1), bat astrovirus (BatAstV), human astrovirus (HumAstV), mink astrovirus (MkAstV), and ovine astrovirus (OvAstV). The NCBI genbank accession number was shown in brackets. The phylogenic tree of a neighbor-joining method is shown performed with 1000 replications. The bootstrap values are shown at the branch knobs
References
E. Mendez, C.F. Arias, in Fields Virology, ed. By D.M. Knipe, P.M. Howley (Lippincott Williams & Wilkins, Philadelphia, 2007), pp. 981–1000
B. Jiang, S.S. Monroe, E.V. Koonin, S.E. Stine, R.I. Glass, Proc. Natl. Acad. Sci. USA 90, 10539–10543 (1993)
B. Marczinke, A.J. Bloys, T.D.K. Brown, M.W. Willcocks, M.J. Carter, I. Brierley, J. Virol. 68, 5588–5595 (1994)
J.M.J. Rebel, F.R.M. Balk, J. Post, S. Van Hemert, B. Zekarias, N. Stockhofe, World Poultry Sci. J. 62, 17–30 (2006)
H. Sellers, E. Linneman, A.H. Icard, E. Mundt, Vaccine 28, 1253–1263 (2010)
R.D. Montgomery, C.R. Boyle, W.R. Maslin, D.L. Magee, Avian Dis. 41, 80–92 (1997)
T. Songserm, J.M. Pol, D. van Roozelaar, G.L. Kok, F. Wagenaar, A.A. ter Huurne, Avian Dis. 44, 556–567 (2000)
T. Songserm, B. Zekarias, D.J. van Roozelaar, R.S. Kok, J.M. Pol, A.A. Pijpers, A.A. ter Huurne, Avian Dis. 46, 87–94 (2002)
E. Mundt, H. Muller, Virology 209, 10–18 (1995)
K. Tamura, J. Dudley, M. Nei, S. Kumar, Mol. Biol. Evol. 24, 1596–1599 (2007)
C.M. Jonassen, T.O. Jonassen, B. Grinde, J. Gen. Virol. 79, 715–718 (1998)
Y. Fu, M. Pan, X. Wang, Y. Xu, X. Xie, N.J. Knowles, H. Yang, D. Zhang, J. Gen. Virol. 90, 1104–1108 (2009)
E. Strain, L.A. Kelley, S. Schultz-Cherry, S.V. Muse, M.D. Koci, J. Virol. 82, 5099–5103 (2008)
T. Imada, S. Yamaguchi, M. Mase, K. Tsukamoto, M. Kubo, A. Morooka, J. Virol. 74, 8487–8493 (2000)
W. Baxendale, T. Mebatsion, Avian Pathol. 33, 364–370 (2004)
M.J. Pantin-Jackwood, K.O. Strother, E. Mundt, L. Zsak, J.M. Day, E. Spackman, Arch. Virol. 156, 235–244 (2011)
Y. Tang, A.M. Murgia, Y.M. Saif, Avian Dis. 49, 514–519 (2005)
M.J. Pantin-Jackwood, J.M. Day, M.W. Jackwood, E. Spackman, Avian Dis. 52, 235–244 (2008)
V.J. Smyth, H.L. Jewhurst, B.M. Adair, D. Todd, Avian Pathol. 38, 293–299 (2009)
D. Todd, J. Trudgett, F. McNeilly, N. McBride, B. Donnelly, V.J. Smyth, H.L. Jewhurst, B.M. Adair, Avian Pathol. 39, 207–213 (2010)
Acknowledgments
This study was supported by the US Poultry and Egg Association’s Harold E. Ford Foundation grant #F036, the Georgia Poultry Federation, and the University of Georgia Research Foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kang, Ki., Icard, A.H., Linnemann, E. et al. Determination of the full length sequence of a chicken astrovirus suggests a different replication mechanism. Virus Genes 44, 45–50 (2012). https://doi.org/10.1007/s11262-011-0663-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-011-0663-z