Abstract
We report the complete genome sequence of a new avastrovirus of goose-origin (FLX). The 7299-nt-long genome consisted of three overlapping open reading frames (ORFs) that were in different reading frames. Pairwise comparisons showed that the FLX genome was 59% identical to its closest relatives and that the levels of amino acid identity shared by FLX with other astroviruses did not exceed 54% in ORF1a, 66% in ORF1b, and 50% in ORF2, respectively. Phylogenetic analysis based on the amino acid sequence of the full-length ORF2 demonstrated that FLX was highly divergent from all other avastroviruses. At the amino acid level the complete capsid region of FLX shared genetic distances of 0.574–0.719 with three official avastrovirus species, suggesting that it can be classified as a member of a novel species in the genus Avastrovirus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Astroviruses are non-enveloped, single-stranded, positive-sense RNA viruses. The polyadenylated genomes of these viruses range in size from 6.1 to over 7.7 kb and are arranged in three open reading frames (ORFs 1a, 1b, and 2), as well as a short 5′ untranslated region (UTR) and a 3′UTR [3]. ORF1a and ORF1b encode the non-structural proteins, including several transmembrane (TM) helical motifs, a serine protease, a nuclear localization signal (NLS), and an RNA-dependent RNA polymerase (RdRp). ORF2 encodes the capsid protein that is required for virion formation [6, 11].
Avian astroviruses are members of the genus Avastrovirus, one of the two genera in the family Astroviridae. As of 2011, the International Committee on Taxonomy of Viruses (ICTV) recognized three species of the genus Avastrovirus. These three species are 1) Avastrovirus 1, including turkey astrovirus 1 (TAstV-1); 2) Avastrovirus 2, including avian nephritis virus (ANV) 1 and 2; and 3) Avastrovirus 3, including turkey astrovirus 2 (TAstV-2) and duck astrovirus 1 (DAstV-1) [4, 15]. Within the genus Avastrovirus, there are also a number of unassigned species [9, 12,13,14,15, 18, 19, 21]. Here, we report the complete genomic sequence of a novel avastrovirus of goose-origin (GAstV).
The FLX strain of GAstV was identified in the liver of a 15-day-old goose with enteritis, which was collected in June 2014 from a commercial goose farm in Hunan province, China. RNA was extracted from the liver sample as described previously [12]. The presence of the astrovirus was demonstrated by amplification of a 720 nt ORF1b–2 sequence using a reverse transcription (RT)-PCR assay with primers Picof and Picor. The astrovirus genome was amplified using RT-PCR with new primers based on conserved regions of chicken, turkey and duck astroviruses [9, 10, 12,13,14], as well as specific sequences from the original 720 nt ORF1b–2 FLX RT-PCR. The 5′ and 3′ ends of the genome were derived using 5′ and 3′ rapid amplification of cDNA ends (RACE) strategies. The initial genome sequence was verified by determination of overlapping DNA fragments amplified with additional primers. The primers applied to amplify the genome sequence are shown in Supplementary Table S1. The complete genomic sequence of FLX has been deposited in GenBank under accession number KY271027.
ORFs in the genome were predicted with DNAMAN 5.2.2 (Lynnon). Sequence similarity searches were conducted by BLASTP in GenBank [17]. Pairwise comparisons were performed using CLUSTALW [16]. Genetic distances were computed by the p-dist method of MEGA6 [20], using parameters as described previously [4]. Neighbor-joining trees were constructed by MEGA6, using Jones-Taylor-Thornton matrix-based model and 1000 bootstrap replications [20]. TM domains in ORF1a were predicted using TMHMM [1].
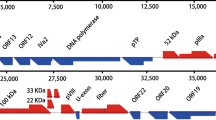
The GAstV FLX genome was 7299 nt in length with three overlapping ORFs (ORF1a, 3283 nt; ORF1b, 1545 nt; and ORF2, 2124 nt) and had a 5′UTR of 22 nt and a 3′UTR of 307 nt (Supplementary Fig. S1). The three ORFs were in different reading frames, as seen in DAstV-1 and duck astrovirus 3 (DAstV-3) [3, 13]. As expected, the overlap region between ORF1a and ORF1b in FLX contained a ribosomal frameshift signal [6, 11], consisting of a heptameric AAAAAAC sequence (nt 3295–3301) and a downstream stem-loop structure (nt 3308–3332). Similar to most avastroviruses [3, 8,9,10, 12,13,14], the conserved CCGAA motif was found at the 5′ end of the genome as well as the 29-nt space between the ORF1b stop codon and the ORF2 start codon. A stem-loop II-like motif (s2m) [7] was detected in the 3′UTR of the genome. The ORF1a and ORF1b amino acid sequences of FLX contained characteristic motifs conserved in other astroviruses [3, 5, 6, 8,9,10,11,12,13,14, 18]. These included motifs typical of: serine proteases (685GNSG688), a NLS (residues 784–815), and a RdRp (264DWTRYD269, 325GNPSG329, 375YGDD378, and 403FGMWVK408). Five TM domains were detected at positions 216–238, 371–388, 401–423, 433–455, and 468–490 of ORF1a.
Amino acid sequences deduced from the three ORFs of FLX by BLAST searches displayed low identity with other astroviruses (ORF1a: <48%; ORF1b: <66%; ORF2: <51%). Low identity at the nucleotide (genome: 51–59%) and amino acid (ORF1a: 27–54%; ORF1b: 52–66%; ORF2: 19–50%) level was also found between FLX and representative members of recognized and unassigned species in the genus Avastrovirus, by pairwise comparisons (Table 1). These analyses suggest that FLX is a novel member of the genus Avastrovirus.
The avastrovirus species are currently defined based on genetic analysis of the complete capsid amino acid sequences. The species demarcation criteria are that mean genetic distances range from 0.576–0.742 between, and 0.204–0.284 within, species, respectively [4]. To classify FLX, we conducted a genetic analysis of the FLX capsid sequence and those of representative members of the classified avastrovirus species as well as unassigned avastroviruses (Table 1). FLX shared a genetic distance of 0.574–0.719 with three official species, suggesting that it could be classified as a member of a novel species in the genus Avastrovirus. FLX also shared high levels of genetic distance (0.605–0.784) with DAstV-3 [13], duck astrovirus 4 (DAstV-4) [14], chicken astrovirus B (CAstV-B) [9, 19], and northern pintail astrovirus (NpAstV) MPJ1332 [2], indicating FLX represents an additional species to the four unassigned avastroviruses. FLX showed relatively low levels of genetic distance (0.468–0.518) with the duck astrovirus 2 (DAstV-2) [12] and the chicken astrovirus A (CAstV-A) [18, 19], indicating that they are closely related, yet distinct from one another.
To gain further insight into the evolutionary relationship of FLX with other avastroviruses where corresponding sequences are available, we performed phylogenetic analyses based on the amino acid sequence of full-length ORF2. Avastroviruses formed three major groups (groups 1, 2, and 3), consistent with those deduced from analyses of the partial ORF1b sequence and the 5′ region of ORF2 [2]. Group 1 contained most avastroviruses (except ANV) previously identified from poultry, which was divided into eight clades as described previously (12–14, 19, 21). FLX formed a distinct clade within group 1, and was most closely related to the DAstV-2 clade (Fig. 1). Phylogenetic analyses of the amino acid sequences of ORF1a and ORF1b demonstrated that FLX was highly divergent from other avastroviruses (Supplementary Fig. S2).
Phylogenetic analysis of avastroviruses based on the amino acid sequence of full-length ORF2. Numbers on the branches indicate bootstrap percentages obtained using 1000 replicates (only values of 70% and above are shown). A human astrovirus (HAstV) isolate was included as an outgroup. GenBank accession numbers of the sequences are indicated in parentheses. FpAstV, feral pigeon astrovirus; GfAstV, guinea fowl astrovirus; PhAstV, pond heron astrovirus; WpAstV, wood pigeon astrovirus. The virus determined in this study is highlighted in bold
Taken together, our data on the full-length genomic sequence suggest that FLX can be identified as a member of a novel species in the genus Avastrovirus. Based on the proposal of Chu et al. (2012) [2], the virus belongs to a novel clade in avastrovirus group 1. The present work contributes to our understanding of the molecular epidemiology and ecology of avastroviruses in domestic geese.
References
Center for biological sequence analysis (2013) Prediction of transmembrane helices in proteins. http://www.cbs.dtu.dk/services/TMHMM. Accessed 27 Nov 2016
Chu DK, Leung CY, Perera HK, Ng EM, Gilbert M, Joyner PH, Grioni A, Ades G, Guan Y, Peiris JS, Poon LL (2012) A novel group of avian astroviruses in wild aquatic birds. J Virol 86:13772–13778
Fu Y, Pan M, Wang X, Xu Y, Xie X, Knowles NJ, Yang H, Zhang D (2009) Complete sequence of a duck astrovirus associated with fatal hepatitis in ducklings. J Gen Virol 90:1104–1108
ICTV Astroviridae Study Group (2011) Reorganize the taxonomy of the genus Avastrovirus (family Astroviridae) into three newly defined species. http://ictvonline.org/proposals/2010.017a-cV.A.v3.Avastrovirus.pdf. Accessed 26 Nov 2016
Imada T, Yamaguchi S, Mase M, Tsukamoto K, Kubo M, Morooka A (2000) Avian nephritis virus (ANV) as a new member of the family Astroviridae and construction of infectious ANV cDNA. J Virol 74:8487–8493
Jiang B, Monroe SS, Koonin EV, Stine SE, Glass RI (1993) RNA sequence of astrovirus: distinctive genomic organization and a putative retrovirus-like ribosomal frameshifting signal that directs the viral replicase synthesis. Proc Natl Acad Sci USA 90:10539–10543
Jonassen CM, Jonassen TØ, Grinde B (1998) A common RNA motif in the 3′ end of the genomes of astroviruses, avian infectious bronchitis virus and an equine rhinovirus. J Gen Virol 79:715–718
Jonassen CM, Jonassen TØ, Sveen TM, Grinde B (2003) Complete genomic sequences of astroviruses from sheep and turkey: comparison with related viruses. Virus Res 91:195–201
Kang KI, Icard AH, Linnemann E, Sellers HS, Mundt E (2012) Determination of the full length sequence of a chicken astrovirus suggests a different replication mechanism. Virus Genes 44:45–50
Koci MD, Seal BS, Schultz-Cherry S (2000) Molecular characterization of an avian astrovirus. J Virol 74:6173–6177
Lewis TL, Greenberg HB, Herrmann JE, Smith LS, Matsui SM (1994) Analysis of astrovirus serotype 1 RNA, identification of the viral RNA-dependent RNA polymerase motif, and expression of a viral structural protein. J Virol 68:77–83
Liu N, Wang F, Shi J, Zheng L, Wang X, Zhang D (2014) Molecular characterization of a duck hepatitis virus 3-like astrovirus. Vet Microbiol 170:39–47
Liu N, Wang F, Zhang D (2014) Complete sequence of a novel duck astrovirus. Arch Virol 159:2823–2827
Liao Q, Liu N, Wang X, Wang F, Zhang D (2015) Genetic characterization of a novel astrovirus in Pekin ducks. Infect Genet Evol 32:60–67
Knowles N (1997) The astrovirus pages. Astrovirus sequence database. http://www.iah-virus.org/astroviridae/avastrovirus/avastrovirus_u_seq.htm. Accessed 27 Nov 2016
Kyoto University Bioinformatics Center (1991) Multiple sequence alignment by CLUSTALW. http://www.genome.jp/tools/clustalw/. Accessed 28 Nov 2016
National Center for Biotechnology Information (2013) Basic local alignment search tool. http://blast.ncbi.nlm.nih.gov/Blast.cgi. Accessed 28 Nov 2016
Sajewicz-Krukowska J, Domanska-Blicharz K (2016) Nearly full-length genome sequence of a novel astrovirus isolated from chickens with ‘white chicks’ condition. Arch Virol 161:2581–2587
Smyth VJ, Todd D, Trudgett J, Lee A, Welsh MD (2012) Capsid protein sequence diversity of chicken astrovirus. Avian Pathol 41:151–159
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Todd D, Smyth VJ, Ball NW, Donnelly BM, Wylie M, Knowles NJ, Adair BM (2009) Identification of chicken enterovirus-like viruses, duck hepatitis virus type 2 and duck hepatitis virus type 3 as astroviruses. Avian Pathol 38:21–30
Acknowledgements
The authors thank Dr. Jiafen Hu from College of Medicine, Pennsylvania State University for proofreading the manuscript.
Author information
Authors and Affiliations
Author notes
L. Yang and D. Zhang contributed equally to this work.
The GenBank accession number determined in this study are KY271027.
Corresponding authors
Ethics declarations
Funding
This study was funded by the China Agriculture Research System (CARS-43).
Conflict of interest
Yuxuan Zhang declares that he has no conflict of interest. Fumin Wang declares that he has no conflict of interest. Ning Liu declares that she has no conflict of interest. Lin Yang declares that he has no conflict of interest. Dabing Zhang declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with animals performed by any of the authors.
Additional information
The GenBank accession number determined in this study are KY271027.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, Y., Wang, F., Liu, N. et al. Complete genome sequence of a novel avastrovirus in goose. Arch Virol 162, 2135–2139 (2017). https://doi.org/10.1007/s00705-017-3297-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-017-3297-1