Abstract
Pathogenic fungi represent one of the major biotic stresses for soybean production across the world. Sclerotinia sclerotiorum, the causal agent of Sclerotinia stem rot, is a devastating fungal pathogen that is responsible for significant yield losses in soybean. In this study, the chitinase gene CmCH1, from the mycoparasitic fungus Coniothyrium minitans, which infects a range of ascomycetous sclerotia, including S. sclerotiorum and S. minor, was introduced into soybean. Transgenic plants expressing CmCH1 showed higher resistance to S. sclerotiorum infection, with significantly reduced lesion sizes in both detached stem and leaf assays, compared to the non-transformed control. Increased hydrogen peroxide content and activities of defense-responsive enzymes, such as peroxidase, superoxide dismutase, phenylalanine ammonia lyase, and polyphenoloxidase were also observed at the infection sites in the transgenic plants inoculated with S. sclerotiorum. Consistent with the role of chitinases in inducing downstream defense responses by the release of elicitors, several defense-related genes, such as GmNPR2, GmSGT-1, GmRAR1, GmPR1, GmPR3, GmPR12, GmPAL, GmAOS, GmPPO, were also significantly upregulated in the CmCH1-expressing soybean after inoculation. Collectively, our results demonstrate that overexpression of CmCH1 led to increased accumulation of H2O2 and up-regulation of defense-related genes and enzymes, and thus enhanced resistance to S. sclerotiorum infection while showing no detrimental effects on growth and development of soybean plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soybean [Glycine max (L.) Merr.] provides one of the primary sources of vegetable oil and protein for both human consumption and use in livestock feed. To meet the continually increasing demand for soybean, many challenges must be overcome to increase soybean production in a sustainable manner. In soybean, more than 30 diseases caused by fungi, bacteria and viruses have been identified. Among these biotic stresses, Sclerotinia stem rot (SSR, also known as white mold) caused by the cosmopolitan soil-borne fungal pathogen, Sclerotinia sclerotiorum, is the second most damaging disease on soybean yield worldwide and accounts for 1.6 billion kilograms of yield loss each year in the US alone (Ranjan et al. 2019), depending on the incidence and severity of the disease (Peltier et al. 2012). Globally, even as high as 60% of yield reductions were reported in some soybean growing regions when climatic factors such as prolonged cool and wet conditions are conducive to disease development (Cunha et al. 2010). Continuous cultivation and canopy density influenced by changes in cultivation practices, such as row spacing, soil fertility, and irrigation, also greatly contribute to disease severity and prevalence (Workneh and Yang 2000). Despite the existence of several tools for SSR management, such as cultural, chemical, and biological control practices, more effective and sustainable control of SSR still largely relies on the genetic resistance of soybean cultivars. However, no available soybean cultivar is completely resistant to SSR, and only moderately resistant cultivars have been identified in some soybean germplasms until now (Collard and Mackill 2008; Guo et al. 2008; Vuong et al. 2008; Zhao et al. 2015; Mccaghey et al. 2017).
S. sclerotiorum has been viewed as a predominately necrotrophic fungus despite the existence of a brief biotrophic phase in pathogenic development (Bolton et al. 2006; Kabbage et al. 2015). It can secrete a repertoire of pathophysiological factors, including oxalic acid (OA), and cell wall degrading enzymes, such as pectinases, cellulases, and hemicellulases, to facilitate plant cell wall degradation and host colonization. Of these factors, OA is usually considered a key virulence factor in the pathogenic success of S. sclerotiorum (Cessna et al. 2000; Dutton and Evans 1996; Godoy et al. 1990; Kabbage et al. 2015; Kim et al. 2008; Rollins 2003; Williams et al. 2011). Moreover, it was reported that other virulence factors targeting host responses may also be involved in the pathogenicity of S. sclerotiorum (Pedras and Ahiahonu 2004; Zhu et al. 2013). Many attempts have been made to confer pathogen resistance by introducing heterologous genes of the OA-metabolizing enzymes, such as oxalate oxidase and oxalate decarboxylase (Cober et al. 2003; Cunha et al. 2010; Dias et al. 2006; Donaldson et al. 2001; Hu et al. 2003; Kumar et al. 2016; Livingstone et al. 2005; Walz et al. 2010; Yang et al. 2019). Silencing of soybean NADPH oxidases (GmRBOHs) has also been shown to enhance resistance to S. sclerotiorum (Ranjan et al. 2017). As an important class of antifungal substances in plants and vertebrates, chitinases can inhibit fungal growth by hydrolyzing chitin, which is a major component of the cell wall in most pathogenic fungi and nematodes. They can also release pathogen-borne elicitors, which can further induce downstream defense response in the host plants (Dana et al. 2006; Zhang et al. 2016). Transgenic plants overexpressing chitinases from several origins have been shown to exhibit improved resistance to a wide range of fungal pathogens and even abiotic stresses (Dana et al. 2006; Kumar et al. 2009; Roslan and Anji 2011; Zhang et al. 2016). Enhanced defense responses by the overexpression of chitinase was reported to be mediated by the rapid accumulation of reactive oxygen species (ROS) and peroxidases as well as the increased expression of several defense-related genes in the transgenic plants (Kumar et al. 2009; Zhang et al.2016).
Coniothyrium minitans is an important mycoparasitic fungus that can parasitize and destroy both hyphae and sclerotia of fungi of the genus Sclerotinia (Ren et al. 2007; Sun et al. 2017). C. minitans has been widely used as a biological agent to control diseases caused by S. sclerotiorum and S. minor in many vegetable crops and rapeseed (Sun et al. 2017). The mechanisms underlying the C. minitans-mediated biocontrol of Sclerotinia include mycoparasitism, degradation of oxalate secreted by S. sclerotiorum, and antifungal activity (Lou et al. 2016). As a biological control agent on S. sclerotiorum and S. minor, C. minitans exhibits strong biocontrol activity against a range of plant fungal pathogens. Extracellular lytic enzymes from C. minitans, such as chitinase, play an essential role in the penetration of the host cells and facilitation of mycoparasitism of C. minitans on S. sclerotiorum (Lou et al. 2016).
In this study, we investigated the effectiveness of the C. minitans chitinase in improving S. sclerotiorum resistance in transgenic plants. A CmCH1 gene from C. minitans was introduced into soybean and transgenic lines were evaluated for their resistance to S. sclerotiorum.
Materials and methods
Vector construction and plant transformation
The chitinase-encoding full-length CmCH1 gene (GenBank accession number AF285086.1) was first isolated from C. minitans and kindly provided by Prof. Daohong Jiang of Huazhong Agricultural University, China. Plasmid pTF101-35S, which contains a phosphinothricin acetyltransferase gene (BAR) expression cassette conferring glufosinate resistance, was used to express CmCH1. For transformation vector construction, the 1.3-kb open reading frame of CmCH1 was amplified from the plasmid pMD18-CmCH1 using the primers CmCH1-F1 (5′-GCTCTAGAATGATGC
TCTTTGCTCTTGTATCAATTCTC-3′, XbaI site underlined) and CmCH1-R1 (5′-GAGAGCTCCTAGTTGTTGGGGAAGCCAGACC-3′, SacI site underlined). The amplified fragment was digested with the endonucleases XbaI and SacI, and then placed under the control of the cauliflower mosaic virus (CaMV) 35S promoter and nopaline synthase terminator (Fig. 1a). The resulting construct pTF101-CmCH1 was introduced into the Agrobacterium tumefaciens EHA101. For genetic transformation experiments, soybean cultivar Williams 82 was used and the transformation protocol was performed as described previously (Yang et al. 2019). The regenerated and healthy plants were grown under greenhouse conditions at 25 ± 2 °C and a 16/8 h light/dark cycle for subsequent molecular characterization.
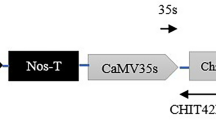
Generation and characterization of transgenic soybean plants. a Schematic diagram of the T-DNA region of the expression vector pTF101-CmCH1 for soybean transformation. The selectable marker BAR gene confers glufosinate resistance for selection of transgenic plants; LB, left border; RB, right border. b T0 transgenic plants grown in a greenhouse. c Southern blot analysis of the T2 transgenic soybean plants. Genomic DNA samples (~30 µg) were digested with XbaI and hybridized against a DIG-labeled CmCH1 probe. M, DNA marker (15 k); Ctl+, positive control; Nt, non-transformed plants; G9, G17, G29, G58, G66, G116, T2 transgenic plants. d RT-PCR analysis of transgenic plants (lines 1–6 transgenic plants) and NT control. The soybean GmACT6 gene was used as an internal control. e Chitinase activity analysis. Crude enzyme extracts were prepared from leaves of the transgenic and NT plants. Chitinase activity was analyzed using a commercial Plant Chitinase ELISA Kit, and the absorbance was measured at 450 nm. The asterisks indicate significant differences at P < 0.01
Molecular analysis of transgenic soybean plants
The regenerated plants were first screened by LibertyLink® strip (EnviroLogix Inc., USA). The presence of the foreign gene was detected by polymerase chain reaction (PCR) using the specific primers CmCH1-F2/CmCH1-R2 which amplified the CmCH1 coding sequences (Table S1). Transgenic progenies of subsequent generations (T1 to T4) were screened by both glufosinate (1500 mg/L) spraying and PCR with the primers mentioned above. Southern blot analysis was conducted to confirm the integration and insertion copy numbers of the foreign gene. Total genomic DNA was extracted from the young leaves of T2 transgenic plants, using a modified cetyltrimethylammonium bromide method described by Telzur et al. (1999). After digestion with XbaI, which cut once in the plasmid pTF101-CmCH1, DNA fragments were separated by agarose gel electrophoresis and blotted onto the positively charged nylon membrane (GE Amersham, USA). The hybridization probe was obtained by amplifying the 1.3-kb fragment that corresponded to the full open reading frame of CmCH1 using the primers CmCH1-F2/CmCH1-R2. The membrane was then hybridized by the digoxigenin (DIG)-labeled CmCH1 probe. Hybridization and subsequent washes were performed at 65 °C using the conditions described in the DIG High Prime DNA Labeling and Detection Starter Kit II (Roche, Germany). Signal visualization was conducted with BCIP/NBT as the substrate at room temperature.
Reverse transcription (RT)-PCR and chitinase activity assays
RT-PCR was used to determine the expression of CmCH1. The leaves of T3 transgenic plants were ground into powder in liquid nitrogen using mortar and pestle. Total RNA was extracted using the EasyPure PlantRNA Kit (Transgen Biotech, Beijing, China) and was reverse transcribed with the TransScript® One-Step gDNA Removal and cDNA Synthesis SuperMix (TansGen Biotech), according to the manufacturer’s instructions. RT-PCR was performed with specific primers CmCH1-F3/CmCH1-R3 (Table S1), with the constitutively expressed native soybean gene GmACT6 (GenBank No. NM_001289231) as the internal control. Chitinase activity was determined using a commercial Plant Chitinase ELISA Kit (Shanghai Enzyme-linked Biotechnology Co. Ltd, Shanghai). Leaf samples were frozen in liquid nitrogen and enzyme extracts were prepared as described by Zhang et al. (2016). Chitinase activity assays were carried out in flat-bottom ELISA plates following the manufacturer’s instructions. The color development of the enzymatic reaction (horseradish peroxidase and 3,3′,5,5′-tetramethylbenzidine) was read at 450 nm with a plate reader (ELx800, BioTek, USA) and chitinase activity (mU/mL) was calculated using the standard curve.
H2O2 and defense-related enzyme activity analysis
H2O2 content was determined in transgenic plants at 2 and 4 h after inoculation with S. sclerotiorum, as described by Kumar et al. (2016). H2O2 was extracted and quantified from the leaves of the transgenic and NT plants using the Micro Hydrogen Peroxide Assay Kit (Solarbio, Beijing, China), according to the instructions of the manufacturer. Absorbance was measured at 415 nm, and a standard curve was used to calculate the levels of total H2O2 (µmol/g · FW). To investigate the influence of CmCH1 expression on activities of the defense-related enzymes including phenylalanine ammonia lyase (PAL), polyphenoloxidase (PPO), peroxidase (POD), and superoxide dismutase (SOD) in transgenic soybean, the crude enzymes were extracted from the leaves of T3 transgenic and NT plants 4 h after S. sclerotiorum inoculation, and enzyme activity was measured as previously reported for PAL, POD, PPO, and SOD (Du et al. 2018). All the samples were analyzed in three biological replicates.
S. sclerotiorum resistance assays
Resistance bioassays of the transgenic lines were performed using detached stems and leaves, as described previously (Yang et al. 2019), using S. sclerotiorum collected from an infected soybean field located in Jilin province, China. Sclerotia were grown at room temperature on potato dextrose agar (PDA) and 5-mm mycelial plugs taken from the actively growing edge were used as the inoculum. For the detached stem bioassay, a single mycelial plug was placed on each detached stem from the six-week-old plants and incubated at 25 °C in the dark. Lesion length (cm) on the main stem was measured at 96 h after inoculation. For detached leaf assays, actively growing S. sclerotiorum agar plugs were placed on the adaxial surface of the detached leaves and incubated at the conditions described above. Necrotic lesions (cm2) were monitored and photographed 5 d after inoculation as described by Cunha et al. (2010). Six detached stems or leaves were inoculated for each transgenic line tested.
Quantitative RT-PCR analysis of the disease-responsive genes
To analyze the relative transcript levels of defense-related genes in the transgenic soybean plants after S. sclerotiorum challenge, quantitative RT-PCR (qRT-PCR) analysis was conducted using gene-specific primers (Table S1). Leaf samples were collected from the T3 transgenic and NT plants 4 h following inoculation with S. sclerotiorum. Total RNA extraction and first-strand cDNA synthesis were carried out as mentioned above. Quantitative RT-PCR was performed with an ABI PRISM 7500 Fast Real-Time PCR System (Applied Biosystems, CA, USA), under the following reaction conditions: 50 °C for 2 min, 95 °C for 10 min, 45 cycles of 95 °C for 2 min, 62 °C for 30 s, and 72 °C for 30 s. The relative transcription levels were calculated using the 2ΔΔCt method with the soybean GmACT6 gene as a reference. All experiments were conducted with three biological replicates.
Agronomic performance of the transgenic lines
CmCH1 transgenic lines were grown at the experimental station in Jilin province, China, without S. sclerotiorum inoculation. At maturity, 20 plants from each transgenic line were randomly sampled and agronomic traits were measured and recorded.
Statistical analysis
Analysis of variance was used to evaluate the data using SPSS software v.17.0 (SPSS Inc., Chicago, IL, USA). Differences between the average values of samples were compared with Student’s t-test.
Results and analysis
Generation and characterization of transgenic soybean plants
The CmCH1 gene was first isolated from C. minitans and the predicted protein possesses 397 amino acids with a calculated molecular weight of 48 kDa and a theoretical isoelectric point of 6.08. The Pfam protein family prediction indicated that CmCH1 belongs to the glyco-8 type II chitinase (data not showed). To test the defense potential of the chitinase CmCH1 for the pathogenic fungi, stable transgenic soybean plants expressing the CmCH1 gene under the CaMV 35S promoter (Fig. 1a) were produced and evaluated for their resistance to S. sclerotiorum. After selection and regeneration, a total of 47 glufosinate-resistant plants were obtained from soybean cultivar Williams 82. Positive transgenic soybean plants were first identified by LibertyLink® strip detection. The expected 1.3-kb PCR product, representing the CmCH1 full coding sequence was observed in the positive plants, whereas no visible amplification was detected in non-transformed plants, confirming the presence of the foreign gene in the regenerated plants (Fig. S1). All transgenic plants were able to flower normally and produce fertile and viable seeds in the greenhouse and exhibited similar phenotypes to the non-transformed (NT) plants (Fig. 1b). For screening of transgenic plants in the subsequent generations, both glufosinate spraying and PCR analysis were conducted to generate homozygous lines. To further investigate the integration and copy numbers of the T-DNA in the genome, Southern blot analysis was carried out using the CmCH1 gene as a probe. The results showed that hybridization signals appeared in all six selected transgenic plants with all of the bands greater than the expected fragment size of 2.0-kb fragment located between the XbaI site and the right border (Fig. 1c). Moreover, transgenic soybean events had different hybridization patterns and lower copy numbers of the T-DNA insertions. Expression of CmCH1 at the mRNA level was further examined by RT-PCR analysis with the presence of a single 332-bp amplified fragment in all six transgenic plants and absent in the NT control (Fig. 1d). To detect the functionality of CmCH1 in the transgenic lines, chitinase activity of the total plant protein extracts was quantified by ELISA analysis. Data showed strong chitinase activity in the transgenic soybean plants were detected corresponding to its own lytic activity, whereas that of control plants was much lower (Fig. 1e).
Enhanced resistance of transgenic lines to S. sclerotiorum
Transgenic lines overexpressing CmCH1 were tested for resistance against S. sclerotiorum by detached stem and detached leaf inoculation assays. The reduction in lesion length on stems and lesion areas on the leaves were calculated to assess disease severity caused by S. sclerotiorum infection. As can be seen in Fig. 2a, at 96 h following inoculation with S. sclerotiorum, the lesion on the stems of the NT plants proceeded aggressively with advanced necrosis and cottony, white mycelia. Whereas transgenic lines exhibited much smaller lesion sizes (15.06–30.27% of those in the NT control) on the detached stems (Fig. 2b). Similar results were also observed in the detached leaf assay. In this case, the untransformed plants showed highly water-soaked lesions across the whole leaves at 5 days after inoculation (Fig. 2c), whereas significantly decreased lesion sizes (43.16–48.47% of that in the NT control) were detected on inoculated leaves of transgenic plants (Fig. 2d). Consistent results seen in the detached stem and detached leaf assays confirmed that CmCH1 expression significantly improved the resistance of transgenic soybean against S. sclerotiorum. Moreover, stable and enhanced resistance in subsequent generations was further observed in subsequent T4 transgenic lines (Fig. 2b). When no S. sclerotiorum inoculation was involved, we did not observe any visible differences in phenotypic or agronomic traits between the CmCH1 transgenic lines and non-transformed soybean plants (Table 1).
Overexpression of CmCH1 enhances soybean resistance to S. sclerotiorum. a Detached stem assays. Six main stems from six-week-old plants were inoculated and lesion length (cm) on the main stem was measured 96 h after inoculation. b Disease resistance evaluation against S. sclerotiorum using detached stem assays in T3 and T4CmCH1 transgenic soybean lines. c Detached leaf assays. Six leaves were inoculated from both the transgenic line and the NT plants and necrotic lesions (cm2) were monitored at 5 d after inoculation. d Disease resistance evaluation of T3 transgenic lines by detached leaf assays. Each experiment was carried out with three independent biological replicates and error bars indicate the standard error. Asterisks indicate a significant difference at P < 0.01.
CmCH1 transgenic plants showed increased H2O2 accumulation and defense responsive enzyme activity
It has previously been reported that chitinases can release elicitors such as reaction oxygen species (ROS) to enhance the plant defense response against pathogens (Dana et al. 2006; Zhang et al. 2016). In this study, we first detected changes in H2O2 levels and defense-related enzymes at inoculation sites of the transgenic plants after infection with S. sclerotiorum. Our results showed that the H2O2 levels were significantly increased in the transgenic plants as compared to the NT controls at 2 and 4 h after inoculation with the pathogen (Fig. 3a). Moreover, we found that S. sclerotiorum infection induced significantly higher activities of the four defense-related enzymes in the transgenic plants compared to the NT plants at 4 h after S. sclerotiorum inoculation (Fig. 3b–d). Among these four defense-related enzymes, the ROS scavenging enzymes, SOD and POD, may increase tolerance to oxidative stress in transgenic plants to diminish the toxic effects of excessive ROS such as H2O2, whereas PAL and PPO may be involved in enhanced resistance against the pathogen. Collectively, these results indicate that the overexpression of CmCH1 increased H2O2 levels and the activities of the defense-related enzymes.
Increased H2O2 levels and defense-responsive enzymes after S. sclerotiorum inoculation. a Determination of H2O2 content in the transgenic soybean plants. At 2 and 4 h after S. sclerotiorum inoculation, H2O2 was extracted and quantified from the leaves of the transgenic and NT plants and absorbance was read at 415 nm. Increased activities of the defense-responsive enzymes PAL (b), PPO (c), POD (d), and SOD (e) in CmCH1 transgenic plants. Fully expanded leaves of 6-week-old soybean plants were inoculated with S. sclerotiorum and collected 4 h after infection. Asterisks indicate significant differences between each transgenic line and NT plants at the level of 0.01. The results shown are the mean of three independent experiments
Enhanced expression of defense-responsive genes in transgenic plants after infection
To further explore the role of CmCH1 overexpression in enhanced soybean resistance to S. sclerotiorum, the transcriptional expression of several defense-related genes, including three hypersensitive response (HR)-related genes, four salicylic acid (SA)-dependent genes, and two jasmonic acid (JA)-dependent genes, were investigated by qRT-PCR analysis. The results showed that transcription levels of all three HR-related genes were higher in the transgenic plants than in the NT control at 4 h after S. sclerotiorum inoculation (Fig. 4a). Higher expression levels of HR-related genes may be related to the elevated release of H2O2, which has been reported to induce a hypersensitive response in host plants. Interestingly, the expression of both SA- and JA-dependent genes were also significantly increased in the transgenic plants relative to the NT plants (Fig. 4b, c), suggesting the involvement of these defense-related genes in S. sclerotiorum resistance. Upregulation of defense-related genes such as GmPPO and GmPAL were also consistent with defense responsive enzyme activity analysis (Fig. 3d, e).
Relative expression of defense-related genes in CmCH1 transgenic soybean plants after S. sclerotiorum inoculation. Relative expression of three HR-related genes (a), four SA-dependent genes (b), and two JA-dependent genes (c) 4 h after S. sclerotiorum inoculation. Relative expression levels were calculated using the formula 2ΔΔCt relative to the internal control GmACT6. Means and standard errors are shown for three independent biological replicates. Asterisks indicate significant differences at the level of 0.01
Discussion
S. sclerotiorum represents one of the major fungal pathogens in many soybean production regions over the world. Traditional breeding for SSR resistance has also been limited by the lack of strong resistance in the soybean germplasm (Cunha et al. 2010; Mccaghey et al. 2017; Yang et al. 2019). Many researchers have attempted to enhance plant protection against pathogens by homologous and heterologous overexpression of disease resistance genes. As a class of antifungal metabolites, chitinases play an important role in plant defense against fungal pathogens by cell wall digestion and releasing pathogen-borne elicitors (Dana et al. 2006; Zhang et al. 2016). Expression of the chitinase genes in plants has proven to be effective in enhancing resistance against a broad range of fungal pathogens (Dana et al. 2006; Zhang et al. 2016). Here, we reported the generation of transgenic soybean plants overexpressing the gene CmCH1 from the mycoparasitic fungus C. minitans. Our results showed that the transgenic plants exhibited significantly increased resistance to S. sclerotiorum as shown in leaf and stem inoculation assay. Compared with the non-transformed control plants, the transgenic plants exhibited a significant reduction in lesion length on the stems and in lesion area on the leaves, which may result from enhanced penetration resistance and limited lesion development of the pathogen at the infection sites. Consistent with enhanced resistance to the pathogen, the chitinase activity of transgenic soybean plants was significantly increased compared to the control plants, suggesting the antimicrobial property of the expressed chitinase in soybean against the pathogen. Similar results were also reported for antifungal activity of chitinase in enhancing resistance against other fungal and bacterial pathogens (Dana et al. 2006; Zhang et al. 2016). We also observed that the expression of the CmCH1 exhibited no unexpected influences on the transgenic plants, consistent with the findings that chitinases are not toxic to plant hosts (Dana et al. 2006).
Previous study reported that the chitinases play a dual role, both by inhibiting fungal growth by cell wall digestion and by releasing pathogen-borne elicitors that induce further defense reactions in the host (Dana et al. 2006). The improved resistance against pathogens in plants overexpressing a chitinase gene is not the only consequence of their enhanced activity to decompose chitin and is very likely accompanied by the triggering of other defense-related mechanisms. In the plant and pathogen interaction, rapid generation of ROS is correlated with plant resistance to pathogens and is also one of the most important determinants leading to pathogen penetration failure in invading plant epidermal cells (Yang et al. 2019). However, in pathogenesis of S. sclerotiorum on the host plant, the secreted OA can inhibit the oxidative burst of the host plant, thus compromising the H2O2-dependent defense responses (Bolton et al. 2010; Cessna et al. 2000; Dutton and Evans 1996; Kumar et al. 2016). In this study, we found that H2O2 content was increased in the transgenic plants after S. sclerotiorum inoculation, suggesting an H2O2-mediated hypersensitive response involved in enhanced resistance to S. sclerotiorum. Consistent with this, transcript levels of three HR-related genes (GmNPR1-2, GmSGT1, and GmRAR1) were significantly upregulated in transgenic plants at 4 h after inoculation compared to the NT plants. Moreover, in addition to the upregulation of the HR-related defense genes, a significant upregulation in the expression of several other known defense-responsive genes involved in SA signaling (GmPR1, GmPR3, GmPR12, and GmPAL) and the JA-dependent genes (GmAOS and GmPPO) were also observed. A rapid induction of SA and JA signaling is associated with an early recognition of the pathogen and involved in resistance in soybean (Ranjan et al. 2019). These results demonstrate that CmCH1 overexpression increased the accumulation of H2O2 and triggered different downstream disease defense-related mechanisms in soybean.
In contrast, although increased H2O2 levels could contribute to the enhancement of disease resistance in plants, excessive H2O2 is also toxic to plant cells. To maintain the balance of the oxidative state in cells, the ROS scavenging enzymes are also increased to counteract the toxic effect of excessive H2O2. Indeed, the redox buffering capacity of the host is essential to counter the oxidative state imposed by pathogens in host-mediated resistance (Ranjan et al. 2019). In the present study, we found a significant increase in levels of ROS scavenging enzymes, such POD and SOD, after S. sclerotiorum inoculation, which might increase tolerance to oxidative stress in transgenic plants and correlate with the improved resistance to plants pathogens (Dana et al. 2006; Wan et al. 2009). The results are consistent with the findings of Karmakar et al., who observed transgenic plants expressing chitinase and oxalate oxidase 4 genes exhibited elevated H2O2 levels and significant increases in activity of ROS scavenging enzymes and reduced membrane damage compared to their wild type counterpart after Rhizoctonia solani infection (Karmakar et al. 2016; Zhang et al. 2016).
In this study, CmCH1-expressing soybean plants showed enhanced, but not complete, resistance to S. sclerotiorum infection. As the likely complex network of responses to the pathogen or its determinants in host plants and multiple factors are involved in the establishment of S. sclerotiorum. Among these factors, OA can weaken plant structures and enhance the activity of cell wall-degrading enzymes for the invasion of the pathogen. Overexpression of CmCH1 can enhance resistance to injury and establishment of fungal infection but cannot completely counteract the effects of OA. It is important to rely on multiple components of defense by pyramiding multiple resistance genes, such as CmCH1 and other OA-degrading genes, to engineer S. sclerotiorum resistance.
References
Bolton MD, Thomma BP, Nelson BD (2006) Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol Plant Pathol 7:1–16
Bolton MD, Thomma BPHJ, Nelson BD (2010) Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol Plant Pathol 7:1–16
Cessna SG, Sears VE, Dickman MB, Low PS (2000) Oxalic acid, a pathogenicity factor for sclerotinia sclerotiorum, suppresses the oxidative burst of the host. Plant Cell 12:2191–2200
Cober ER, Rioux S, Rajcan I, Donaldson PA, Simmonds DH (2003) Partial resistance to white mold in a transgenic soybean line. Crop Sci 43:92–95
Collard BC, Mackill DJ (2008) Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos Trans R Soc Lond 363:557–572
Cunha WG, Tinoco MLP, Pancoti HL, Ribeiro RE, Aragão FJL (2010) High resistance to Sclerotinia sclerotiorum in transgenic soybean plants transformed to express an oxalate decarboxylase gene. Plant Pathol 59:654–660
Dana M, Pintor-Toro T, Cubero B (2006) Transgenic tobacco plants overexpressing chitinases of fungal origin show enhanced resistance to biotic and abiotic stress agents. Plant Physiol 142:722–730
Dias BBA, Cunha WG, Morais LS, Vianna GR, Rech EL, Capdeville G, Aragao FJL (2006) Expression of an oxalate decarboxylase gene from Flammulina sp. in transgenic lettuce (Lactuca sativa) plants and resistance to Sclerotinia sclerotiorum. Plant Pathol 55:187–193
Donaldson PA, Anderson T, Lane BG, Davidson AL, Simmonds DH (2001) Soybean plants expressing an active oligomeric oxalate oxidase from the wheat gf-2.8 (germin) gene are resistant to the oxalate-secreting pathogen Sclerotina sclerotiorum. Physiol Mol Plant Pathol 59:297–307
Du Q, Yang XD, Zhang JH, Zhong XF, Kim KS, Yang J, Xing GJ, Li XY, Jiang ZY, Li QY, Dong YS, Pan HY (2018) Over-expression of the Pseudomonas syringae harpin-encoding gene hrpZm confers enhanced tolerance to Phytophthora root and stem rot in transgenic soybean. Transgenic Res 27:277–288
Dutton MV, Evans CS (1996) Oxalate production by fungi: its role in pathogenicity and ecology in the soil environment. Can J Microbiol 42:881–895
Godoy G, Steadman JR, Dickman MB, Dam R (1990) Use of mutants to demonstrate the role of oxalic acid in pathogenicity of Sclerotinia sclerotiorum on Phaseolus vulgaris. Physiol Mol Plant Pathol 37:179–191
Guo X, Wang D, Gordon SG, Helliwell E, Smith T, Berry SA, St. Martin SK, Dorrance AE (2008) Genetic mapping of QTLs underlying partial resistance to Sclerotinia sclerotiorum in soybean PI 391589A and PI 391589B. Crop Sci 48:1129–1139
Hu X, Bidney DL, Yalpani N, Duvick JP, Crasta O, Folkerts O, Lu GH (2003) Overexpression of a gene encoding hydrogen peroxide-generating oxalate oxidase evokes defense responses in sunflower. Plant Physiol 133:170–181
Kabbage M, Yarden O, Dickman MB (2015) Pathogenic attributes of Sclerotinia sclerotiorum: switching from a biotrophic to necrotrophic lifestyle. Plant Sci 233:53–60
Karmakar S, Molla KA, Chanda PK, Sarkar SN, Datta SK, Datta K (2016) Green tissuespecific co-expression of chitinase and oxalate oxidase 4 genes in rice for enhanced resistance against sheath blight. Planta 243:115–130
Kim KS, Min JY, Dickman MB (2008) Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Mol Plant Microbe Interact 21:605–612
Kumar V, Parkhi V, Kenerley CM, Rathore KS (2009) Defense-related gene expression and enzyme activities in transgenic cotton plants expressing an endochitinase gene from Trichoderma virens in response to interaction with Rhizoctonia solani. Planta 230:277–291
Kumar V, Chattopadhyay A, Ghosh S, Irfan M, Chakraborty N, Chakraborty S, Datta A (2016) Improving nutritional quality and fungal tolerance in soya bean and grass pea by expressing an oxalate decarboxylase. Plant Biotechnol J 14:1394–1405
Livingstone DM, Hampton JL, Phipps PM, Grabau EA (2005) Enhancing resistance to Sclerotinia minor in peanut by expressing a barley oxalate oxidase gene. Plant Physiol 137:1354–1362
Lou Y, Han Y, Yang L, Wu M, Zhang J, Cheng J, Wang M, Jiang D, Chen W, Li G (2016) CmpacC regulates mycoparasitism, oxalate degradation and antifungal activity in the mycoparasitic fungus Coniothyrium minitans. Environ Microbiol 17:4711–4729
Mccaghey M, Willbur J, Ranjan A, Grau CR, Chapman S, Diers B, Groves C, Kabbage M, Smith DL (2017) Development and evaluation of Glycine max germplasm lines with quantitative resistance to Sclerotinia sclerotiorum. Front Plant Sci 8:1495. doi:https://doi.org/10.3389/fpls.2017.01495
Pedras MSC, Ahiahonu PWK (2004) Phytotoxin production and phytoalexin elicitation by the phytopathogenic fungus Sclerotinia sclerotiorum. J Chem Ecol 30:2163–2179
Peltier AJ, Bradley CA, Chilvers MI, Malvick DK, Mueller DS, Wise KA, Esker PD (2012) Biology, yield loss and control of Sclerotinia stem rot of soybean. J Integr Pest Manag 3:1–7
Ranjan A, Jayaraman D, Grau C, Hill JH, Whitham SA, Ané JM, Smith DL, Kabbage M (2017) The pathogenic development of Sclerotinia sclerotiorum in soybean requires specific host NADPH oxidases. Mol Plant Pathol 19:700–714
Ranjan A, Westrick NM, Jain S, Piotrowski JS, Ranjan M, Kessens R, Stiegman L, Grau CR, Conley SP, Smith DL, Kabbage M (2019) Resistance against Sclerotinia sclerotiorum in soybean involves a reprogramming of the phenylpropanoid pathway and up-regulation of antifungal activity targeting ergosterol biosynthesis. Plant Biotechnol J 17:1567–1581
Ren L, Li GQ, Han YC, Jiang DH, Huang HC (2007) Degradation of oxalic acid by Coniothyrium minitans and its effects on production and activity of β-1,3-glucanase of this mycoparasite. Biol Control 43:1–11
Rollins JA (2003) The Sclerotinia sclerotiorum pac1 gene is required for sclerotial development and virulence. Mol Plant Microbe Interact 16:785–795
Roslan HA, Anji SB (2011) Characterization of inflorescence-predominant chitinase gene in Metroxylon sagu via differential display. Biotech 1:27–33
Sun XP, Zhao Y, Jia JC, Xie JT, Cheng JS, Liu HQ, Jiang DH, Fu YP (2017) Uninterrupted expression of cmsit1 in a sclerotial parasite coniothyrium minitans leads to reduced growth and enhanced antifungal ability. Front Microbiol 8:2208. https://doi.org/10.3389/fmicb.2017.02208
Telzur N, Abbo S, Myslabodski D, Mizrahi Y (1999) Modified CTAB procedure for DNA isolation from Epiphytic Cacti of the Genera Hylocereus and Selenicereus (Cactaceae). Plant Mol Biol Report 17:249–254
Vuong TD, Diers BW, Hartman GL (2008) Identification of QTL for resistance to Sclerotinia stem rot in soybean plant introduction 194639. Crop Sci 48:2209–2214
Walz A, Zingen-Sell I, Loeffler M, Sauer M (2010) Expression of an oxalate oxidase gene in tomato and severity of disease caused by Botrytis cinerea and Sclerotinia sclerotiorum. Plant Pathol 57:453–458
Wan XQ, Tan JL, Lu SY, Lin CY, Hu YH, Guo ZF (2009) Increased tolerance to oxidative stress in transgenic tobacco expressing a wheat oxalate oxidase gene via induction of antioxidant enzymes is mediated by H2O2. Physiol Plant 136:30–44
Williams B, Kabbage M, Kim HJ, Britt R, Dickman MB (2011) Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog 7(6):e1002107
Workneh F, Yang XB (2000) Prevalence of Sclerotinia stem rot of soybeans in the north-central United States in relation to tillage, climate, and latitudinal positions. Phytopathology 90:1375–1382
Yang X, Yang J, Wang Y, He H, Niu L, Guo D, Xing G, Zhao Q, Zhong X, Sui L, Li Q, Dong Y (2019) Enhanced resistance to sclerotinia stem rot in transgenic soybean that overexpresses a wheat oxalate oxidase. Transgenic Res 28(1):103–114
Zhang FL, Ruan XL, Wang X, Liu ZH, Hu LZ, Li CW (2016) Overexpression of a chitinase gene from Trichoderma asperellum increases disease resistance in transgenic soybean. Appl Biochem Biotechnol 180:1–17
Zhao X, Han YP, Li YH, Liu DY, Sun MM, Zhao Y, Lv CM, Li DM, Yang ZJ, Huang L, Teng WL, Qiu LJ, Zheng HK, Li WB (2015) Loci and candidate gene identification for resistance to Sclerotinia sclerotiorum in soybean (Glycine max L. Merr.) via association and linkage maps. Plant J Cell Mol Biol 82:245–255
Zhu WJ, Wei W, Fu YP, Cheng JS, Xie JT, Li GQ, Yi XH, Kang ZS, Dickman MB, Jiang DH (2013) A secretory protein of necrotrophic fungus Sclerotinia sclerotiorum that suppresses host resistance. PLoS ONE 8(1):e53901
Acknowledgements
This work was supported by grants from the Jilin Provincial Agricultural Science & Technology Innovation Project (CXGC2017JQ013) and China National Novel Transgenic Organisms Breeding Project (2016ZX08004-004). We thank Prof. Daohong Jiang of Huazhong Agricultural University for providing the CmCHI1 gene. We also thank Editage (www.editage.cn) for English language editing.
Author information
Authors and Affiliations
Contributions
YD and QL designed the experiments. XY and JY conducted the experiments and drafted the manuscript. GX, DG, and QZ conducted the Agrobacterium-mediated transformation experiments. WX and HL conducted the inoculation assay. LN and YZ participated in the qRT-PCR analyses. All authors participated in the manuscript revision.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiangdong Yang and Jing Yang have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11248_2020_190_MOESM1_ESM.pptx
M, DNA marker (2k); Ctl+, positive control; Nt, non-transformed plants Williams 82; Bk; Blank control; 1–7, T0 transgenic plants (PPTX 2921 kb)
Rights and permissions
About this article
Cite this article
Yang, X., Yang, J., Li, H. et al. Overexpression of the chitinase gene CmCH1 from Coniothyrium minitans renders enhanced resistance to Sclerotinia sclerotiorum in soybean. Transgenic Res 29, 187–198 (2020). https://doi.org/10.1007/s11248-020-00190-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-020-00190-2