Abstract
Quantitative real-time PCR (qPCR) is a sensitive method used to investigate relevant changes in gene expression during somatic embryogenesis. Understanding its regulatory network might be helpful to the process of induction of embryos and facilitate the development of efficient plant regeneration procedures. In this study, a set of 12 genes was selected and their stability was assessed in different tissues of somatic embryogenesis-related cultures of Coffea arabica. Analyses of gene expression stability were performed using the RefFinder tool that integrates the geNorm, NormFinder, BestKeeper and Delta-Ct algorithms. Among the all candidate reference genes studied, APRT/EF1a, UBQ/ACT, ACT/24S, RPL39/24S, PP2A/RPL39, PP2A/AP47, emerged as the most stable for normalization of qPCR analyses of embryogenic cell suspensions, non-embryogenic calli, embryogenic calli, combined embryogenic and non-embryogenic calli, somatic embryos and plantlet, respectively. A combination of two genes, 24S and PP2A, was identified as most suitable reference genes across all samples for the C. arabica tissues studied. The commonly employed reference gene encoding glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was found to be inappropriate as a reference gene for embryogenic tissues of C. arabica. In addition, Baby boom (BBM) gene expression was investigated to confirm the validity of the selected reference genes, the transcript levels of gene were overestimated when unsuitable reference genes were used for normalization. The results shown herein will permit a more precise and reliable normalization of qPCR in experiments involving somatic embryogenesis of C. arabica.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Coffee is one of the main commodities traded worldwide, with Coffea arabica L. accounting for about 60% of global production (International Coffee Organization 2014). However, to maintain production levels and to satisfy the expectations of demanding markets that best remunerate the producer, it is necessary to sustain programs aiming at coffee breeding. The release of elite genotypes that are disease resistant and produce top quality beans can be accelerated by combining conventional breeding and biotechnological techniques (Gatica-Arias et al. 2008). In this context, somatic embryogenesis is a technique with considerable potential since it not only provides the possibility of mass clonal propagation of genetically improved varieties and maintenance of germplasm, but also serves as an efficient regeneration system for the genetic transformation process (Pathi et al. 2013; Ribas et al. 2011; Winkelmann 2010).
Quantitative polymerase chain reaction (qPCR) is a rapid and sensitive technique that has frequently been used to study gene expression during somatic embryogenesis and embryo germination (Gruszczyńska and Rakoczy-Trojanowska 2011; Ma et al. 2012; Silva et al. 2014; Zhang et al. 2014). Although the procedure offers high reproducibility, precision and throughput (Bustin et al. 2005; Logan et al. 2009), the reliability of the technique depends on various factors including the integrity of RNA, the quality of cDNA synthesis, the number of repetitions, the efficiency of amplifications and the appropriate selection of reference genes used as internal controls for normalizing and monitoring sample-to-sample and run-to-run variations (Pfaffl et al. 2004; Santis et al. 2011; Vandesompele et al. 2002). Reference genes must exhibit moderate levels of expression and be stably expressed in different cell types and experimental conditions, but must not be associated with pseudogenes (Ling et al. 2014; Lland et al. 2006; Wan et al. 2010).
A great number of reference genes have been validated for gene expression analysis in plants, including those encoding β-actin, β-tubulin, ubiquitin, glyceraldehyde-3-phosphate dehydrogenase and elongation factors (Kumar et al. 2011). However, there appears to be no universal reference gene that could be used for all plant tissues and/or experimental conditions (Chen et al. 2011; Cheng et al. 2013; Gutierrez et al. 2008a, b; Imai et al. 2014; Lin et al. 2013; Rodrigues et al. 2014; Zeng et al. 2014), hence it is important to assess the stability of the selected reference gene under the specific experimental conditions employed in the expression analysis (Vandesompele et al. 2009). Indeed, the selection of suitable reference genes is a key step in qPCR analysis, since inappropriate choices will negatively affect the reliability of the results (Carvalho et al. 2013b; Docimo et al. 2013; Fan et al. 2013; Kong et al. 2014; Nolan et al. 2006).
RefFinder is a computational tool for evaluating and screening candidate reference genes from experimental datasets in order to infer their suitability for normalization of qPCR data. The web-based application assigns appropriate weights to individual candidate genes based on data obtained using the algorithms geNorm (Vandesompele et al. 2002), NormFinder (Andersen et al. 2004), BestKeeper (Pfaffl et al. 2004) and Delta-Ct (Silver et al. 2006), and ranks the genes according to the geometric mean of their weights (RefFinder 2016).
In the case of C. arabica, various reference genes have already been described for expression analysis (Barsalobres-Cavallari et al. 2009; Carvalho et al. 2013b; Cruz et al. 2009; Figueiredo et al. 2013; Goulao et al. 2012), but none have been validated in samples related to somatic embryogenesis. Considering the increasing interest in functional genomics of coffee that has arisen in response by the recent availability of genome and transcriptome data, the requirements for appropriate reference genes for expression normalization have become much more stringent. In order to address this issue, 12 candidate reference genes were selected and evaluated in 18 different embryogenic and non-embryogenic samples obtained from C. arabica explants to identify those suited for normalization of qPCR analyses and, therefore, appropriate for application in expression studies involving embryogenic tissues of this species. Also, the best combination of reference genes determined for all tissue types was used to further assess the expression of the Baby boom (BBM) gene—plant-specific transcription factor belonging to the AP2/ERF superfamily—which activates developmental pathways associated with cell proliferation and growth (Passarinho et al. 2008), and is involved in the acquisition of embryogenic competence (Namasivayam 2007). The results of the present study will be valuable in future research for defining reference genes that are appropriate for evaluation of target gene expression during the process of somatic embryogenesis in C. arabica.
Materials and methods
Somatic embryogenesis
Embryogenic and non-embryogenic calli were established from leaves of greenhouse-grown plants of C. arabica cv. Catuaí Amarelo IAC 62 according to the protocol described by Teixeira et al. (2004). Embryogenic cell suspensions were obtained by transferring calli to Erlenmeyer flasks containing liquid multiplication medium T3 (Van Boxtel and Berthouly 1996) at an inoculum density of 10 g callus L−1 (Zamarripa et al. 1991). The flasks were maintained in the dark under constant agitation at 100 rpm in a growth room at 25 °C, and the medium was replaced every 15 days. Embryos were regenerated in 2 months after transferring cell suspensions to RR medium (Carvalho et al. 2013a) at an inoculum density of 1 g L−1. Maturation and germination of somatic embryos was accomplished following the method described by Teixeira et al. (2004).
Experimental design
The experiment was performed according to a totally randomized design and each sample comprised three biological replicates. The samples evaluated included: (i) embryogenic and non-embryogenic calli: each repetition encompassed a set of ten calli obtained from different leaf explants; (ii) two cell lines of embryogenic cell suspensions with different culture times (60, 90, 120, 150, 180 and 210 days): each repetition consisted of 200 mg of cell agglomerates for each cell line (iii) somatic embryos at different stages of development: each repetition included 275 globular embryos, 25 cordiform/torpedo embryos and 25 cotyledonary embryos; (iv) coffee plantlets: each repetition included 25 plantlets. All samples were stored at −80 °C until required for the RNA extraction.
Extraction of total RNA and cDNA synthesis
Total RNA was extracted from embryogenic calli, embryogenic suspension cells and globular embryos using Macherey Nagel (Düren, Germany) NucleoSpin® kits, and from non-embryogenic calli, cordiform/torpedo embryos, cotyledonary embryos and plantlets using Invitrogen™ (Life Technologies, Carlsbad, CA, USA) Concert™ Plant RNA reagent. RNA extracts were treated with Ambion® (Life Technologies) Turbo DNA-free kit reagents in order to remove any contaminating genomic DNA. The quantity and purity of total RNA was assessed with an ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, NC, USA), while quality and integrity were verified using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) to ensure an RNA integrity number (RIN) ≥ 7.0. The synthesis of cDNA from 1000 ng aliquots of RNA was carried out using Applied Biosystems (Life Technologies) High-Capacity cDNA Reverse Transcription kits according to the recommendations of the manufacturer.
Selection of candidate reference genes and design of primers
A set of 12 potential reference genes that had been reported in other crops or frequently used in coffee were selected. These included ribosomal protein 24S (24S), β-actin (ACT), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), cyclophilin (CYCL), elongation factor 1α (EF1a), β-tubulin (TUB), protein phosphatase 2A (PP2A), clathrin adaptor complex medium subunit (AP47), 60S ribosomal protein L39 (RPL39), adenine phosphoribosyltransferase (APRT), ubiquitin (UBQ) and protein 14-3-3 (14-3-3).
In the case of 24S, ACT, GAPDH, CYCL, EF1a, TUB, PP2A, RPL39, AP47, APRT, UBQ, and 14-3-3 genes were selected from C. arabica EST sequences developed by the Brazilian Coffee Genome Project Consortium (http://www.lge.ibi.unicamp.br/cafe/) (Vieira et al. 2006) (Table 1). For BBM gene, we used primers pairs already reported in previous studies in Coffea (Silva et al. 2015). Coffee homologue sequences with the best matches were retrieved and submitted to Primer Express software version 3.0 (Applied Biosystems) for primer design. The specificity of each pair of primers was verified by analysis of the dissociation (melting) curves. PCR amplification efficiencies (E) and regression coefficients (R2) were determined during the validation of primers according to the standard curve method using a set of all cDNA samples with 5× serial dilution. The specifications of the selected reference genes and primer pairs are shown in Table 1.
qPCR amplification
qPCR analyses were performed using a Qiagen (Venlo, Netherlands) Rotor Gene-Q® thermal cycler with a reaction mix containing 7.5 μL of 2× Rotor-Gene SYBR® Green PCR Master Mix (Qiagen), 5 ng of cDNA, optimized concentrations of primers (see Table 1) and RNase-free water to a total volume of 15 μL. Amplification conditions involved an initial activation at 95 °C for 5 min and 40 cycles of denaturation at 95 °C for 5 s and combined annealing/extension at 60 °C for 10 s. In order to confirm the specificity of primers, melting curves were recorded after the 40 amplification cycles had been completed by increasing the temperature from 60 to 95 °C. All qPCR assays were carried out in technical and biological triplicate.
Analysis of expression stability of candidate reference genes
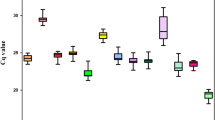
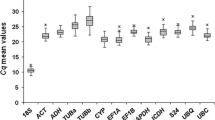
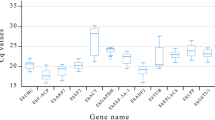
In qPCR, amplified DNA bears a fluorescent label and the amount of fluorescence detected during the reaction is directly proportional to the amount of amplified DNA present. The levels of expression of candidate reference genes were determined on the basis of the quantification cycle (Cq), also known as the threshold cycle (Ct), which is defined as the cycle at which the fluorescence from amplification exceeds that of the background. The Cq values of samples were determined using Qiagen Rotor Gene-Q Series software with the fluorescence threshold set at 0.2, and corrected according to the efficiency of each pair of primers with the aid of GenEx Enterprise software (MultiD Analyses, Göteberg, Sweden; http://genex.gene-quantification.info/). Box plot-type diagrams were made using Systat Software (San Jose, CA, USA) SigmaPlot version 12.0 to illustrate levels and variations in expression of the tested reference genes.
The RefFinder tool (http://fulxie.0fees.us) was employed to assess the stability of the tested genes in seven sample sets: (i) embryogenic cell suspensions at different culture times, (ii) non-embryogenic calli, (iii) embryogenic calli (iv) combined embryogenic and non-embryogenic calli; (v) globular, cordiform/torpedo and cotyledonary embryos, (vi) plantlets, and (vii) a pool of biological samples representing all tissue types from (i) to (vi). The RefFinder tool evaluates the rankings of stability according to the geNorm, NormFinder, BestKeeper and Delta-Ct algorithms and provides an overall ranking for the tested reference genes (RefFinder 2016). The geNorm algorithm calculates an M-value describing the variation of each candidate gene in comparison with all other candidates, and eliminates the gene with the highest M-value. This process is repeated until only two genes remain and these are then ranked as the best possible pair of reference genes. A low M-value, i.e. one that is below the cut-off point of 1.5, indicates transcriptional stability (Mamo et al. 2007; Rodrigues et al. 2014; Spinsanti et al. 2006; Vandesompele et al. 2002). NormFinder calculates both intra- and inter-sample variance and the stability value (SV) for each of the candidate reference genes. A low SV value matches a gene with high transcriptional stability (Andersen et al. 2004). The BestKeeper algorithm assesses the transcriptional stability of all candidate reference genes based on calculated variation parameters, namely standard deviation (SD), correlation coefficient (r) and coefficient of covariance (CV). The genes are then ranked according to variability from those most stably expressed (lowest variation) to the least stable (highest variation) (Pfaffl et al. 2004). The Delta-Ct algorithm calculates the differences between the Cq values of the tested reference genes in which the smaller the value the more stable is the gene transcript (Silver et al. 2006).
Validation of reference genes by BBM expression analysis
Coffea arabica baby boom (CaBBM) was screened from EST library in the Brazilian Coffee Genome Project data (Vieira et al. 2006). In order to validate the reference genes, the expression levels of CaBBM were quantified in the same different tissue types for reference genes tested using both the most-stable reference genes and the most-unstable ones, to demonstrate how the different reference genes adoption can affect the expression of a specific gene of interest. To determine the optimal number of reference genes for normalization in each experimental condition, pairwise variation (Vn/Vn + 1) was calculated using geNorm. Vandesompele et al. (2002) usually used 0.15 as a cutoff value to determine the optimal number of reference genes, below which the inclusion of additional reference genes is not required. The transcriptional activity of BBM was calculated by applying Pfaffl formula (Pfaffl 2001).
Results
Specificity and efficiency of primers
The amplification efficiencies (E) and correlation coefficients (R2) of the 12 candidate genes and BBM gene were generated using the slopes of the standard curves obtained by serial dilutions. Values of E varied from 80 to 100%, while those of R2 obtained from standard curves were ≥0.947 (Fig. S1 and Table 1), indicating that the amount of product was doubled at the end of each cycle. The specificity of each of the tested primer pairs was confirmed by the presence of a single peak corresponding to one amplicon in the respective melt curves (Fig. S1).
Levels of expression of candidate reference genes
The expression profiles of all qPCR products for all genes and all sample sets are shown in Fig. 1. The mean Cq values of the 12 candidate reference genes ranged from 17 to 28, indicating a wide variation in the levels of expression. The results obtained with the pool of biological samples showed that UBQ presented the lowest level of expression (mean Cq = 25.3) while EF1a exhibited the highest (mean Cq = 18.7). The coefficients of variation (CV) (lower values represent lower variability) of 12 reference genes were 1.80% (24S), 2.69% (ACT), 3.47% (GAPDH), 2.76% (CYCL), 2.61% (EF1a), 2.26% (TUB), 1.93% (PP2A), 2.48% (AP47), 2.39% (RPL39), 1.84% (APRT), 2.40% (UBQ), 2.86% (14-3-3).
When expression was evaluated in embryogenic cell suspensions at different culture times, GAPDH presented the highest CV (2.72%) and APRT the lowest (1.52%). For non-embryogenic calli, APRT presented the highest CV (0.70%) while UBQ exhibited the lowest (0.05%). Regarding embryogenic calli, EF1a presented the highest CV (2.43%) and RPL39 the lowest (0.24%). With the combined embryogenic and non-embryogenic calli, the highest CV was observed for GAPDH (4.21) and the lowest for RPL39 (0.42%). Samples from somatic embryos at different stages of development exhibited for TUB higher CV (5.17%), while APRT exhibited lower value (1.77%). Regarding plantlet samples, highest CV was observed for CYCL (0.60%) and the lowest for PP2A (0.06%).
Expression stability of candidate reference genes
According to M-values calculated by the geNorm algorithm using data obtained from the pool of biological samples (Table 2), all candidate genes exhibited acceptable transcriptional stability (M ≤ 1.5) although the stabilities of 24S and RPL39 (M = 0.273) were higher than those of the other genes. On the other hand, PP2A and 24S were designated as the most stable genes by NormFinder (SV = 0.290 and 0.390, respectively) and Delta-Ct (ΔCt = 0.592 and 0.628, respectively), while BestKeeper assigned 24S and APRT as most stable (SD = 0.425 and 0.435, respectively). All four algorithms identified GAPDH as the least stable gene (M = 0.706, SV = 0.711, SD = 0.562 and ΔCt = 0.850), although none of the stability values exceeded the recommended cut-off points. The overall ranking established by RefFinder revealed that the most stable candidate genes were 24S followed by PP2A.
In embryogenic cell suspension samples (Table 3), the most stable genes were identified as 24S and RPL39 by geNorm (M = 0.249), APRT by NormFinder and Delta-Ct (SV = 0.257 and ΔCt = 0.494, respectively) and EF1a by BestKeeper (SD = 0.342). All four algorithms identified AP47 as the least stable gene (M = 0.583, SV = 0.665, SD = 0.643 and ΔCt = 0.755). The overall ranking established by RefFinder revealed that the most stable candidate genes were APRT followed by EF1a, whereas AP47 was the least stable.
In non-embryogenic calli samples (Table 4), the geNorm algorithm identified UBQ and ACT as the most stable genes (M = 0.042), while BestKeeper and Delta-Ct algorithms both designated UBQ (SD = 0.013 and ΔCt = 0.098) as the most stable. In contrast, NormFinder assigned RPL39 as the most stable gene (SV = 0.029) followed by UBQ (SV = 0.032). All four algorithms identified APRT as the least stable gene (M = 0.127, SV = 0.172, SD = 0.160 and ΔCt = 0.188). In the overall ranking, UBQ and ACT were classified as the most stable genes, although all candidate genes presented relatively high transcriptional stability in this system.
In embryogenic calli samples (Table 5), geNorm identified 24S and UBQ as the most stable genes (M = 0.094), whereas NormFinder designated ACT and TUB as most stable (SV = 0.040). In contrast, BestKeeper assigned RPL39 and 24S (SD = 0.047 and 0.106, respectively) as most stable, while Delta-Ct identified ACT as showing the greatest transcriptional stability (ΔCt = 0.253) similar to the finding of NormFinder. In the overall ranking, ACT followed by 24S were classified as the most stable genes with EF1a as the least stable.
In combined embryogenic and non-embryogenic calli samples (Table 6), the genes RPL39 and 24S were identified as the most stable by all four algorithms (M = 0.106; SV = 0.053 and 0.094, respectively; SD = 0.083 and 0.145, respectively; ΔCt = 0.489 and 0.488, respectively). According to the overall ranking, the most stable genes were RPL39 followed by 24S, while UBQ constituted the least stable.
In samples from somatic embryos at different stages of development (Table 7), geNorm classified PP2A and RPL39 as the most stable genes (M = 0.200), a finding that was in agreement with those of NormFinder (SV = 0.074 and 0.099, respectively) and Delta-Ct (ΔCt = 0.470 and 0.475, respectively). According to BestKeeper, however, APRT and 14-3-3 were the most stable genes (SD = 0.421 and 0.427, respectively). In the overall ranking, PP2A and RPL39 were considered the most stable genes while TUB was the least stable.
Finally, in samples of C. arabica plantlets (Table 8), PP2A and APRT were indicated as the most stable genes by geNorm (M = 0.024) and BestKeeper (SD = 0.014 and 0.024, respectively), while NormFinder and Delta-Ct classified AP47 as the most stable (SV = 0.017 and ΔCt = 0.069, respectively). According to the overall ranking, PP2A followed by AP47 were the most stable genes and CYCL was the least stable, although all candidate genes presented relatively high stability.
Optimal number of reference genes
To generate accurate and reliable results, a combination of stable reference genes is paramount to obtaining accurate results when using several reference genes (Liu et al. 2015). Normalization with an inadequate number of reference gene can produce significant analyses errors (Vandesompele et al. 2002). Results showed that pairwise variation values for V2/3 were below the cut-off value of 0.15 in all sample sets (Fig. 2), indicating that the combination of two stable reference genes would be sufficient for the gene expression normalization.
Reference genes validation
To assess the impact of the selection of reference genes on gene expression calculations, we analyzed the BBM expression by employing two normalization strategies (Fig. 3). The comparative analysis of relative expression profiles for target gene in samples related the process of somatic embryogenesis were very similar when normalized with different internal reference genes. BBM transcripts showed higher relative expression levels in embryogenic calli and embryogenic cell suspensions. However, the relative transcript abundance for target gene was dependent on the reference genes used for normalization, BBM expression levels were obviously overestimated when unsuitable reference genes were used for normalization. Moreover, the utilization of the newly identified normalization controls resulted in significantly lower standard deviations underlining the higher reproducibility of the results. This indicates that questionable results would be produced by using unstable reference genes. These results reinforce the importance of validating reference genes prior to experimental applications.
Differential gene expression of BBM using the selected reference genes. Relative gene expression quantification was performed using two different normalization strategies: the combination of the two top ranked genes and combination two most unstable genes. The columns represent the gene expression in different materials (NEC non-embryogenic calli, EC embryogenic calli, ECS embryogenic cell suspensions with 60, 90, 120, 150, 180 and 210 days culture, GLO globular embryos, TOR cordiform/torpedo embryos, COT cotyledonary embryos, PLA plantlets) of C. arabica. Error bars indicate standard deviation (SD). Letters denote statistically significant differences (Student’s t test, P < 0.05)
Discussion
Coffee is the world’s favorite beverage, plays an important role in industry. Studies are necessary to produce quality coffee or protect the coffee supply chain from economic, climate, or pest and disease threats. Despite the rapid exploration of the coffee genome and the growing requirement for the deep biological study of gene function in published papers that employ—OMIC, to our knowledge, very limited information is available on the expression stability of reference genes in Coffea spp. during somatic embryogenesis. The expression pattern, a reflection of the biological function of a target gene, is preferably detected by qPCR method, in which reference gene is used for normalization (Lin et al. 2014). The expression patterns of reference genes are expected to be stable irrespective of experimental conditions (Chen et al. 2011; Cheng et al. 2013; Gutierrez et al. 2008a, b; Imai et al. 2014; Lin et al. 2013; Rodrigues et al. 2014; Zeng et al. 2014). It is assumed that genes encoding proteins involved in the primary metabolism and structural integrity of cells are uniformly expressed regardless of the experimental conditions and cell type (Vandesompele et al. 2002). However, studies have shown that there is no universal reference gene appropriate for all cells and conditions because these genes can participate in other cell functions (Chen et al. 2011; Cheng et al. 2013; Gutierrez et al. 2008a, b; Imai et al. 2014; Lin et al. 2013; Rodrigues et al. 2014; Zeng et al. 2014). Therefore, it is necessary to perform a systematic validation of candidate reference genes for the specific tissues to be evaluated.
RefFinder ranks candidates genes on the basis of output from different algorithms, namely geNorm, NormFinder, BestKeeper and Delta-Ct. When we consider the stability for some genes in the four analyzes, it was observed discrepant results in the same sample set because the algorithms use dissimilar mathematical models to calculate the gene expression stability. Volland et al. (2016) recommended to employ a minimum of two experimental validation procedures, as individual algorithms can produce variable results, and no consensus can be reported. The use of the RefFinder tool can be an alternative for a global ranking. However, when raw Cq values are used as input in RefFinder, the results are not faithful to those generated by the software packages (Spiegelaere et al. 2015). Before the data entry in RefFinder, the raw data should be converted assuming specific primer efficiency for each gene. Thereafter, similar rankings to those of the original algorithms can be verified, as demonstrated by Li et al. (2016).
Real time qPCR data are frequently normalized without validation of the reference genes (Gutierrez et al. 2008a, b). In present study, it is of interest to note, the commonly employed reference gene GAPDH (Cardoso et al. 2014; Ivamoto et al. 2015; Marraccini et al. 2011; Ságio et al. 2014; Silva et al. 2014, 2015) exhibited very high levels of expression (i.e. very low Cq values) but was ranked as one of the most unstable of the candidate genes by all four of the algorithms accessed by RefFinder. One explanation for this finding is that the criterion for selection of a reference gene requires gene expression at a moderate level (Ling et al. 2014). Moreover, the protein encoded by GAPDH performs alternative metabolic roles (Zaffagnini et al. 2013), hence expression levels are likely to be highly variable. Instability in GAPDH expression has been reported for coffee hypocotyls inoculated with Colletotrichum kahawae (Figueiredo et al. 2013), although the gene exhibited stable expression in other tissues/organs of Coffea arabica (namely, roots, stems, leaves and fruits) (Barsalobres-Cavallari et al. 2009) and in leaves and roots that had been submitted to abiotic stress (Carvalho et al. 2013b). In soybean under water stress (Stolf-Moreira et al. 2011), strawberry fruits (Galli et al. 2015), maize grains (Galli et al. 2013), lettuce (Borowski et al. 2014), among others, the GAPDH gene was also considered less stable.
Expression analysis of 12 candidate different reference genes in a pool of C. arabica samples comprising embryogenic and non-embryogenic calli, embryogenic cell suspensions with different culture times, somatic embryos at different developmental stages and plantlets, revealed that 24S and PP2A were the most stably expressed. However, distinct analyses of the different categories of samples revealed that other genes exhibited transcriptional stability. For instance, UBQ emerged as the most stable gene in non-embryogenic calli, while ACT and APRT were the most stable in embryogenic calli and cell suspensions, respectively. In contrast, when combined samples of embryogenic and non-embryogenic calli were analyzed, RPL39 emerged as the most stable. Moreover, the highest level of transcriptional stability in somatic embryos and plantlet samples was attained by PP2A. Variation in the expression stability of reference genes in different tissues has been frequently reported (Barsalobres-Cavallari et al. 2009; Carvalho et al. 2013b; Chao et al. 2012; Imai et al. 2014; Lin et al. 2013; Yeap et al. 2014), hence the discrepancies observed in our study are not surprising and reinforce the importance of caution in the selection of suitable reference genes for the normalization of qPCR analyses.
In summary, even though GAPDH and ACT have been previously employed for normalization of expression data in calli and cell suspensions of coffee (Silva et al. 2014, 2015), the present study revealed that GAPDH was not appropriate as a reference gene for any embryogenic tissues of coffee, while ACT was appropriate only for embryogenic calli. The arbitrary choice of reference genes leads to inadequate normalization, imprecision in the qPCR technique and improper quantification of the target gene (Carvalho et al. 2013b; Fan et al. 2013; Kong et al. 2014; Nolan et al. 2006). The results obtained in this study assist to emphasize the necessity of validating reference genes for the specific tissues to be evaluated.
To demonstrate the need for accurate relative quantification using suitable reference genes, the expression of the C. arabica BBM gene was studied. A clear role for CaBBM in the embryogenic process is none-the-less evident based on high expression embryogenic calli but not in the non-embryogenic calli. The previous reports showed baby boom (BBM) gene is related to cell proliferation and morphogenesis during embryogenesis (Boutilier et al. 2002; Florez et al. 2015; Kulinska-Lukaszek et al. 2012; Passarinho et al. 2008). However, results this work suggest that BBM gene in C. arabica can be only associated with cell proliferation and acquisition of embryogenic capacity because high levels of transcription of the target gene were not verified during the transition from embryos to plantlets.
In summary, among the 12 candidate reference genes studied, 24S/PP2A emerged as the most appropriate for normalization of qPCR analyses of all somatic embryogenesis-related cultures of C. arabica. We recommend APRT/EF1a, UBQ/ACT, ACT/24S, RPL39/24S, PP2A/RPL39, PP2A/AP47 for embryogenic cell suspensions, non-embryogenic calli, embryogenic calli, combined embryogenic and non-embryogenic calli, somatic embryos, plantlet, respectively. The results provide guidelines for reference genes selection towards a more accurate use in normalization of qPCR in future Coffea transcriptomic studies involving embryogenesis-related tissues.
References
Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64(15):5245–5250. doi:10.1158/0008-5472.CAN-04-0496
Barsalobres-Cavallari CF, Severino FE, Maluf MP, Maia IG (2009) Identification of suitable internal control genes for expression studies in Coffea arabica under different experimental conditions. BMC Mol Biol 10:1–11. doi:10.1186/1471-2199-10-1
Borowski JM, Galli V, da Silva Messias R, Perin EC, Buss JH, dos Anjos e Silva SD et al (2014) Selection of candidate reference genes for real-time PCR studies in lettuce under abiotic stresses. Planta 239(6):1187–1200. doi:10.1007/s00425-014-2041-2
Boutilier K, Offringa R, Sharma VK, Kieft H, Ouellet T, Zhang L et al (2002) Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14(8):1737–1749. doi:10.1105/tpc.001941
Bustin SA, Benes V, Nolan T, Pfaffl MW (2005) Quantitative real-time RT-PCR—a perspective. J Mol Endocrinol 34(3):597–601. doi:10.1677/jme.1.01755
Cardoso DC, Martinati JC, Giachetto PF, Vidal RO, Carazzolle MF, Padilha L et al (2014) Large-scale analysis of differential gene expression in coffee genotypes resistant and susceptible to leaf miner–toward the identification of candidate genes for marker assisted-selection. BMC Genomics 15:66. doi:10.1186/1471-2164-15-66
Carvalho CHS, Paiva ACRS, Silva EQ, Custódio AA (2013a) Custo de produção de mudas clonais de café arábica produzidas por embriogênese somática. Circular Técnica 3, Embrapa, Brasília. http://www.sapc.embrapa.br/arquivos/consorcio/circulartecnica/circular_tecnica_03.pdf. Accessed 1 June 2015
Carvalho K, Bespalhok Filho JC, Dos Santos TB, De Souza SGH, Vieira LGE, Pereira LFP et al (2013b) Nitrogen starvation, salt and heat stress in coffee (Coffea arabica L.): identification and validation of new genes for qPCR normalization. Mol Biotechnol 53(3):315–325. doi:10.1007/s12033-012-9529-4
Chao WS, Doğramaci M, Foley ME, Horvath DP, Anderson JV (2012) Selection and validation of endogenous reference genes for qRT-PCR analysis in leafy spurge (Euphorbia esula). PLoS One 7:e42839. doi:10.1371/journal.pone.0042839
Chen L, Zhong H-Y, Kuang J-F, Li J-G, Lu W-J, Chen J-Y (2011) Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta 234(2):377–390. doi:10.1007/s00425-011-1410-3
Cheng D, Zhang Z, He X, Liang G (2013) Validation of reference genes in Solenopsis invicta in different developmental stages, castes and tissues. PLoS One 8(2):e57718. doi:10.1371/journal.pone.0057718
Cruz F, Kalaoun S, Nobile P, Colombo C, Almeida J, Barros LMG et al (2009) Evaluation of coffee reference genes for relative expression studies by quantitative real-time RT-PCR. Mol Breed 23(4):607–616. doi:10.1007/s11032-009-9259-x
Docimo T, Schmidt GW, Luck K, Delaney SK, D’Auria JC (2013) Selection and validation of reference genes for quantitative gene expression studies in Erythroxylum coca. F1000Res 2:37. doi:10.3410/f1000research.2-37.v1
Fan C, Ma J, Guo Q, Li X, Wang H, Lu M (2013) Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis). PLoS One 8(2):e56573. doi:10.1371/journal.pone.0056573
Figueiredo A, Loureiro A, Batista D, Monteiro F, Várzea V, Pais MS et al (2013) Validation of reference genes for normalization of qPCR gene expression data from Coffea spp. hypocotyls inoculated with Colletotrichum kahawae. BMC Res Notes 6:388. doi:10.1186/1756-0500-6-388
Florez SL, Erwin RL, Maximova SN, Guiltinan M, Curtis WR (2015) Enhanced somatic embryogenesis in Theobroma cacao using the homologous BABY BOOM transcription factor. BMC Plant Biol 15:121. doi:10.1186/s12870-015-0479-4
Galli V, da Silva Messias R, dos Anjos e Silva e SD, Rombaldi CV (2013) Selection of reliable reference genes for quantitative real-time polymerase chain reaction studies in maize grains. Plant Cell Rep 32(12):1869–1877. doi:10.1007/s00299-013-1499-x
Galli V, Borowski JM, Perin EC, da Silva Messias R, Labonde J, Pereira IS et al (2015) Validation of reference genes for accurate normalization of gene expression for real time-quantitative PCR in strawberry fruits using different cultivars and osmotic stresses. Gene 554(2):205–214. doi:10.1016/j.gene.2014.10.049
Gatica-Arias AM, Arrieta-Espinoza G, Espinoza Esquivel AM (2008) Plant regeneration via indirect somatic embryogenesis and optimisation of genetic transformation in Coffea arabica L. cvs. Caturra and Catuaí. Electron J Biotech 11(1):1–11. doi:10.2225/vol11-issue1-fulltext-9
Goulao LF, Fortunato AS, Ramalho JC (2012) Selection of reference genes for normalizing quantitative real-time PCR gene expression data with multiple variables in Coffea spp. Plant Mol Biol Rep 30(3):741–759. doi:10.1007/s11105-011-0382-6
Gruszczyńska A, Rakoczy-Trojanowska M (2011) Expression analysis of somatic embryogenesis-related SERK, LEC1, VP1 and NiR ortologues in rye (Secale cereale L.). J Appl Genet 52(1):1–8. doi:10.1007/s13353-010-0015-z
Gutierrez L, Mauriat M, Guénin S, Pelloux J, Lefebvre JF, Louvet R et al (2008a) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J 6(6):609–618. doi:10.1111/j.1467-7652.2008.00346.x
Gutierrez L, Mauriat M, Pelloux J, Bellini C, Van Wuytswinkel O (2008b) Towards a systematic validation of references in real-time RT-PCR. Plant Cell 20(7):1734–1735. doi:10.1105/tpc.108.059774
Imai T, Ubi BE, Saito T, Moriguchi T (2014) Evaluation of reference genes for accurate normalization of gene expression for real time-quantitative PCR in Pyrus pyrifolia using different tissue samples and seasonal conditions. PLoS One 9(1):e86492. doi:10.1371/journal.pone.0086492
International Coffee Organization World coffee trade (1963–2013) (2014) A review of the markets, challenges and opportunities facing the sector. London, International Coffee Council. http://www.ico.org/news/icc-111-5-r1e-world-coffee-outlook.pdf. Accessed 1 June 2015.
Ivamoto ST, Domingues DS, Vieira LGE, Pereira LFP (2015) Identification of the transcriptionally active cytochrome P450 repertoire inCoffea arabica. Genet Mol Res 14(1):2399–2412. doi:10.4238/2015.March.27.25
Kong Q, Yuan J, Niu P, Xie J, Jiang W, Huang Y et al (2014) Screening suitable reference genes for normalization in reverse transcription quantitative real-time PCR analysis in melon. PLoS One 9(1):e87197. doi:10.1371/journal.pone.0087197
Kulinska-Lukaszek K, Tobojka M, Adamiok A, Kurczynska EU (2012) Expression of the BBM gene during somatic embryogenesis of Arabidopsis thaliana. Biol Plant 56(2):389–394. doi:10.1007/s10535-012-0105-
Kumar V, Sharma R, Trivedi PC, Vyas GK, Khandelwal V (2011) Traditional and novel references towards systematic normalization of qRT-PCR data in plants. Aust J Crop Sci 5(11):1455–1468
Li J, Han J, Hu Y, Yang J (2016) Selection of reference genes for quantitative real-time PCR during flower development in tree peony (Paeonia suffruticosa Andr.). Front Plant Sci 7:516. doi:10.3389/fpls.2016.00516
Lin L, Han X, Chen Y, Wu Q, Wang Y (2013) Identification of appropriate reference genes for normalizing transcript expression by quantitative real-time PCR in Litsea cubeba. Mol Genet Genomics 288(12):727–737. doi:10.1007/s00438-013-0785-1
Lin Y, Zhang C, Lan H, Gao S, Liu H, Liu J et al (2014) Validation of potential reference genes for qPCR in maize across abiotic stresses, hormone treatments, and tissue types. PLoS One 9(2):e95445. doi:10.1371/journal.pone.0095445
Ling H, Wu Q, Guo J, Xu L, Que Y (2014) Comprehensive selection of reference genes for gene expression normalization in sugarcane by real time quantitative RT-PCR. PLoS One 9(5):e97469. doi:10.1371/journal.pone.0097469
Liu M, Wu Z, Jiang F (2015) Selection and validation of garlic reference genes for quantitative real-time PCR normalization. Plant Cell Tiss Organ Cult 122:435. doi:10.1007/s11240-015-0780-9
Lland H, Hertzberg M, Marlton P (2006) Myeloid leukemia: methods and protocols. Humana Press, Totowa
Logan J, Edwards K, Saunders N (2009) Real-time PCR: current technology and applications. Poole. Caister Academic Press, Wymondham
Ma J, He Y, Wu C, Liu H, Hu Z, Sun G (2012) Cloning and molecular characterization of a SERK gene transcriptionally induced during somatic embryogenesis in Ananas comosus cv. Shenwan. Plant Mol Biol Rep 30(1):195–203. doi:10.1007/s11105-011-0330-5
Mamo S, Gal AB, Bodo S, Dinnyes A (2007) Quantitative evaluation and selection of reference genes in mouse oocytes and embryos cultured in vivo and in vitro. BMC Dev Biol 7:14. doi:10.1186/1471-213X-7-14
Marraccini P, Freire LP, Alves GSC, Vieira NG, Vinecky F, Elbelt S et al (2011) RBCS1expression in coffee: Coffea orthologs, Coffea arabica homeologs, and expression variability between genotypes and under drought stress. BMC Plant Biol 11:85. doi:10.1186/1471-2229-11-85
Namasivayam P (2007) Acquisition of embryogenic competence during somatic embryogenesis. Plant Cell Tiss Organ Cult 90:1. doi:10.1007/s11240-007-9249-9
Nolan T, Hands RE, Bustin SA (2006) Quantification of mRNA using real-time PCR. Nat Protoc 1(3):1559–1582. doi:10.1038/nprot.2006.236
Passarinho P, Ketelaar T, Xing M, Van Arkel J, Maliepaard C, Hendriks MW et al (2008) BABY BOOM target genes provide diverse entry points into cell proliferation and cell growth pathways. Plant Mol Biol 68(3):225–237. doi:10.1007/s11103-008-9364-y
Pathi KM, Tula S, Tuteja N (2013) High frequency regeneration via direct somatic embryogenesis and efficient Agrobacterium-mediated genetic transformation of tobacco. Plant Signal Behav 8:e24354. doi:10.4161/psb.24354
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29(9):e45–e45. doi:10.1093/nar/29.9.e45
Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol Lett 26(6):509–515. doi:10.1023/B:BILE.0000019559.84305.47
RefFinder (2016). Evaluating reference genes expression. Greenville, B. Zhang, East Carolina University. http://fulxie.0fees.us. Accessed 9 Nov 2016
Ribas AF, Dechamp E, Champion A, Bertrand B, Combes MC, Verdeil JL et al (2011) Agrobacterium-mediated genetic transformation of Coffea arabica (L.) is greatly enhanced by using established embryogenic callus cultures. BMC Plant Biol 11:92. doi:10.1186/1471-2229-11-92
Rodrigues TB, Khajuria C, Wang H, Matz N, Cardoso DC, Valicente FH et al (2014) Validation of reference housekeeping genes for gene expression studies in western corn rootworm (Diabrotica virgifera virgifera). PLoS One 9(10):e109825. doi:10.1371/journal.pone.0109825
Ságio SA, Barreto HG, Lima AA, Moreira RO, Rezende PM, Paiva LV et al (2014) Identification and expression analysis of ethylene biosynthesis and signaling genes provides insights into the early and late coffee cultivars ripening pathway. Planta 239(5):951–963. doi:10.1007/s00425-014-2026-1
Santis C, Smith-Keune C, Jerry DR (2011) Normalizing RT-qPCR data: are we getting the right answers? An appraisal of normalization approaches and internal reference genes from a case study in the finfish Lates calcarifer. Mar Biotechnol 13(2):170–180. doi:10.1007/s10126-010-9277-z
Silva AT, Barduche D, Livramento KG, Ligterink W, Paiva LV (2014) Characterization of a putative serk-like ortholog in embryogenic cell suspension cultures of Coffea arabica L. Plant Mol Biol Rep 32(1):176–184. doi:10.1007/s11105-013-0632-x
Silva AT, Barduche D, Livramento KG, Paiva LV (2015) A putative BABY BOOM -like gene (CaBBM) is expressed in embryogenic calli and embryogenic cell suspension culture ofCoffea arabica L. In Vitro Cell Dev Biol 51(1):93–101. doi:10.1007/s11627-014-9643-z
Silver N, Best S, Jiang J, Thein SL (2006) Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol 7:33. doi:10.1186/1471-2199-7-33
Spiegelaere W, Dern-Wieloch J, Weigel R, Schumacher V, Schorle H, Nettersheim D et al (2015) Reference gene validation for RT-qPCR, a note on different available software packages. PLoS One 10(3):e0122515. doi:10.1371/journal.pone.0122515
Spinsanti G, Panti C, Lazzeri E, Marsili L, Casini S, Frati F et al (2006) Selection of reference genes for quantitative RT-PCR studies in striped dolphin (Stenella coeruleoalba) skin biopsies. BMC Mol Biol 7:32. doi:10.1186/1471-2199-7-32
Stolf-Moreira R, Lemos EGM, Abdelnoor RV, Beneventi MA, Rolla AAP, Pereira SS, et al (2011) Identification of reference genes for expression analysis by real-time quantitative PCR in drought-stressed soybean. Pesq Agropec Bras 46(1):58–65. doi:10.1590/S0100-204X2011000100008
Teixeira JB, Junqueira CS, Costa Pereira AJC, Mello RIS, Silva APD, Mundim DA (2004) Multiplicação clonal de café (Coffea arabica L.) via embriogênese somática. Documentos 121. Brasília, Embrapa. http://www.embrapa.br/documents/1355163/2021185/doc121.pdf/8b7b0ff2-9308-4485-a32d-34cb4bdcda4d. Accessed 1 June 2015
Van Boxtel J, Berthouly M (1996) High frequency somatic embryogenesis from coffee leaves. Factors influencing embryogenesis and subsequent proliferation and regeneration in liquid medium. Plant Cell Tiss Org Cult 44(1):4–17. doi:10.1007/BF00045907
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy R, De Paepe A et al (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. doi:10.1186/gb-2002-3-7-research0034
Vandesompele J, Kubista M, Pfaffl MW (2009) Reference gene validation software for improved normalization. In: Logan J, Edwards K, Saunders N (eds) Real-time PCR: current technology and applications. Poole, Caister Academic Press, pp 47–64
Vieira LGE, Andrade AC, Colombo CA, Moraes AHA, Metha A, Oliveira AC et al (2006) Brazilian coffee genome project: an EST-based genomic resource. Braz J Plant Physiol 18(1):95–108
Volland M, Blasco J, Hampel M (2016) Validation of reference genes for RT-qPCR in marine bivalve ecotoxicology: systematic review and case study. BioRxiv. doi:10.1101/074542
Wan H, Zhao Z, Qian C, Sui Y, Malik AA, Chen J (2010) Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal Biochem 399(2):257–261. doi:10.1016/j.ab.2009.12.008
Winkelmann T (2010) Clonal propagation of Cyclamen persicum via somatic embryogenesis. Methods Mol Biol 589:281–290. doi:10.1007/978-1-60327-114-1_26
Yeap W, Loo JM, Wong YC, Kulaveerasingam H (2014) Evaluation of suitable reference genes for qRT-PCR gene expression normalization in reproductive, vegetative tissues and during fruit development in oil palm. Plant Cell Tiss Organ Cult 116:55. doi:10.1007/s11240-013-0382-3
Zaffagnini M, Fermani S, Costa A, Lemaire SD, Trost P (2013) Plant cytoplasmic GAPDH: redox post-translational modifications and moonlighting properties. Front Plant Sci 4:450. doi:10.3389/fpls.2013.00450
Zamarripa A, Ducos JP, Bollon H, Dufour M, Pétiard V (1991) Production d’embryons somatiques de caféier en milieu liquide: effets densité d’inoculation et renouvellement du milieu. Café Cacao Thé 35(4): 233–244
Zeng S, Liu Y, Wu M, Liu X, Shen X, Liu C et al (2014) Identification and validation of reference genes for quantitative real-time PCR normalization and its applications in Lycium. PLoS One 9(5):e97039. doi:10.1371/journal.pone.0097039
Zhang Y, Clemens A, Maximova SN, Guiltinan MJ (2014) The Theobroma cacao B3 domain transcription factor TcLEC2 plays a duel role in control of embryo development and maturation. BMC Plant Biol 14(1):106. doi:10.1186/1471-2229-14-106
Acknowledgements
We thank especially MSc Wesley Pires Flausino Máximo for helpful comments on this manuscript. The authors also would like to thank Fundação de Amparo à Pesquisa de Minas Gerais (FAPEMIG), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for their financial support. These funding programs did not play a role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
Conceived and designed the experiments NCF, HGB, LVP, ACJ. Performed the experiments NCF, CNF, ROM. Analyzed and interpreted the data NCF, CNF. Contributed with reagents and materials LVP, ACJ. Wrote the paper NCF. Made critical revisions of the content of the paper LVP, HGB, ACJ.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Freitas, N.C., Barreto, H.G., Fernandes-Brum, C.N. et al. Validation of reference genes for qPCR analysis of Coffea arabica L. somatic embryogenesis-related tissues. Plant Cell Tiss Organ Cult 128, 663–678 (2017). https://doi.org/10.1007/s11240-016-1147-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-016-1147-6