Abstract
Soil salinity can be a major limiting factor for productivity in agriculture and forestry and in order to fully utilize saline lands productively in plantation forestry for fig production, the genetic modification of tree species for salt tolerance may be required. Na+/H+ antiporters have been suggested to play important roles in salt tolerance in plants. Here, we isolated AgNHX1 a vacuolar Na+/H+ antiporter from a halophytic species Atriplex gmelini and introduced it into fig (Ficus carica L.) cv. Black Mission via Agrobacterium-mediated transformation. Leaf discs explants of fig were co-cultivated for 2 days with Agrobacterium tumefaciens strain LBA 4404 harboring the binary vector pBI121 containing the AgNHX1 gene and the hpt selectable marker gene which encodes hygromycin phosphotransferase. Explants were cultured on MS medium containing 30 mg L−1 hygromycin, 3 % sucrose, 0.2 mg L−1 kinetin and 2.0 mg L−1 2,4-dichlorophenoxyacetic acid solidified with 2.5 g L−1 phytagel in darkness for callus formation. The calli were cultured on MS medium containing 2.0 mg L−1 zeatin riboside in combination with 0.4 mg L−1 indole acetic acid in the light for plant regeneration. Putative regenerated transformant shoots were confirmed by polymerase chain reaction (PCR) and Southern hybridization for the AgNHX1 gene. Reverse transcriptase polymerase chain reaction analysis indicated that the gene was highly expressed in transgenic plants, but the degree of this expression varied among transformants. Overexpression of the AgNHX1 gene conferred high tolerance to salt stress and transgenic fig plants overexpressing AgNHX1 developed normally under salinity conditions compared to those of non-transgenic plants. Salt treated transgenic plants contained high proline and K+ but less Na+ compared to non-transgenic control plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The common fig (Ficus carica L.) of the family Moraceae, has been cultivated for its fruit and saleable produce commercially for many years. Fig is native to western Asia and has subsequently been transplanted and cultivated throughout Mediterranean region and in other similar climatic zones around the world. The main producers of edible figs include Turkey, Egypt, Morocco, Spain, Greece, California, Italy, Brazil and elsewhere in regions with mild winters that are usually dry and with summers that are hot often requiring irrigation (Küden 1996). As a perennial plantation crop the use of continuous groundwater irrigation is commonplace but can lead to build-up mineral salts in the rooting zone reducing the production potential (Tóth et al. 2008). Plant breeding of fig is laborious and time consuming often with variable results and successful genetic transformation of commercial fig cultivars provides a promising tool for the introduction of desirable genes to improve current fig cultivars (Flaishman et al. 2008a; Akdemir et al. 2012). Genetic and transformation methodologies offer the opportunity for the production of transgenic cultivars with improved agronomic characteristics, such as disease resistance, fruit storability, and enhanced fruit quality and flavour (Azafadi 2012; Meneses and Orellana 2013; Bakhsh and Hussain 2015). In addition, transformation of figs could provide the means for the production of desirable proteins in the edible parts of fig leading to enhanced nutritional and/or pharmaceutical composition (Yancheva et al. 2005; Flaishman et al. 2008b). The cultivar “Black Mission” is the most renowned fig in agricultural production and it produces two crops a year, with big and sweet fruits with distinctive flavour figs that can eaten fresh or dried.
Plants having the genetic ability to grow on salty land are classified as salt tolerant (glycophytes) or salt resistant (halophytes) on the basis of coping with excess Na+ in the cytocel. Halophytes have evolved the ability to grow normally under high salinity (Flowers and Yeo 1986; Khan and Duke 2001; Roohi et al. 2011) but glycophytes have the ability to develop a metabolic steady state to growth under salt stress and often restrict the intake of Na+ into the cytoplasm (Niu et al. 1993; Zhu 2007; Flowers and Colmer 2008). Some glycophytes can reduce Na+ ions out of the cytosol by transporting them into the vacuolar lumen or out of the cell using Na+/H+ exchangers localized in the vacuolar and plasma membranes (Apse and Blumwald 2007). Bassil et al. (2012) confirmed that Na+/H+ antiporters play a vital part in the homeostatic mechanism and are found in the plasma membrane, where they assist in moving Na+ from cells or sequester it into the vacuole. Given these hypothesis and results, Borsani et al. (2003) and Khan (2011) indicated that to enhance the salt tolerance in a salt sensitive plant, genetic engineering with sodium and hydrogen (Na+/H+) antiporters could lead to an improvement in salt tolerance.
Hamada et al. (2001) have successfully isolated an assumed Na+/H+ antiporter gene (AgNHX1) from Atriplex gmelini and made this available for transformation. The isolated cDNA is 2607 bp in length and contains one open reading frame, which comprises 555 amino acid residues with a predicted molecular mass of 61.9 kDa. The amino acid sequence of the AgNHX1 gene showed more than 75 % identity with those of the previously isolated NHX1 genes from glycophytes, Arabidopsis thaliana and Oryza sativa. There is evidence that overexpression of AtNHX1 in wild type Arabidopsis thaliana plants conferred higher salt tolerance (Apse et al. 1999). The transformed gene for a putative plant vacuolar Na+/H+ antiporter (AtNHX1) was isolated from Arabidopsis and the activity of Na+/H+ antiporter increased upon treatment with NaCl and showed more increase plant tolerance to NaCl in transformants plants compared to wild types (Yu et al. 2007; Dorani-Uliaie et al. 2012). Also, overexpression of AtNHX1 in tomato resulted in transgenic plants that were able to grow, flower and set fruits at higher salt concentrations (Zhang and Blumwald 2001; Razzaque et al. 2014).
This work described here concerns the in vitro regeneration and transformation of fig (Ficus carica L.) cv. Black Mission based on the infection of leaf sections with Agrobacterium tumefaciens harboring the AgNHX1 gene and to determine whether overexpression of AgNHX1 under 300 mM NaCl improve the mRNA and salt tolerance. These results are the first recorded that test the efficiency of cloning AgNHX1 in the fig (Ficus carica L.) genome.
Materials and methods
In vitro regeneration of fig (Ficus carica L.)
The leaves of Ficus carica L. cv. Black Mission were taken from in vitro formed plants produced from shoot tips and the in vitro leaves were used as explants. Explants were cut into two pieces longitudinally and cultured on MS medium containing 100 mg L−1 myo-inositol, 30 g L−1 sucrose, 0.2 mg L−1 kinetin and 2.0 mg L−1 2,4-dichlorophenoxyacetic acid (2,4-D) for 4 weeks to induce calli. The explants with induced calli were transferred to MS medium supplemented with 100 mg L−1 myo-inositol, 30 g L−1 sucrose, 1.0–3.0 mg L−1 zeatin riboside combined with 0.1–0.5 mg L−1 indole acetic acid (IAA) and 1–5 mg L−1 thiadiazuron (TDZ) combined with 0.2–0.5 mg L−1 naphthaleneacetic acid (NAA) at 4 weeks for shoot induction. The small new shoots were transferred to MS medium supplemented with 100 mg L−1 myo-inositol, 30 g L−1 sucrose and 1 mg L−1 N-6-(Δ2-isopentenyl adenine (2ip) for shoot elongation. The developed shoots were transferred to MS medium with 2.0 mg L−1 benzyl amino purine (BAP) at 4 weeks for shoot multiplication. The elongated shoots were transferred to MS medium supplemented with 30 g L−1 sucrose, 1.0 mg L−1 indole-3-butyric acid (IBA), 0.5 mg L−1 NAA and 15 mg L−1 hygromycin in the presence of 2.0 mg L−1 activated charcoal for rooting. The rooted plantlets were transferred to soil and grown on in a growth chamber for 4 weeks under a photoperiod of 16/8 h (light/dark) and finally transferred to the greenhouse. The applied treatments were taken from the literature of Ficus carica as described by Soliman et al. (2010). Ten Petri dishes, each containing five explants were used per treatment. The sterilized explants were cultured on the media under complete aseptic conditions in a laminar Air Flow Hood. Petri dishes and jars were then placed in an incubation room at 25 ± 2 °C under 16 h photoperiod of 35 μmol m−2 s−1 supplied with cool white fluorescent lamps.
Plasmid construction and Agrobacterium preparation
Total RNA was isolated from the halophyte Atriplex gmelini. First strand cDNA was synthesized with a SMART RACE cDNA Amplification Kit (Clontech). Primers for 3´-RACE and 5′-RACE were 5′-CAT CAG TGT CAA TTC GAG AAA CAA CAG-3′ and 5′-CTA TGT TCT GTC TAC CAA ATT GTT GTT GCT-3′. After obtaining the full length of AgNHX1, we amplified its coding region with primers 5′-CTA ACG TCA ACG AGG CAC CGG TAG AAG-3′and 5′-CGC TAC TCT CTA GGC TAC AGC ACC TAC-3′, which contain BamH1 and Sac1 sites for cloning, respectively. The vector for plant transformation was constructed by replacing AgNHX1of the pBI-121 vector (Fig. 1). This binary plasmid contains hpt selectable marker gene which encodes hygromycin phosphotransferase within the TDNA region, under the nos promoter, and the CaMV 35S promoter. The pBI-121-AgNHX1 plasmid transferred to Agrobacterium tumefaciens strain LBA4404 by electroporation and was used for transformation. The bacteria was grown overnight at 28 °C in 5 mL Lauria broth (LB) liquid medium supplemented with 50 mg L−1 rifampicin and 30 mg L−1 hygromycin on a shaker at 150 rpm to mid-log phase (OD600 = 0.8–1). Then, the bacterial cells were collected by centrifugation at 2500×g for 5 min and re-suspended in liquid Luria broth (LB) medium to a final OD600 of 0.2 for use in transfection.
Diagram of T-DNA of the recombinant binary vector pBI121 carrying the AgNHX1 gene driven by CaMV 35S promoter. LB left border, nopaline synthase gene terminator, hpt selectable marker gene, 35S cauliflower mosaic virus 35S promoter, AgNHX1 fig Na+ antiporter gene, nopaline synthase gene terminator, RB right border
Hygromycin sensitivity test
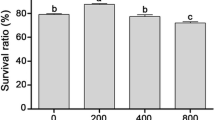
In order to test the transformed explants, a reliable selection method is required; this was achieved by using the hygromycin resistance gene. To determine the least lethal dose of hygromycin it was added to MS medium in different concentrations (0, 5, 10, 15, 20, 25, 30, 35, 40 mg L−1 hygromycin) and ten leaf explants for each concentration was assessed. The hygromycin was sterilized by filtration through disposable micropore filters (0.22 μm) and incorporated into precooled (45–50 °C) autoclaved medium. The percentages of explant survival (hygromycin resistant) were recorded after 4 weeks from culturing.
Agrobacterium-mediated transformation of (Ficus carica L.)
Leaf explants were then inoculated with A. tumefaciens strain LBA4404 harboring the plasmid pBI121 contains the AgNHX1 and hpt genes. Explants were co-cultivated with Agrobacterium for 2 days on MS medium-free hormones. Following co-cultivation, the explants were transferred to the callus medium but supplemented with 300 mg L−1 carbinicillin and 30 mg L−1 hygromycin to select for transformed cells. After 4 weeks of incubation, callus was transferred to regeneration medium containing the selection antibiotics. The cultures were reincubated under the same conditions. At least five Petri dishes, each containing four explants were used per treatment and all experiments were repeated at least three times.
Detection of transgenic plants
Polymerase chain reaction (PCR) analysis
Genomic DNA was extracted from leaf tissue of putatively transformed and non-transformed control tissues using the acetyl tri-methyl-ammonium bromide (CTAB) method. The presence of the AgNHX1 transgene in putative transgenic fig plants was detected by polymerase chain reaction (PCR) using Sigma RED Extract-N-Amp Tissue PCR Kit following the manufacturers’ instructions. Primers were forward primer, 5′-TCC CGT GTA CTT GGG AAT GC-3′; reverse primer, 5′-CGC CCA CAA TAC CAA ACA CC-3′, which amplified a 2600 bp segment. The PCR reactions were preheated to 95 °C for 2 min, followed by 35 cycles with denaturation at 95 °C for 30 s, annealing at 58 °C for 30 s, extension at 72 °C for 1 min, and a final extension step at 72 °C for 10 min.
Southern blot hybridization
Genomic DNA was isolated from 5 weeks-old in vitro propagated plants from all transformed and untransformed control plants. It was then digested with HindIII. RT-PCR-amplified fragment (448 bp) of AgNHX1 was labelled with [α-32P] and used as a probe. Hybridization was conducted in the hybridization buffer (6 × SSC, 0.5 % SDS, 0.2 mg mL−1 salmon sperm DNA) at 65 °C for 24 h. The membrane was washed once at room temperature for 15 min in 0.2 SSC, 0.5 % SDS and twice at 55 °C for 20 min in 0.1 SSC, 0.5 % SDS. The DNA probe was then used in Southern blot hybridization as described in Sambrook et al. (1989). The DNA samples which were blotted onto the membrane were the PCR products of Ficus carica L. genomic DNA amplified using AgNHX1 conserved region primer pair. Hybridization was conducted in the hybridization buffer (6 × SSC, 0.5 % SDS, 0.2 mg mL−1 salmon sperm DNA) at 65 °C for 24 h. The membrane was washed once at room temperature for 15 min in 0.2 SSC, 0.5 % SDS and twice at 55 °C for 20 min in 0.1 SSC, 0.5 % SDS. The DNA probe was then used in Southern blot hybridization as described in Sambrook et al. (1989). The DNA samples which were blotted onto the membrane were the PCR products of Ficus carica L. genomic DNA amplified using AgNHX1 conserved region primer pair.
RT-PCR analysis of AgNHX1 expression in transgenic fig plants
Total RNA was extracted using RNA-plus solution as described previously (Fukuda et al. 1991). Northern blot analysis was performed according to the method described by Hayakawa et al. (1992). RT-PCR was performed using total RNA pre-treated with primers forward (′5-CTT TAT GTT GGT ATT GAT GCC TTG G-3′) and reverse (5′-ATT GAT TTT GGA GTG GTT GGT TCT G-3′) and cDNA was synthesized under conditions of 94 °C for 5 min; then 33 cycles of 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 45 s; with a final step of 72 °C for 7 min. DNA probes were made by a multiprime labelling system (Amersham Pharmacia Biotech, Cleveland, OH) with a portion of the cloned cDNA fragment and 32P-dCTP. For assessing the relative quantities, the loaded RNAs were stained with ethidium bromide after electrophoresis.
Measurement of fresh, dry weight and Na+, K+contents
Transgenic and non-transgenic fig plants were grown in greenhouse for 7 months and then treated with 100–300 mM L−1 NaCl. Ten individual plants of each treatment for each condition were used. After 3 months under NaCl treatment, plants were assessed by measuring fresh weight, dry weight and Na+, K+ content. Dry weight was measured after 48 h incubation at 80 °C. Also, dried samples were ground using a pestle and mortar for determination of mineral composition according to Abdel-Wanis et al. (2012). Cation contents were determined by atomic absorption spectrophotometry according to Jackson (1985).
Determination of proline content
Proline content was measured as described by Bates et al. (1973). 100 mg of frozen plant material was homogenized in 10 mL of 3 % sulphosalicylic acid and the residue was removed by centrifugation. 200 μl of supernatant was reacted with 200 μl glacial acetic acid and 200 μl acid ninhydrin (1.25 g ninhydrin warmed in 30 mL glacial acetic acid and 20 mL 6 M phosphoric acid until dissolved) for 30 min at 100 °C. The reaction was then terminated in an ice bath. The reaction mixture was extracted with 600 μl toluene. The chromophore containing toluene was warmed to room temperature and its optical density was measured at 520 nm.
Statistical analysis
Data was analysed using ANOVA and the differences among means for all treatments were tested for significance at 5 % level using Duncan (1955) new multiple range test as described by Snedecor and Cochran (1967). Means followed by the same letter are not significantly different at p ≤ 0.05.
Results and discussion
In vitro regeneration of fig (Ficus carica L.)
Leaf explants were excised from in vitro plants, and cultured on MS medium supplemented with 100 mg L−1 myo-inositol, 30 g L−1 sucrose, 0.2 mg L−1 kinetin and 2.0 mg L−1 2, 4-D solidified with 2.5 g L−1 phytagel (Fig. 2a). Then the calli were cultured on MS medium containing different concentrations and combinations of zeatin riboside in combination with IAA or TDZ in combination with NAA to establish the best condition for plant regeneration.
Regeneration of transgenic plants of Ficus carica L. cv. Black Mission via indirect somatic embryogensis from leaf explants. a Callus induction in transformed tissue; b transgenic shoot regeneration on MS medium supplemented with 2.0 mg L−1 zeatin riboside and 0.2 mg L−1 IAA in presence of 20 mg L−1 hygromycin; c shoot multiplication of transgenic plants on MS medium supplemented with 2.0 mg L−1 BAP; and d rooting of transgenic plants on MS medium supplemented with 1.0 mg L−1 IBA and 0.5 mg L−1 NAA
The results showing shoot regeneration from the range of concentrations of growth regulators tested are presented in Table 1. Various ranges of regeneration efficiencies (46–70 %), mean number of shoots per explant (1.58–4.85) and mean length of shoots formed per explant (1.00–1.72 cm.) were obtained when leaf explants were grown in media containing different concentrations of growth regulators. The best results of the shoot formation percentage (78 %) and mean number of shoots per explant (5.25) was obtained on MS medium supplemented with 2.0 mg L−1 zeatin riboside in combination with 0.4 mg L−1 IAA compared to the other treatments. While, the best results of mean length of shoots formed per explant were produced on 4.0 mg L−1 TDZ and 0.4 mg L−1 NAA; reaching 1.75 cm compared to the other treatments (Fig. 2b). After 4–6 weeks, the shoots were transferred to MS medium with 2.0 mg L−1 BAP for shoot multiplication (Fig. 2c). The elongated shoots were transferred to MS medium supplemented with 30 g L−1 sucrose, 1.0 mg L−1 IBA, 0.5 mg L−1 NAA and 2.0 mg L−1 activated charcoal for rooting (Fig. 2d). The rooted plantlets were transferred to soil and grown under a photoperiod of 16/8 h (light/dark) in a plant growth chamber for 4 weeks and finally transferred to greenhouse. These results agree with those obtained by Soliman et al. (2010) who reported a method for callus induction and regeneration from leaf explants of Ficus carica L. cv. Sultani on MS medium supplemented with 2.0 mg L−1 2,4-D and 0.2 kinetin for callus induction and MS medium supplemented with 2.0 mg L−1 TDZ and 4 mg L−1 2iP for shoot formation. Wounded leaf explants cultured on MS medium supplemented with TDZ in combination with IBA produced more multiple shoots than did other combinations of auxin and cytokinin (Kim et al. 2007). Yakushiji et al. (2003) and Dhage et al. (2012) also reported a method for the induction of organogenesis from leaf explants of F. carica on MS medium supplemented with different combinations of 2,4-D, TDZ and phloroglucinol (PG), and the frequency of adventitious bud differentiation from leaf fragments was relatively low (22 %), and no adventitious buds were observed without PG.
Transformation of fig (Ficus carica L.) cv. Black Mission
Hygromycin sensitivity
Hygromycin is necessary to study the sensitivity of growth and differentiation of fig tissues and it is important selection of transformed shoots. To determine the optimum concentration of hygromycin for the selection of transformed fig shoots, a kill curve experiment was carried out using cultured non-transformed fig explants. The selective media were prepared by adding filter sterilized with different concentrations 5, 10, 15, 20, 25, 30, 35, 40 mg L−1 hygromycin. Results showed that increasing the hygromycin concentrations was accompanied by a decrease in the percentage of the survival of explants with the same conditions for regeneration stage. The lethal dose of hygromycin was estimated at 30 mg L−1 (Fig. 3). Therefore, this concentration was chosen as a selection threshold for transformed fig tissues. In agreement with our results, Lifang et al. (2001) demonstrated that the concentrations of the hygromycin used in the regeneration and subculture media for wheat plants was 20 and 30 mg L−1, respectively. Sjahril et al. (2006) reported that 25 mg L−1 hygromycin was used in the regeneration medium for selection of putative transgenic Phalaenopsis orchid plantlets. The results are also consistent with those obtained by Angenon et al. (1994) and Cosson et al. (2015) as who reported that the selection agent should fully inhibit growth of untransformed plant cells, therefore the lowest concentration of the selection agent that suppresses growth of untransformed cells is generally used. They also demonstrated that the sensitivity of plant cells to the selection agent depends upon the genotype, the explant type and the developmental stage. Most of studies have reported that hygromycin can be used as a more efficient selectable marker, compared to kanamycin, and that it is an effective agent in plant transformation, very efficient in terms of regeneration, does not inhibit regeneration and does not affect the subsequent fertility of transgenic plants (Reynaerts et al. 1988; Twyman et al. 2002; Htwe et al. 2014).
Transformation via Agrobacterium
Leaves of F. carica were used as explants, and incubated with A. tumefaciens strain LBA4404 harboring the plasmid pBI121 contains the AgNHX1 and hpt genes for 20 min before to co-cultivation media. Then, explants were co-cultivated with Agrobacterium for 48 h on the MS medium-free hormones. Following co-cultivation, the explants were cultured on MS medium supplemented with 100 mg L−1 myo-inositol, 30 g L−1 sucrose, 0.2 mg L−1 kinetin and 2.0 mg L−1 2, 4-D solidified with 2.5 g L−1 phytagel for 4 weeks. After that, the explants were transferred to the regeneration medium containing 2 mg L−1 zeatin riboside in combination with 0.4 mg L−1 IAA with 300 mg L−1 carbinicillin and 30 mg L−1 hygromycin to select for transformed cells for 4 weeks. Adventitious shoots were excised when approximately 2–3 cm in length, and transferred to MS medium containing 2 mg L−1 BAP for shoot multiplication, then hygromycin resistant shoots were transferred to rooting medium containing 1 mg L−1 IBA, 0.5 mg L−1 NAA and 2 mg L−1 activated charcoal (Fig. 2d).
Molecular analysis of transgenic fig plants
PCR detection of transformed plants
Successful introduction of the transgenes (AgNHX1 gene) was confirmed by PCR using specific primers for each of the transgenes. A fragment of 2600 bp (Fig. 4) was amplified from the DNA of the hygromycin resistant fig plants by using the AgNHX1 specific primers. These bands were not amplified with non-transformed plants. It was observed that PCR-positive plants (38 %) for Ag NHX1 gene, indicating that the gene was successfully transferred to the explants by using the Agrobacterium-mediated transformation method. The polymerase chain reaction is a very powerful technique that is now used in many areas of biology. A simple and obvious use of PCR in plant transformation studies is to use it to confirm the presence of a foreign gene in transgenic plant tissues (Lassner et al. 1989; Yi et al. 2013; Singh et al. 2014). Polymerase chain reaction as indicator for the presence of gus intron gene into transformed fig plants cv. Sultani grown on selective media by obtaining the expected product size was detected by Soliman et al. (2010).
Southern blot analysis
PCR-positive plants were further analyzed by Southern blot analysis to study confirm the presence of the fig transgenic plants and to study the integration patterns using Ag NHX1 as a probe. Genomic DNA was digested with HindIII and data revealed that only one band was obtained but was the integration of AgNHX1 gene into the genomic DNA of the fig transformed plants (Fig. 5).
RT-PCR demonstrating AgNHX1 and expression in fig (Ficus carica L.) cv. Black Mission. PCR products are electrophoresed on 1 % agarose gel and stained with ethidium bromide. Lanes T1, T2, T3, T4, T5, T6, T7 and T8 transformed lines, Lane C; non-transformed plant. M: DNA marker (1 kb plus DNA ladder)
RT-PCR detection of transgenic fig plants
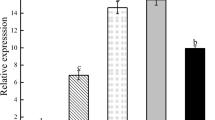
Transgenic fig positive in their PCR identification were selected for expression analysis of the AgNHX1 gene. RT-PCR and primers specific for their cDNAs were used, based on relatively high salt tolerances in those plants and the expression of AgNHX1 corresponding to be measured after 200, 250 and 300 mM NaCl treatments. An AgNHX1-specific band with expected size of 448-bp was amplified from DNase-treated RNA prepared from the leaves of transgenic fig plants (Fig. 6). Relative amounts of the mRNA in the leaves increased up to 4 and 7 times in response to treatment with 200 and 300 mM NaCl, respectively. In examining the accumulation of mRNA for the introduced AgNHX1, transgenic plants were detected by RNA gel blot analysis (Fig. 6).
Salt tolerance of transgenic plants and analysis of proline content
The survival rates of control fig plants decreased with increasing salt concentration from 100 to 250 mM and at the high salt concentrations (300 mM) all control plants died. Transgenic fig plants however had a high survival rate when exposed to 300 mM NaCl. The data showed that the Na+ content was significantly lower in the transgenic AgNHX1 plants than in non-transformed plants, and the K+ content of transgenic plants was higher that in the non-transformed plants (Table 2). Also, the results indicated that the proline content in transgenic leaves was higher than in non-transformed leaves. Transgenic plants stressed with 300 mM NaCl displayed a four fold increase in proline content compared with non-transgenic plants (Fig. 7). The accumulation of solutes such as glycine and proline has been linked to water stress, salinity and other abiotic plant stresses (Ashraf and Harris 2004; Munns and Tester 2008; Lu et al. 2009; Bhauso et al. 2014), indicating an essential role for these solutes in tolerance to these stresses. Proline accumulates under salt stress and acts as a reserve source of carbon, nitrogen and energy during recovery from stress (Watanabe et al. 2000; Chen et al. 2007; Kumar et al. 2010; Anjum et al. 2011; Lum et al. 2014; Jariteh et al. 2015).
In the present study, it was shown that the salt tolerance of fig (Ficus carica L.) cv. Black Mission was improved by the introduction of the AgNHX1 gene. The transgenic fig plants expressing AgNHX1 exhibited improved shoot growth under the salt condition of 300 mM NaCl in the greenhouse (Fig. 8). A significant difference in shoot growth was observed between the AgNHX1 transgenic plants and non-transgenic plants. A comparative analysis on Na+ accumulation revealed that transgenic plants accumulated significantly less Na+ in leaves than the non-transgenic control after 35 days of salt treatment. Similar results have been reported for vacuolar antiporter overexpressing plants such as tomato (Gisbert et al. 2000), wheat (Xuea et al. 2004), fescue (Zhao et al. 2007), Arabidopsis (Li et al. 2007), sugar beet (Liu et al. 2008) and apple rootstock (Li et al. 2010). Hamada et al. (2001) successfully isolated a Na+/H+ antiporter gene from a halophytic plant, Atriplex gmelini, and named it AgNHX1. The isolated cDNA is 2607 bp in length and contains one open reading frame, which comprises 555 amino acid residues with a predicted molecular mass of 61.9 kDa. The amino acid sequence of the AgNHX1 gene showed more than 75 % identity with those of the previously isolated NHX1 genes from glycophytes, Arabidopsis thaliana and Oryza sativa. The sensitivity to salt of cytosolic enzymes is similar in both glycophytes and halophytes, indicating that the maintenance of a high cytosolic K+/Na+ concentration ratio is a key requirement for plant growth in high salt (Greenway and Munns 1980; Maris and Eduardo 2002; Rajabi and Vazan 2013). The compartmentation of Na+ ions into vacuoles provides an efficient mechanism to avert the toxic effects of Na in the cytosol (Li et al. 2010). The transport of Na into the vacuoles is mediated by a Na+/H+ antiporter that is driven by the electrochemical gradient of protons generated by the vacuolar H+-translocating enzymes, the H+-ATPase and the H+-PPase (Blumwald 1987; Yamaguchi et al. 2003; Bassil et al. 2011; Hui et al. 2012; Bakhsh and Hussain 2015).
Conclusion
Adverse abiotic stress environmental affects yield and quality of most crops more than biotic stress. Thus, the major goal of scientists in the field of plant breeding, genetics and biotechnology is to alleviate abiotic stress on the plant and maintain high productivity and quality under abiotic stress. Plant biotechnology has the potential to address various problems in agriculture such as salt stress. The overexpression of a vacuolar Na+/H+ antiporter gene (AgNHX1) appears to hold great promise in improving agricultural productivity under abiotic salt stress. Thus, regeneration and transformation systems have to optimize as a necessary requirement for the introduction of the desired gene into plants. Given this observation, the hypothesis of this research was to enhance salt tolerance in fig (Ficus carica L.) by the transfer of Na+/H+ antiporter gene (AgNHX1) using Agrobacterium tumefaciens. To achieve this goal, we succeeded first to establish a useful protocol for production of a sufficient number of regenerated in vitro plants, which then allowed routine introduction of AgNHX1 gene into the fig genome by by a co-cultivation technique. Second, we demonstrated that expression of AgNHX1 gene in fig putative plantlets conferred tolerance to salt stress in the transgenic plantlets. The physiological mechanisms responsible for enhanced tolerance to salt stress can be accounted for by enhanced accumulation of proline, inhibition of Na+ accumulation and retention of K+ via up-regulating of the Na+/H+ antiporter gene (AgNHX1). The results are the first recorded to assess the effectiveness of transformation method of cloning Na+/H+ antiporter gene (AgNHX1) using Agrobacterium tumefaciens and to enhance the salt tolerance in fig (Ficus carica L.).
Abbreviations
- 2,4-D:
-
2,4-Dichlorophenoxyacetic acid
- 2ip:
-
N-6-(Δ2-isopentenyl) adenine
- BAP:
-
Benzylaminopurine
- CTAB:
-
Cetyltrimethylammonium bromide
- Hpt :
-
Hygromycin phosphotransferase
- IAA:
-
Indole acetic acid
- IBA:
-
Indole-3-butaric Acid
- LB:
-
Luria broth medium
- NAA:
-
α-Naphthaleneacetic acid
- OD:
-
Optical density
- ORF:
-
Opening reading frame
- PCR:
-
Polymerase chain reaction
- RT-PCR:
-
Reverse transcriptase polymerase chain reaction
- TDZ:
-
Thiadiazuron (N-phenyl-N-1,2,3,-thiadiasol-5-ylurea)
- ZR:
-
Zeatin riboside
References
Abdel-Wanis A, Abdel-Baky M, Salman S (2012) Effect of grafting and salt stress on the growth, yield and quality of cucumber grown in NFT system. J Appl Sci Res 8:50–59
Akdemir H, Gago J, Gallego P, Ciftci Y (2012) Recent advances in fruit species transformation, transgenic plants—advances and limitations, PhD. Yelda Ozden Çiftçi (Ed.), ISBN: 978-953-51-0181-9, In Tech. doi:10.5772/35139. http://www.intechopen.com/books/transgenic-plants-advances-and-limitations/recent-advances-in-fruit-species-transformation
Angenon G, Dillen W, Montagu MV (1994) Antibiotic resistance markers for plant transformation. Plant Mol Biol Manual C1:1–13
Anjum S, Xie X, Wang L, Saleem M, Man C, Lei W (2011) Morphological, physiological and biochemical responses of plants to drought stress. Afr J Agric Res 6:2026–2032
Apse M, Blumwald E (2007) Na transport in plants. FEBS Lett 581:2247–2254
Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285:1256–1258
Ashraf M, Harris P (2004) Potential biochemical indicators of salinity tolerance in plants. Plant Sci 166:3–16
Azafadi MA (2012) Genetic and biochemical properties of apples that affect storability and nutritional value. PhD Thesis, Swedish University of Agricultural Sciences, Swedish
Bakhsh A, Hussain T (2015) Engineering crop plants against abiotic stress: current achievements and prospects. Emir J Food Agric 27:24–39
Bassil E, Tajima H, Liang Y, Ohto M, Ushijima K, Nakano R, Esumi T, Coku A, Belmonte M, Blumwald E (2011) The arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell 23:3482–3497
Bassil E, Coku C, Blumwald E (2012) Cellular ion homeostasis: emerging roles of intracellular NHX Na+/H+ antiporters in plant growth and development. J Exp Bot 16:5727–5740
Bates L, Waldren R, Teare I (1973) Rapid determination of free proline for water stress studies. Plant Soil 39:205–207
Bhauso T, Radhakrishnan T, Kumar A, Mishra G, Dobaria J, Patet K, Rajam M (2014) Over-expression of bacterial mtlD gene in peanut improves drought tolerance through accumulation of mannitol. Sci World J 2014; Article ID 125967. doi:10.1155/2014/125967
Blumwald E (1987) Tonoplast vesicles for the study of ion transport in plant vacuoles. Physiol Plant 69:731–734
Borsani O, Valpuesta V, Botella M (2003) Developing salt tolerant plants in a new century: a molecular biology approach.). Plant Cell Tiss Org Cult 73:101–115
Chen Z, Cuin TA, Zhou M, Twomey A, Naidu BP, Shabala S (2007) Compatible solute accumulation and stress mitigating effects in barley genotypes contrasting in their salt tolerance. J Exp Bot 58:4245–4255
Cosson V, Eschstruth A, Pascal Ratet (2015) Medicago truncatula transformation using leaf explants. Methods Mol Biol 1223:43–56
Dhage S, Pawar B, Chimote V, Jadhav A, Kale A (2012) In vitro callus induction and plantlet regeneration in fig (Ficus carica L.). J Cell Tissue Res 12:1–6
Dorani-Uliaie E, Ghareyazie B, Farsi M, Kogel K, Imani J (2012) Improved salt tolerance in canola (Brasica napus) plants by overexpression of arabidopsis Na+/H+ antiporter gene AtNHX1. J Plant Mol Breed 1:34–42
Duncan DB (1955) Multiple range and multiple “F” test. Biometrics 11:1–42
Flaishman M, Rodov V, Stover E (2008a) The fig: botany, horticulture and breeding. In: Janick J (ed) Horticulture review. Wiley, New York; ISBN:9780470171530
Flaishman M, Yablovich Z, Golobovich S, Salamon A, Cohen Y, Perl A, Yancheva S, Kerem Z, Haklay E (2008b) Molecular breeding in fig (Ficus carica) by the use of genetic transformation. Acta Hortic 798:151–158
Flowers T, Colmer T (2008) Salinity tolerance in halophytes. New Phytol 179:945–963
Flowers T, Yeo A (1986) Ion relations of plants under drought and salinity. Aust J Plant Physiol 13:75–91
Fukuda Y, Ohme M, Shinshi H (1991) Gene structure and expression of a tobacco endochitinase gene in suspension-cultured cells. Plant Mol Biol 16:1–10
Gisbert C, Rus M, Bolarin C, Lopez-Coronado M, Arrillaga I, Montesinos C, Caro M, Serrano R, Moreno V (2000) The yeast HAL1 gene improves salt tolerance of transgenic tomato. Plant Physiol 123:393–402
Greenway H, Munns R (1980) Mechanisms of salt tolerance in nonhalohytes. Ann Rev Plant Physiol 31:149–190
Hamada A, Shono M, Xia T, Ohta M, Hayashi Y, Tanaka A, Hayakawa T (2001) Isolation and characterization of a Na+/H+ antiporter gene from the halophyte Atriplex gmelini. Plant Mol Biol 46:35–42
Hayakawa T, Zhu Y, Itoh K, Kimura Y, Izawa T, Shimamoto K, Toriyama S (1992) Genetically engineered rice resistant to rice stripe virus, an insect-transmitted virus. Proc Natl Acad Sci USA 89:9865–9869
Htwe N, Ling H, Zaman F, Mazizh M (2014) Plant genetic transformation efficiency of selected Malaysian rice based on selectable marker gene (hptII). Pak J Biol Sci 17:472–481
Hui Z, Yaxin L, Yuan X, Sean C, Andrew L, Tao X (2012) A newly isolated Na+/H+ antiporter gene, DmNHX1, confers salt tolerance when expressed transiently in Nicotiana benthamiana or stably in Arabidopsis thaliana. Plant Cell Tiss Org Cult 110:189–200
Jackson ML (1985) Soil chemical analysis. Prentic-Hall, Englewood Cliffs
Jariteh M, Ebrahimzadeh H, Niknam V, Mirmasoumi M, Yahdati K (2015) Developmental changes of protein, proline and some antioxidant enzymes activities in somatic and zygotic embryos of Persian walnut (Juglans regia L.). Plant Cell Tiss Org Cult 122:101–115
Khan MS (2011) Role of sodium and hydrogen (Na+/H+) antiporters in salt tolerance of plants: present and future challenges. Afr J Biotechnol 10:13693–13704
Khan M, Duke N (2001) Halophytes—a resource for the future. Wetl Ecol Manag 6:455–456
Kim K, Kim M, Yun P, Chandrasekhar T, Lee H, Song P (2007) Production of multiple shoots and plant regeneration from leaf segments of fig tree (Ficus carica L.). J Plant Biol 50:440–446
Küden AB (1996) Plant genetic resources of Fig, Mediterranean Selected Fruits Intercountry Network (MESFIN) under the aegis of FAO, Rome, Italy
Kumar V, Shriram V, Kavi-Kishor P, Jawali N, Shitole M (2010) Enhanced proline accumulation and salt stress tolerance of transgenic indica rice by over-expressing P5CSF129A gene. Plant Biotechnol Rep 4:37–48
Lassner M, Peterson P, Yoder J (1989) Simultaneous amplification of multiple DNA fragments by polymerase chain reaction in the analysis of transgenic plants and their progeny. Plant Mol Biol Rep 7:116–128
Li J, Jiang G, Huang P, Ma J, Zhang F (2007) Overexpression of the Na+/H+ antiporter gene from Suaeda salsa confers cold and salt tolerance to transgenic Arabidopsis thaliana. Plant Cell Tiss Org Cult 90:41–48
Li Y, Zhang Y, Feng F, Liang D, Cheng L, Ma F, Shi S (2010) Overexpression of a Malus vacuolar Na+/H+ antiporter gene (MdNHX1) in apple rootstock M.26 and its influence on salt tolerance. Plant Cell Tiss Org Cult 102:337–345
Lifang W, Hong L, Huiyun F, Lijun W, Zengliang Y (2001) Introduction of rice chitinase gene into wheat via low energy Ar+ beam implantation. Chin Sci Bull 46:318–322
Liu H, Wang Q, Yu M, Zhang Y, Wu Y, Zhang H (2008) Transgenic salt- tolerance sugar beet (Beta vulgaris L.) constitutively expressing an Arabidopsis thaliana vacuolar Na+/H+ antiporter gene, Atnhx3, accumulate more soluble sugar but less salt in storage roots. Plant Cell Environ 31:1325–1334
Lu SY, Chen CH, Wang ZC, Guo ZF, Li HH (2009) Physiological responses of somaclonal variants of triploid bermudagrass (Cynodon transvaalensis x Cynodon dactylon) to drought stress. Plant Cell Rep 28:517–526
Lum A, Hanafi M, Rafii Y, Akmar A (2014) Effect of drought stress on growth, proline and antioxidant enzyme activities of upland rice. J Anim Plant Sci 24:1487–1493
Maris P, Eduardo B (2002) Engineering salt tolerance in plants. Curr Opin Biotechnol 13:146–150
Meneses C, Orellana A (2013) Using genomics to improve fruit quality. Biol Res 46:347–352
Munns R, Tester M (2008) Mechanisms of salt tolerance. Ann Rev Plant Biol 59:651–681
Niu X, Narasimhan M, Salzman R, Bressan R, Hasegawa P (1993) NaCl regulation of plasma membrane H+-ATPase gene expression in a glycophyte and a halophyte. Plant Physiol 103:713–718
Rajabi F, Vazan S (2013) Effect of salinity on Na+ and K+ compartmentation in salt tolerant and sensitive wheat genotypes. Sch J Agric Sci 3:358–366
Razzaque S, Chakraborty D, Tammil R, Elias S, Seraj Z, Islam A (2014) Cloning of three aniporter genes from arabidopsis and rice for over-expressing them in farmer popular tomato varieties of Bangladesh. Am J Plant Sci 5:3957–3963
Reynaerts A, De Block M, Hernalsteens J, Van Montagu M (1988) Selectable and screenable markers. In: Gelvin SB, Schilperoort RA (eds) Plant molecular biology manual. Kluwer, Dordrecht, pp A9:1–16
Roohi A, Nazish B, Nabgha-e-Amen Maleeha M, Waseem S (2011) A critical review on halophytes: salt tolerant plants. J Med Plants Res 5:7108–7118
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning, vol 2. Gold Spring Harbor Laboratory Press, New York
Singh H, Bhattacharyya N, Agarwala N, Bhagawati P, Deka M, Das S (2014) Exogenous gene transfer in Assam tea [Camellia assamica (Masters)] by Agrobacterium-mediated transformation using somatic embryo. Eur J Exp Biol 4:166–175
Sjahril R, Chin DP, Khan RS, Yamamura S, Nakanmura I, Amemiya Y, Mii M (2006) Transgenic Phalaenopsis plants with resistance to Erwinia carotovora produced by introducing wasabi defensin gene using Agrobacterium method. Plant Biotechnol 23:191–194
Snedecor G, Cochran W (1967) Statistical methods, 6th edn. Iowa State University Press, Ames
Soliman H, Gabr M, Abdallah N (2010) Efficient transformation and regeneration of fig (Ficus carica L.) via somatic embryogenesis. GM Crops Food 1:42–53
Tóth G, Montanarella L, Rusco E (2008) Update map of salt affected soils in the European Union. In: Tóth G, Montanarella L, Rusco E (eds) Threats to soil quality in Europe. Office for Official Publications of the European Communities, Luxembourg, pp 61–74
Twyman R, Stöger E, Kholi A, Capell T, Christou P (2002) Selectable and screenable markers for rice transformation. In: Jackson J, Linskens H, Inman R (eds) Molecular methods of plant analysis. Springer, Heidelberg, pp 1–17
Watanabe A, Kojima K, Ide Y, Sasaki S (2000) Effects of saline and osmotic stress on proline and sugar accumulation in Populus euphratica in vitro. Plant Cell Tiss Org Cult 63:199–206
Xuea Z, Zhi D, Xueb G, Zhang H, Zhaoc Y, Xia G (2004) Enhanced salt tolerance of transgenic wheat (Triticum aestivum L.) expressing a vacuolar Na+/H+ antiporter gene with improved grain yields in saline soils in the field and a reduced level of leaf Na+. Plant Sci 167:849–859
Yakushiji H, Mase N, Sato Y (2003) Adventitious bud formation and plantlet regeneration from leaves of fig (Ficus carica L.). J Hort Sci Biotechnol 78:874–878
Yamaguchi T, Apse M, Shi H, Blumwald E (2003) Topological analysis of a plant vacuolar N+/H +antiporter reveals a luminal C terminus that regulates antiporter cation selectivity. Proc Natl Acad Sci USA 100:12510–12515
Yancheva S, Golubowicz S, Yablowicz Z, Perl A, Flaishman M (2005) Efficient Agrobacterium-mediated transformation and recovery of transgenic fig (Ficus carica L.) plants. Plant Sci 168:1433–1441
Yi D, Cui L, Wang L, Liu Y, Zhuang M, Zhang Y, Zhang J, Lang Z, Zhang Z, Fang Z, Yang L (2013) Pyramiding of Bt cry1Ia8 and cry1Ba3 genes into cabbage (Brassica oleracea L. var. capitata) confers effective control against diamondback moth. Plant Cell Tiss Organ Cult 115:419–428
Yu JN, Huang J, Wang ZN, Zhang JS, Chen SY (2007) An Na+/H+ antiporter gene from wheat plays an important role in stress tolerance. J Bio Sci 32:1153–1161
Zhang H, Blumwald E (2001) Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat Biotechnol 19:765–768
Zhao J, Zhi D, Xue Z, Liu H, Xia G (2007) Enhanced salt tolerance of transgenic progeny of tall fescue (Festuca arundinacea) expressing a vacuolar Na+/H+ antiporter gene from Arabidopsis. J Plant Physiol 164:1377–1383
Zhu JK (2007) Plant salt stress. eLS. doi:10.1002/9780470015902.a0001300.pub2
Acknowledgments
This project was funded by the Deanship of Scientific Research (DSR), King Abdulaziz University, Jeddah, under grant number (3-965-35-RG). The authors, therefore, acknowledge with thanks DSR technical and financial support. The authors are thankful to the Tissue culture and Biotechnology Labs., Marout Research Station, Desert Research Center, Egypt.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Metwali, E.M.R., Soliman, H.I.A., Fuller, M.P. et al. Molecular cloning and expression of a vacuolar Na+/H+ antiporter gene (AgNHX1) in fig (Ficus carica L.) under salt stress. Plant Cell Tiss Organ Cult 123, 377–387 (2015). https://doi.org/10.1007/s11240-015-0842-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-015-0842-z