Abstract
A binary vector carrying Bacillus thuringiensis cry1Ia8 and cry1Ba3 genes was introduced into an inbred line of white cabbage by Agrobacterium tumefaciens-mediated transformation for control of diamondback moth (DBM, Plutella xylostella), and 14 kanamycin-resistant plantlets were obtained. Presence and expression of the cry1Ia8 and cry1Ba3 genes in five transformed plants were confirmed by polymerase chain reaction (PCR), Southern blot, reverse transcription-polymerase chain reaction (RT-PCR), and Western blot analyses. Insect bioassays showed that these transgenic plants were able to effectively control both susceptible and Cry1Ac-resistant DBM larvae as compared to non-transformed counterparts. Ten homozygous insect-resistant cabbage lines were obtained in the T2 generation through self-pollination, and molecular methods and insect bioassays. After natural infestation under greenhouse and field conditions, the pyramided lines exhibited excellent efficacy against DBM. Furthermore, data from field trials indicated that there were no significant differences in most agronomic traits between most the homozygous lines and the original variety. These transgenic lines may allow field study of resistance management strategies involving gene pyramiding and serve as novel insect-resistant resources in cabbage breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cabbage (Brassica oleracea L. var. capitata) is an extremely important cruciferous crop imparting a significant contribution to global vegetable supply. Sustainable production of cabbage is subject to diverse biotic and abiotic stresses including various insect pests. Diamondback moth (DBM, Plutella xylostella), one of the most destructive lepidopteran insects of crucifers worldwide, causes severe damage to the quality and production of cabbage (Dutta et al. 2012; Macharia et al. 2005). At present, insecticides are the primary method of controlling insect pests; however, chemical insecticides pollute land and water, are toxic to non-target organisms, and have adverse effects on human and animals (Frutos et al. 1999; Tu et al. 2000). Furthermore, some DBM populations have evolved resistance to many insecticides (Cheng 1988; Furlong et al. 2013; Talekar and Shelton 1993). The use of insect-resistant cultivars is the most economical and effective method of controlling DBM (Furlong et al. 2013). However, the availability of suitable insect-resistance genes in the cabbage germplasm base is limited. Therefore, introducing foreign insect-resistance genes into cabbages through genetic engineering may provide effective germplasm resources for breeding.
Insect-resistant transgenic crops that produce insecticidal toxins from Bacillus thuringiensis (Bt) were first cultivated commercially in 1996 (James 2012). The utilization of Bt crops has significantly reduced the use of chemical insecticides and brought great economic benefits to growers (Yang et al. 2011). Although Bt crops show excellent control against major insect pests, there are concerns that efficacy of Bt proteins would be lost because the extensive adoption of Bt crop places strong selection pressure on the target pest populations, which could result in development of resistance to Bt toxin. So far, control failure or reduced efficacy of Bt crops due to insect resistance development under field conditions has been clearly documented in four cases in the world: Busseola fusca to Bt corn producing Cry1Ab in South Africa (Van Rensburg 2007), Spodoptera frugiperda to Bt corn producing Cry1F in Puerto Rico (Matten et al. 2008), Pectinophora gossypiella to Bt cotton producing Cry1Ac in India (Bagla 2010), and Diabrotica virgifera virgifera to Bt maize producing Cry3Bb1 in Iowa (Gassmann et al. 2011). All the cases of field-evolved resistance have emerged from systems where the Bt crops expressed only a single Bt protein. The risk of resistance to single-gene Bt crops has caused concern in many pest management systems because the target insects may be able to more rapidly adapt to a single toxin than to multiple toxins (Roush 1998; Zhao et al. 2003). To delay the development of pest resistance, several insect resistance management (IRM) strategies have been adopted in the United States and Australia, including gene stacking (Bates et al. 2005; Yang et al. 2011).
Gene stacking, pyramiding of two or more Bt genes into one variety, is currently considered to be one of the most effective strategies to inhibit pest resistance evolution. Computer simulations predicted that the likelihood of pests evolving simultaneous resistance to two Bt toxins would decrease if the toxins had different binding sites (Roush 1998). Therefore, many efforts have been made to develop and deploy transgenic crops expressing two or more Bt toxins. Maqbool et al. (2001) reported that transgenic rice stacked with gna, cry1Ac, and cry2A genes showed higher levels of insect pests (rice leaf folder, yellow stemborer, and brown planthopper) resistance compared to the transformants expressing single genes. Broccoli plants pyramided with cry1Ac and cry1C genes were able to effectively control diamondback moths resistant to either single Bt protein (Cao et al. 2002) and significantly postpone insect resistance evolution (Zhao et al. 2003). Second generation transgenic cottons, Bollgard II (cry1Ac + cry2Ab) and Widestrike (cry1Ac + cry1F), producing different Bt toxins have been developed to raise the level of resistance against cotton bollworm (Gahan et al. 2005; Jackson et al. 2003). Thus, it is desirable and practical to develop insect-resistant crops with two or more exogenous insecticidal genes.
In our previous studies, we produced transgenic cabbage plants carrying either a cry1Ia8 gene (Cui et al. 2009) or a cry1Ba3 gene (Yi et al. 2011). Therefore, cry1Ia8 and cry1Ba3 gene could be pyramided into cabbage by conventional cross-pollination. However, the biennial nature of cabbage is a significant limitation of this strategy because no flowers or seeds are produced without vernalization. Moreover, cross-pollination is labour intensive and time consuming. Co-transformation has been identified to be one of the most promising strategies employed to date for pyramiding of multiple exotic genes (Halpin 2005). The method is simple and involves Agrobacterium tumefaciens-mediated transformation with one plasmid carrying different genes. To date, however, there are no reports of production of transgenic cabbages carrying two or more Bt genes by A. tumefaciens-mediated co-transformation and evaluation of Bt cabbages in the field.
Our goal is to introduce two Bt genes (cry1Ia8 and cry1Ba3) with different modes of action into white cabbage for an effective control of DBM and maintenance of a sustainable pest resistance management. As initial step toward the goal, a binary vector carrying Bt cry1Ia8 and cry1Ba3 genes was introduced into an inbred line of white cabbage by A. tumefaciens-mediated transformation. In this paper, we report the development, molecular characterization, insect bioassays, insecticidal activity, and agronomic performance of these transgenic cabbage plants.
Materials and methods
Genes and binary vector
Cry1Ba3 gene, encoding a 140 kD insecticidal Cry1Ba3 protein, has been cloned from the Bt strain UV17 (Wang et al. 2008). The cry1Ia8 gene was isolated from the Bt strain Btc008. It encodes the 81 kD Cry1Ia8 protein (Cui et al. 2009). The genes were kindly provided by Institute of Plant Protection, Chinese Academy of Agricultural Sciences (CAAS). Bioassays showed that the Cry1Ia8 and Cry1Ba3 toxins possessed highly toxic against DBM and predominantly Lepidopteran larvae, and there was no cross resistance with the Cry1Ac (Cui et al. 2009; Wang et al. 2008).
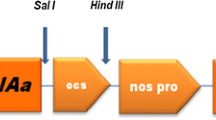
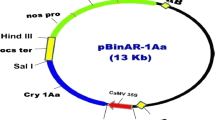
The binary vector pCSIaBaN, which contains cry1Ia8, cry1Ba3, and the neomycin phosphotransferase-II (nptII) genes, was constructed by Biotechnology Research Institute, CAAS. The T-DNA region of the binary vector is shown in Fig. 1. Cry1Ia8 and cry1Ba3 were both driven by the enhanced cauliflower mosaic virus 35S promoter and terminated by nopaline synthase terminator. The nptII gene was used as a selectable marker allowing transformed shoots to grow and develop in Murashige and Skoog (MS) salts (Murashige and Skoog 1962) regeneration medium containing kanamycin. The cry1Ia8 and cry1Ba3 fragments were introduced into expression vector pCAMBIA2300, which carried the nptII gene with CaMV35S promoter, and then transformed into Escherichia coli strain JM110. The resulting plasmid was transferred to A. tumefaciens strain LBA4404 by electroporation (Mersereau et al. 1990).
The T-DNA region of the binary vector pCSIaBaN used for cabbage transformation. LB is T-DNA left border, RB is T-DNA right border, eCaMV35 is enhanced CaMV35S promoter, Tnos is nopaline synthase terminator. The Hind III sites were used for DNA digestion in Southern blot. A 0.769 kb PCR fragment containing part of the cry1Ia8 gene was used as a probe for DNA hybridization
Plant materials and transformation
An elite inbred line of cabbage (Brassica oleracea L. var. capitata) ‘A21-3’ with superior quality and high yield was used as transformation target. ‘A21-3’ was derived from recurrent self-pollination and selection of a landrace ‘heiyexiaopingtou’.
A protocol optimized in our laboratory (Yi et al. 2011) was adopted for the routine transformation of cabbage. Full-strength MS salts and 28 mg L−1 sucrose were present in all the following media except Luria–Bertani (LB) medium. Cabbage seeds were surface-sterilized in 70 % ethanol for 2 min, and then in 10 % (v/v) Clorox for 10 min. The seeds were then rinsed five times with sterilized water and placed on hormone-free MS medium for germination at 25 °C under 16/8 h light (150 umol m−2 s−1)/dark regime. Five-day-old hypocotyls were cut into 10 mm segments and laid on MS medium for 2 day pre-culture at 25 °C under 16/8 h light (150 umol m−2 s−1)/dark regime. For transformation, A. tumefaciens cells, derived from a single colony harboring pCSIaBaN, were suspended in liquid LB medium containing 100 mg L−1 kanamycin and cultured at 28 °C with shaking at 250 rpm until the OD600 reached to 0.6–0.9. Pre-cultured explants were immersed in the Agrobacterium suspension for 10 min, and then rinsed once with MS liquid medium. Inoculated explants were placed on MS regeneration medium containing 2 mg L−1 6-benzyladenine[BA, Sigma (Saint Louis, Missouri, USA)]and 0.1 mg L−1 α-naphthaleneacetic acid (NAA, Sigma) for co-cultivation at 28 °C in dark for 3 days. Explants were first transferred to and cultured on MS regeneration medium supplemented with 150 mg L−1 Timentin (Biomed-tech Company, Beijing, China) for 7 days to eliminate bacterial cells, and then on MS regeneration medium supplemented with 150 mg L−1 Timentin and 20 mg L−1 kanamycin for selection of resistant plants. The explants were sub-cultured once every two weeks. Initially emerged green plantlets were excised from the calli and further cultured on MS regeneration medium containing a higher concentration (25 mg L−1) of kanamycin to reduce the number of escapes. Plantlets that remained green were transferred to rooting medium supplemented with 0.1 mg L−1 NAA and 0.1 mg L−1 3-indolebutyric acid (IBA, Sigma). Rooted plants were transplanted into soil and grown in a greenhouse.
Molecular analysis
PCR and southern blot analyses
Genomic DNA was isolated from the fresh leaves of putative transformants and untransformed plants using a modified CTAB method (Doyle 1990; Liu et al. 2011). PCR was performed employing primer pairs specific for the coding sequence of cry1Ia8 (forward: 5′-AGCCGTTTGTTAGTGCCT-3′, reverse: 5′-ACTTGGATGCGGATGGAC-3′) and cry1Ba3 (forward: 5′-CCAGAGACTACTCCGACTATTGCG-3′, reverse: (5′-TACACTTCCCCATTGCCACTAA-3′). Reactions were carried out in a 20 μL volume containing 300 ng DNA, 1 × Buffer, 1.2 mM MgCl2, 160 μmol dNTP, 4 μmol of each primer, and 1 unit of Taq DNA polymerase. PCR was conducted in a thermal cycler with the following amplification program: 1 cycle of 4 min at 94 °C; 30 cycles of 60 s at 94 °C, 60 s at 55 °C (cry1Ia8) or 60 °C (cry1Ba3), and 90 s at 72 °C; and a final cycle of 10 min at 72 °C. The amplified products were electrophoresed in 1 % agarose gel, stained with ethidium bromide, and visualized under ultraviolet light.
For Southern blot analysis, approximately 15 μg genomic DNA per sample was digested with Hind III, electrophoresed in 1 % agarose gel, and then transferred onto Hybond N+ nylon membrane. The DNA probe was prepared with a non-radioactive digoxigenin-labeled 0.769 kb fragment covering part of the cry1Ia8 gene. DNA gel blots hybridization was performed according to the manufacture’s instruction with a digoxigenin (DIG) high prime DNA labeling and detection kit II from Roche Molecular Biochemicals to verify the presence of T-DNA and check the number of T-DNA insertions in transgenic plants.
RT-PCR analysis
Total RNA was extracted from fresh leaves of transgenic plants and non-transformed counterparts using a total RNA extraction ksit from Biomed-tech Company (Beijing, China) and the manufacturer’s protocol. Synthesis of the cDNA was performed with revert AID™ first strand cDNA synthesis kit from Fermentas Company (Burlington, Ontario, Canada), and then cDNA was used as the template for PCR to amplify cry1Ia8 (769 kb) and cry1Ba3 (1.222 kb), respectively. β-actin (forward primer: 5′-CGGAATGGTCAAGGCTGGTTTC-3′ and reverse primer: 5′-GCTCGTTGTAGAAAGTGTGATGCC-3′) was used as an internal reference. Conditions of the PCR reaction described in the previous section were followed. The PCR products were electrophoresed in 1 % agarose gel.
Western blot analysis
Total proteins were extracted from fresh leaves of transgenic and non-transformed plants in the buffer (200 mM Tris·HCl, 100 mM·NaCl,400 mM sucrose,14 mM β-Mercaptoethanol,1 mM phenylmethylsulfonyl fluoride, and 0.05 % Tween-20) as described by Sambrook et al. (1989). The mixture was centrifuged at 5000 × g for 20 min at 4 °C, and the supernatant was collected. Protein concentrations in the supernatant were determined using a Lowry protein assay (Larson et al. 1986). For immunoblots, Protein samples were subjected to 10 % SDS-PAGE, and then the separated proteins were electro-transferred onto polyvinylidene-fluoride membrane. Western blot analysis was carried out according to the enhanced chemiluminescence Western blotting protocol described by Bradd and Dunn (1993). The membrane was probed with polyclonal rabbit anti-Cry1Ia8 or anti-Cry1Ba3 serum (1:500 dilution). Secondary antibody against rabbit IgG (whole molecule) produced in goat was purchased from Sigma Company (Saint Louis, Missouri, USA).
Diamondback moth and insect bioassays
Second instar larvae of susceptible and Cry1Ac-resistant DBM were used for insect bioassays. The DBM populations were started with material collected from American in 2005 and since then maintained in the laboratory of Entomology, Institute of Vegetables and Flowers, CAAS. Both susceptible and Cry1Ac-resistant DBM were reared on cabbage plants and kept in an environmental chamber at (27 ± 1) °C,(35 ± 2) % relative humidity, and photoperiod of 16/8 h light (150 umol m−2 s−1)/dark. At the time of the bioassays, the Cry1Ac-resistant strain was approximately 714.33-fold resistant to Cry1Ac protoxin compared to the susceptible one (Yang et al. 2012). For the bioassays of transgenic cabbage plants, a detached leaf bioassay was applied. Two leaves were collected from each transgenic and untransformed plant and cut into 70 mm leaf discs. One leaf was used for bioassay with susceptible DBM larvae and the other with Cry1Ac-resistant ones. Each leaf section was placed on a moist filter paper in a 90 mm petri dish. Ten 2nd instar DBM larvae were put on the surface of each leaf section, and then the petri dishes were sealed with parafilm. All insect bioassays were carried out in triplicate and maintained at 25 °C under 16/8 h light (150 umol m−2 s−1)/dark regime. Leaf damage (visual estimate) and mortality of larvae were scored after 6 days.
Selection of homozygous transgenic lines
Morphologically normal T0 plants were transferred to greenhouse under normal operating conditions, followed by vernalization for 2 months at 4 °C to induce flowering. Each T0 transformant confirmed to have single-copy cry1Ia8-cry1Ba3 genes by Southern blot was selfed and advanced to the T2 generation. After seeds were harvested, each T0 and T1 plant was subjected to natural infestation of insect pests under greenhouse conditions. The presence and expression of cry1Ia8 and cry1Ba3 in T1 and T2 plants were confirmed by PCR and bioassays. The homozygous lines were used in the subsequent field evaluation.
Evaluation of agronomic performance and insecticidal activity in the field
The agronomic performance and insecticidal activity of the homozygous cabbage lines were evaluated at the Langfang experimental farm affiliated with CAAS in Hebei province, China, in 2012. To evaluate the agronomic performance in the field, seeds of pure lines and the untransformed control were sown in a seedling bed in July 12, and the seedlings were transplanted to field in August 22. The arrangement of the seedlings followed a completely randomized block design with three replicates. Each replicate consisted of 14 plants in two rows, 45 cm apart within a row and 55 cm between rows. Each plot was bordered by two rows of non-transgenic cabbage plants. Normal cultural practices for growing cabbage were followed during the entire growing season. The 5 plants in the middle of each plot were harvested to evaluate the agronomic traits. The plant height, plant breadth (two measurements taken 90° across the plant), number of outer leaves, vertical and transverse diameter of the head, core length, and head weight were measured at maturity. Statistical analysis was conducted with SAS v.8.0, and Duncan’s multiple range test was used to determine the significance of differences in main agronomic traits between transgenic lines and original variety.
Four homozygous lines were selected for evaluation of insecticidal activity. Transgenic and non-transgenic lines were grown in the same way as for the agronomic performance evaluation described above except that no insecticides were applied during the entire growing season. The DBM surviving on each plant were investigated and counted every 7 days up to 10 weeks after planting.
Results
Recovery of T0 transformants
The 2 days pre-cultured hypocotyls were inoculated with Agrobacterium suspension (Fig. 2a) and co-cultivated for 3 days in the dark. Subsequently, the explants were transferred onto MS regeneration medium allowing cell division and callus formation within the next 7 days, and then onto MS regeneration medium containing kanamycin for selection of resistant plantlets. Kanamycin-resistant shoots emerged about 5 weeks after transformation (Fig. 2b). The majority of the initially green shoots gradually turned white or brown and some became necrotic after subsequent subcultures. A total of fourteen plantlets with kanamycin resistance were obtained from 300 hypocotyls (Fig. 2c) and they were allowed to form root for 2–3 weeks (Fig. 2d). Morphologically normal plants were transplanted into plastic pots and grown in a greenhouse (Fig. 2e).
Integration and expression of Bt genes in T0 transformants
In initial PCR screening, nine out of the 14 putative transformants and the plasmid pCSIaBaN showed amplification of the expected 0.769 kb band representing the cry1Ia8 fragment and 1.222 kb band representing the cry1Ba3 fragment. The PCR-positive plants were designated as T0-1, T0-2, T0-3, T0-4, T0-5, T0-6, T0-7, T0-8, T0-9, respectively. Neither band was amplified from the genomic DNA of non-transformed plants. Representative results of PCR analyses were shown in Fig. 3a, b. Lanes 4, 9, and 10 indicated that neither Bt gene was inserted into the genome of the kanamycin-resistant plants.
a and b PCR analysis of genomic DNA from cry1Ia8 + cry1Ba3 cabbage plants. 1 2 kb molecular weight maker, 2 pCSIaBaN carrying cry1Ia8 and cry1Ba3, 3 untransformed cabbage, 4–11 transformants. c Southern blot analysis of cry1Ia8 + cry1Ba3 cabbages. Genomic DNA isolated from transgenic and untransformed plants was digested with HindIII and hybridized with probe from the PCR products of cry1Ia8 gene 1 plasmid pCSIaBaN, 2 non-transformed cabbage, 3 T0-1, 4 T0-2, 5 T0-3, 6 T0-4, 7 T0-5, 8 T0-6, 9 T0-7, 10 T0-8, 11 T0-9
Southern blot hybridization of the nine PCR-positive plants was conducted to further confirm the integration of cry1Ia8 and cry1Ba3. Genomic DNA isolated from the non-transformed and PCR-positive plants was digested with Hind III and hybridized with probe from the PCR products of the cry1Ia8 gene. The results showed that two of the nine tested plants had only one insertion site (T0-1 and T0-9), three contained two insertion sites (T0-3, T0-6, and T0-7), and no hybridization signal was detected for the rest of the plants (T0-2, T0-4, T0-5, and T0-8) (Fig. 3c).
Transgenic cabbage plants (T0-1, T0-3, T0-6, T0-7, and T0-9) confirmed by Southern blot hybridization were analyzed by RT-PCR and Western blot to evaluate expression of cry1Ia8 and cry1Ba3. In RT-PCR analyses, all the transgenic cabbages showed amplification of the expected cry1Ia8 (0.769 kb), cry1Ba3 (1.222 kb), and β-actin (0.196 kb) fragments while only the 0.196 kb internal reference band was amplified in non-transformed cabbage (Fig. 4a, b). Western blot results (Fig. 4c, d) indicated the presence of the expected 81 kD Cry1Ia8 protein and 140 kD Cry1Ba3 protein in all the five transgenic plants but not in the untransformed control.
a and b RT-PCR analysis of cry1Ia8 + cry1Ba3 plants. 1 2 kb molecular weight maker, 2 pCSIaBaN carrying cry1Ia8 and cry1Ba3, 3 untransformed cabbage, 4 T0-1, 5 T0-3, 6 T0-6, 7 T0-7, 8 T0-9. c and d Western blot analysis of cry1Ia8 + cry1Ba3 plants. 1 protein marker, 2 Cry1Ia8/Cry1Ba3 protein, 3 T0-1, 4 T0-3, 5 T0-6, 6 T0-7, 7 T0-9, 8 non-transformed cabbage
Insect bioassays of T0 plants
To assess the effectiveness of Bt protein in T0 transformants, the five transgenic plants (T0-1, T0-3, T0-6, T0-7, and T0-9) confirmed through western blot analyses were tested for insect resistance by exposing in vitro leaves to susceptible and Cry1Ac-resistant DBM, respectively. The results showed that the transgenic cabbages were able to effectively control both susceptible and Cry1Ac-resistant DBM larvae as compared to non-transformed counterparts (Fig. 5). In the first 48 h, most of larvae began to die on transgenic leaves; the others ceased feeding and became stunted. All transgenic cabbages exhibited no leaf damage and caused complete mortality of larvae within 3 days. Six days later, there was no mortality of larvae on the control leaves, and leaf disks from control cabbages were completely consumed.
In-vitro bioassay of the non-transgenic and transgenic cabbages 3 days after inoculation with DBM larvae. Left figure is assays with DBM susceptible to Cry1Ac, 1 non-transformed control, 2 T0-1, 3 T0-3, 4 T0-6, 5 T0-7, 6 T0-9. Right figure is assays with DBM resistant to Cry1Ac, 1 non-transformed control, 2 T0-1, 3 T0-3, 4 T0-6, 5 T0-7, 6 T0-9
Production of homozygous transgenic lines
The single-copy T0 transformants (T0-1 and T0-9) were self-pollinated to obtain T1 seeds. In the T1 population derived from T0-1, seventeen of 23 plants amplified both cry1Ia8 and cry1Ba3 fragments, as shown through PCR analysis. The T1 population derived from T0-9 also showed a 3:1 segregation ratio (Table 1), suggesting a single-copy insertion in the parental lines. Fifteen PCR-positive T1 plants developed from each line (T0-1 and T0-9) were then vernalized to produce T2 seeds by self-pollination. Thirty T2 plants derived from each line were screened for presence of cry1Ia8 and cry1Ba3 by PCR as well as bioassays (data not shown). If the T1 line was homozygous, all T2 progeny were expected to inherit the target genes. On this basis four T1 plants of the line T0-1 (T0-1-1, T0-1-4, T0-1-6, and T0-1-14) and six T1 plants of the line T0-9 (T0-9-2, T0-9-3, T0-9-5, T0-9-7, T0-9-12, and T0-9-13) were identified as homozygous lines.
In addition, each T0 and T1 plant was subjected to natural infestation of insect pests after harvesting seeds, and no insecticides were applied during the entire growing season under greenhouse conditions. The transgenic plants exhibited excellent efficacy against insect pests as compared to non-transformed counterparts (Fig. 6a).
a Insect-resistant performance of transgenic and non-transgenic cabbages after harvesting seeds in the greenhouse. The left row is Cry1Ia8 + Cry1Ba3 cabbages and the right row is non-transgenic plants. b Insect-resistant performance of transgenic and non-transgenic cabbage lines under natural infestation conditions in the field. The left row is Cry1Ia8 + Cry1Ba3 cabbages and the right row is non-transgenic plants
Agronomic traits and insecticidal activity of pyramided lines in the field
Ten homozygous lines (T0-1-1, T0-1-4, T0-1-6, T0-1-14, T0-9-2, T0-9-3, T0-9-5, T0-9-7, T0-9-12, and T0-9-13) were selected for agronomic traits evaluation in Langfang experimental farm in 2012. The untransformed line ‘A21-3’ were used as control for agronomic traits comparison. The data showed that the pyramided lines performed as well as the control for most traits except core length of T0-1-14 and head weight of T0-9-3 (Table 2). These observations imply that the introduced transgenes have little or no effects on the main agronomic traits.
Four homozygous lines (T0-1-1, T0-1-6, T0-9-5, and T0-9-13) were chosen to evaluate the insecticidal activity of pyramided lines. The results (Fig. 7) showed that the number of DBM on the control ‘A21-3′ increased rapidly after August 22, reaching 25.3 heads plot−1 on 13 September. Then the number of DBM decreased gradually as the temperature dropped. No DBM survived on control plants after October 25. Almost no DBM were observed on the transgenic lines during the entire season. By September 13, the control ‘A21-3’ were eaten severely but the pyramided lines showed no or little leaf damage (Fig. 6b).
Discussion
Many approaches can be adopted by researchers for introduction of multiple foreign genes into one plant, such as sexual crossing between parental lines containing single gene, sequential retransformation of plants with new genes, and co-transformation with one plasmid carrying different genes (Halpin 2005). Retransformation and sexual crossing strategies have been used to develop pyramided genetically modified (GM) cruciferous crops such as broccoli (Cao et al. 2002), collard (Cao et al. 2005), canola (Liu et al. 2011), Indian mustard (Cao et al. 2008), etc. The sexual crossing strategy needs more time and larger progeny populations to enable selection of target traits. A drawback of the accompanied with re-transformation strategy is that it requires a variety of selectable marker genes to be available so that a different one can be utilized with each sequential transformation. In addition, GM crops with an accumulation of such genes would encounter significant hurdles to regulatory approval and consumer acceptance. Co-transformation is considered to be one of the most viable methodologies employed to date for pyramiding of multiple foreign genes. The advantage of this method is obvious. The co-introduced foreign genes tend to co-integrate at the same chromosomal position, and this ensures that target genes are unlikely to segregate apart in subsequent generations (Halpin 2005). A handful of impressive examples have been reported in pyramiding GM maize (Halpin 2005) and rice (Maqbool et al. 2001; Ye et al. 2000) developed by co-transformation but very few in cruciferous crops. To our knowledge, this paper is the first successful report of introducing two Bt genes into cabbage using A. tumefaciens-mediated co-transformation.
The target genes, cry1Ia8 and cry1Ba3, were successfully introduced into an elite inbred line of cabbage, ‘A21-3’, and 10 homozygous cabbage lines harboring the two Bt genes were obtained in the T2 generation by means of repeated self-pollination and molecular-assisted selection of two T0 transformants. The resulting homozygous lines exhibited excellent efficacy against DBM as compared to non-transformed counterparts in the natural infestation situation, while there were no significant differences in main agronomic traits between most homozygous lines and the original variety. These findings indicated that the pyramided genes in the cabbage plants functioned normally without altering the main agronomic traits and yield to a large degree. In contrast, previous studies of transgenic rice Taipei 309 with nptII and IR72/Koshihikari with bar indicated poor field performance, which limited their potential commercial value (Tu et al. 2000).
The use of cabbage heterosis is now becoming an important technology applied all over the world. In previous reports (Bhattacharya et al. 2002; Jin et al. 2000; Metz et al. 1995; Rafat et al. 2010), transformed materials were predominantly commercial hybrids of cabbage, which may limit their practical uses to a great extent. Obtaining plants homozygous for all genes may be virtually impossible because the complexity of segregation of the independent genes. The successful expressions of cry1Ia8 (Cui et al. 2009), cry1Ba3 (Yi et al. 2011), and cry1Ia8 + cry1Ba3 in the genome of an elite inbred line in our laboratory will provide a good resource for development of Bt hybrid cabbage and management of cabbage pests in China.
Besides the commercial value of Bt plants, Bt brassicas have served as excellent research tools. For example, Bt broccoli plants have been used in empirical tests of various resistance management strategies designed to inhibit the evolution of resistance insects (Zhao et al. 2005, 2003). Simulation modeling studies (Roush 1998) have predicted that adoption of cultivars with pyramided insect-resistance genes was in general more durable than the sequential deployment of cultivars with single insect-resistance genes. Availability of the Cry1Ia8 (Cui et al. 2009), Cry1Ba3 (Yi et al. 2011), and Cry1Ia8 + Cry1Ba3 cabbage plants will allow us to test these models experimentally. Using these plants, we can select for Cry1Ia8R, Cry1Ba3R, and Cry1Ia8R + Cry1Ba3R DBM populations in controlled settings, and learn modes and dynamics of the evolution of resistance to Cry1Ia8 and Cry1Ba3 during selection. The results from these studies may provide valuable information relevant to effective control of Bt-resistant insects.
References
Bagla P (2010) Hardy cotton-munching pests are latest blow to GM crops. Science 327:1439
Bates SL, Zhao J-Z, Roush RT, Shelton AM (2005) Insect resistance management in GM crops: past, present and future. Nat Biotechnol 23:57–62
Bhattacharya R, Viswakarma N, Bhat S, Kirti P, Chopra V (2002) Development of insect-resistant transgenic cabbage plants expressing a synthetic cryIA (b) gene from Bacillus thuringiensis. Curr Sci India 83:146–150
Bradd SJ, Dunn MJ (1993) Analysis of membrane proteins by western blotting and enhanced chemiluminescence. Biomembrane Protocols. Springer, Berlin, pp 211–218
Cao J, Zhao JZ, Tang J, Shelton A, Earle E (2002) Broccoli plants with pyramided cry1Ac and cry1C Bt genes control diamondback moths resistant to Cry1A and Cry1C proteins. Theor Appl Genet 105:258–264
Cao J, Shelton AM, Earle ED (2005) Development of transgenic collards (Brassica oleracea L. var. acephala) expressing a cry1Ac or cry1C Bt gene for control of the diamondback moth. Crop Prot 24:804–813
Cao J, Shelton AM, Earle ED (2008) Sequential transformation to pyramid two Bt genes in vegetable Indian mustard (Brassica juncea L.) and its potential for control of diamondback moth larvae. Plant Cell Rep 27:479–487
Cheng E (1988) Problems of control of insecticide-resistant Plutella xylostella. Pestic Sci 23:177–188
Cui L, Yang LM, Liu N, Lang ZH, Liu YM, Zhuang M, Zhang YY, Zhang YJ, Huang D, Fang ZY (2009) Transformation and expression of Bt gene cry1Ia8 in cabbage. Acta Horticulturae Sinica 36:1161–1168
Doyle JJ (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Dutta D, Niwas R, Gopal M (2012) Comparative persistence of thiacloprid in Bt-transgenic cabbage (Brassica oleracea cv. capitata) vis-a-vis non-transgenic crop and its decontamination. B Environ Contam Tox 89:1027–1031
Frutos R, Rang C, Royer M (1999) Managing insect resistance to plants producing Bacillus thuringiensis toxins. Crit Rev Biotechnol 19:227–276
Furlong MJ, Wright DJ, Dosdall LM (2013) Diamondback moth ecology and management: problems, progress, and prospects. Annu Rev Entomol 58:517–541
Gahan LJ, Ma YT, MacGregor Coble ML, Gould F, Moar WJ, Heckel DG (2005) Genetic basis of resistance to Cry1Ac and Cry2Aa in Heliothis virescens (Lepidoptera: Noctuidae). Journal Econ Entomol 98:1357–1368
Gassmann AJ, Petzold-Maxwell JL, Keweshan RS, Dunbar MW (2011) Field-evolved resistance to Bt maize by western corn rootworm. PLoS ONE 6:1–7
Halpin C (2005) Gene stacking in transgenic plants–the challenge for 21st century plant biotechnology. Plant Biotechnol J 3:141–155
Jackson R, Bradley J, Van Duyn J (2003) Field performance of transgenic cottons expressing one or two Bacillus thuringiensis endotoxins against bollworm, Helicoverpa zea (Boddie). J Cotton Sci 7:57–64
James C (2012) Global status of commercialized biotech/GM Crops: 2012. ISAAA Brief No. 44. ISAAA, Ithaca
Jin RG, Liu YB, Tabashnik B, Borthakur D (2000) Development of transgenic cabbage (Brassica oleracea var. capitata) for insect resistance by Agrobacterium tumefaciens-mediated transformation. In Vitro Cell Dev-Pl 36:231–237
Larson E, Howlett B, Jagendorf A (1986) Artificial reductant enhancement of the Lowry method for protein determination. Anal Biochem 155:243–248
Liu H, Guo X, Naeem MS, Liu D, Xu L, Zhang W, Tang G, Zhou W (2011) Transgenic Brassica napus L. lines carrying a two gene construct demonstrate enhanced resistance against Plutella xylostella and Sclerotinia sclerotiorum. Plant Cell Tiss Org Cult 106:143–151
Macharia I, Löhr B, De Groote H (2005) Assessing the potential impact of biological control of Plutella xylostella (diamondback moth) in cabbage production in Kenya. Crop Prot 24:981–989
Maqbool SB, Riazuddin S, Loc NT, Gatehouse AM, Gatehouse JA, Christou P (2001) Expression of multiple insecticidal genes confers broad resistance against a range of different rice pests. Mol Breeding 7:85–93
Matten SR, Head GP, Quemada HD (2008) How governmental regulation can help or hinder the integration of Bt crops within IPM programs. Integration of Insect-Resistant Genetically Modified Crops within IPM Programs. Springer, Berlin, pp 27–39
Mersereau M, Pazour GJ, Das A (1990) Efficient transformation of Agrobacterium tumefaciens by electroporation. Gene 90:149–151
Metz TD, Dixit R, Earle ED (1995) Agrobacterium tumefaciens-mediated transformation of broccoli (Brassica oleracea var. italica) and cabbage (B. oleracea var. capitata). Plant Cell Rep 15:287–292
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plantarum 15:473–497
Rafat A, Aziz MA, Rashid AA, Abdullah SNA, Kamaladini H, Sirchi M, Javadi M (2010) Optimization of Agrobacterium tumefaciens-mediated transformation and shoot regeneration after co-cultivation of cabbage (Brassica oleracea subsp. capitata) cv. KY Cross with AtHSP101 gene. Sci Hortic 124:1–8
Roush R (1998) Two–toxin strategies for management of insecticidal transgenic crops: can pyramiding succeed where pesticide mixtures have not? Philos Trans R Soc Lond B Biol Sci 353:1777–1786
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Talekar N, Shelton A (1993) Biology, ecology, and management of the diamondback moth. Annu Rev Entomol 38:275–301
Tu J, Zhang G, Datta K, Xu C, He Y, Zhang Q, Khush GS, Datta SK (2000) Field performance of transgenic elite commercial hybrid rice expressing Bacillus thuringiensis δ-endotoxin. Nat Biotechnol 18:1101–1104
Van Rensburg J (2007) First report of field resistance by the stem borer, Busseola fusca (Fuller) to Bt-transgenic maize. South African Journal of Plant and Soil 24:147–151
Wang G, Zhang J, Song F, Gu A, Uwais A, Shao T, Huang D (2008) Recombinant Bacillus thuringiensis strain shows high insecticidal activity against Plutella xylostella and Leptinotarsa decemlineata without affecting nontarget species in the field. J Appl Microbiol 105:1536–1543
Yang Z, Chen H, Tang W, Hua H, Lin Y (2011) Development and characterisation of transgenic rice expressing two Bacillus thuringiensis genes. Pest Manag Sci 67:414–422
Yang ZX, Wu QJ, Wang SL, Chang XL, Wang JH, Guo ZJ, Lei YY, Xu BY, Zhang YJ (2012) Expression of cadherin, aminopeptidase N and alkaline phosphatase genes in Cry1Ac-susceptible and Cry1Ac-resistant strains of Plutella xylostella (L.). J Appl Entomol 136:539–548
Ye X, Al-Babili S, Klöti A, Zhang J, Lucca P, Beyer P, Potrykus I (2000) Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287:303–305
Yi DX, Cui L, Liu YM, Zhuang M, Zhang YY, Fang ZY, Yang LM (2011) Transformation of cabbage (Brassica oleracea L. var. capitata) with Bt cry1Ba3 gene for control of diamondback moth. Agr Sci China 10:1693–1700
Zhao JZ, Cao J, Li Y, Collins HL, Roush RT, Earle ED, Shelton AM (2003) Transgenic plants expressing two Bacillus thuringiensis toxins delay insect resistance evolution. Nat Biotechnol 21:1493–1497
Zhao JZ, Cao J, Collins HL, Bates SL, Roush RT, Earle ED, Shelton AM (2005) Concurrent use of transgenic plants expressing a single and two Bacillus thuringiensis genes speeds insect adaptation to pyramided plants. P Natl Acad Sci USA 102:8426–8430
Acknowledgments
We are grateful to Prof. Dafang Huang, Prof. Jie Zhang, and Dr. Zhihong Lang for providing Bt genes and vector. We thank Prof. Youjun Zhang for their help in sending us the DBM larvae. Thanks are due to Prof. Elizabeth Earle (Department of Plant Breeding & Genetics, Cornell University, USA) for constructive advice on writing the manuscript and to Dr. Jun Cao (Athenix Corporation, USA) for providing the Brassica genetic transformation protocol. This work was supported by grants from the National High Technology Research and Development Program of China (863 Program, No. 2008AA10Z155), the National Natural Science Foundation of China (No. 31071697), and the earmarked fund for Modern Agro-industry Technology Research System (No. nycytx-35-gw01).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yi, D., Cui, L., Wang, L. et al. Pyramiding of Bt cry1Ia8 and cry1Ba3 genes into cabbage (Brassica oleracea L. var. capitata) confers effective control against diamondback moth. Plant Cell Tiss Organ Cult 115, 419–428 (2013). https://doi.org/10.1007/s11240-013-0373-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-013-0373-4