Abstract
A catalytic single chain variable antibody (scFv), 3D8 scFv, which has DNase activities, was functionally expressed in Nicotiana tabacum. The subcellular localization of the GFP-fused 3D8 indicated that the 3D8 protein was expressed in the cytosol of the N. tabacum protoplasts. Progenies of the transgenic tobacco plants exhibited complete resistance against two single stranded (ss) DNA geminiviruses, including the Beet curly top virus and the Beet severe curly top virus, without viral accumulation or disease symptoms. We presented a novel strategy for targeting the viral DNA itself in a sequence non-specific manner, rather than the viral proteins or RNAs, in order to generate virus-resistant transgenic plants. No noticeable adverse effects on the growth and reproduction of the transgenic plants were observed. Our results demonstrated that targeting viral DNA is an effective strategy for protecting plants from ssDNA viruses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants are hosts to thousands of infectious diseases caused by pathogenic viruses, fungi, bacteria, mycoplasmas, insects, and nematodes (Lucas 1998). Plant viruses have an enormous negative impact on agricultural crop production throughout the world (Scholthof et al. 1993; Baker et al. 1997). In order to secure plant resistance to numerous viruses, pathogen-derived resistance genes, various viral genes, and other antiviral factors have been engineered into plants (Baulcombe 1996). However, there is little to report about the complete acquired resistance against plant virus infection.
The plants in the family Geminiviridae have circular single-stranded DNA genomes (Fauquet et al. 2003) and are currently classified into four genera (begomovirus, curtovirus, mastrevirus, and topocuvirus) on the basis of their genome organization, insect vectors, and host range (Fauquet et al. 2003). Geminiviruses are the cause of devastating plant diseases (beet curly top, cassava mosaic, and maize streak diseases) recognized as serious problems for world agriculture (Varma and Malathi 2003; Mansoor et al. 2006). Economic losses by Geminivirus infections are estimated to be US$1300–2300 million in Africa (Thresh et al. 1998), US$5000 million in Pakistan (Briddon and Markham 2000), and US $140 million in Florida (Moffat 1999). Therefore, the geminivirus-resistant crop development is necessary.
Plants have evolved two primary defense mechanisms in transgenic plants against DNA viruses: replication associated protein (Rep)-mediated resistance and RNA-mediated gene silencing resistance (Prins et al. 2008). The Rep protein-mediated resistance is a resistance mechanism by the overexpression of the modified Rep protein in transgenic plants. Hong and Stanley demonstrated resistance with the African cassava mosaic virus (ACMV) Rep protein in the tobacco cell line BY-2 (Hong and Stanley 1995). In addition, Noris showed that the truncated Rep protein transgenic Nicotiana benthamiana has resistance against the tomato yellow leaf curl geminivirus (Noris et al. 1996). RNA-mediated gene silencing is a sequence-specific defense mechanism that targets the degradation of both cellular and viral mRNAs in eukaryotes (Vance and Vaucheret 2001). When challenged with the African cassava mosaic virus (ACMV), the accumulation of viral DNA was reduced by up to 98 % compared to the controls in short-interfering RNAs specific to the AC1 transgenic plants (Chellappan et al. 2004). Another strategy was reported using Pac1, a RNase isolated from the yeast Schizosaccharomyces pombe. Challenging a transgenic potato plant that expresses Pac1 with potato spindle tuber viroid (PSTVd) resulted in the suppression of the PSTVd infection and accumulation without significant undesired effects on the plant. This results showed that the anti-viroid effect was primarily caused by RNase activity (Sano et al. 1997). These findings revealed that nuclease activity could be applied for antiviral effects.
Previously, we isolated the anti-DNA monoclonal antibody 3D8 scFv from the spleen cells of a MRL-lpr/lpr mouse, which spontaneously developed an auto-immune syndrome that resembles human systemic lupus erythematosus (Kwon et al. 2002). The 3D8 scFv has catalytic activity supporting the hydrolysis of both single stranded- (ss-) and double stranded- (ds-) DNAs in the presence of Mg2+ without significant sequence specificity (Kim et al. 2006). While the catalytic activity of 3D8 scFv for DNAs required Mg2+ (Kim et al. 2006), 3D8 scFv’s efficient degradation of RNAs in the presence of EDTA demonstrated that 3D8 scFv did not need divalent metal ions for its RNase activity, which is similar to a standard RNase such as RNase A (Blackburn et al. 1977; Kwon et al. 2002; Jang et al. 2009). We also reported that 3D8 scFv demonstrated antiviral activity against the classical swine fever virus (CSFV) in the porcine kidney cell line PK-15 (Jun et al. 2010).
In this study, we characterized 3D8 scFv as a new antiviral protein that is capable against DNA viruses. We devised a novel antibody-based strategy targeting viral DNA without any sequence-specificity or binding/degradation bias to DNA, in order to generate transgenic plants resistant to viruses with ss-DNA as a genome.
Materials and methods
Plant transformation
The gene encoding 3D8 scFv was cloned into the pBI121 vector (BD Biosciences Clontech, Mountain View, CA, USA) using the HindIII/EcoRI site in order to generate pBI121-35S-3D8 scFv and pBI121-35Smini-3D8 scFv (Benfey and Chua 1990) (Fig. 1a) for cytoplasmic expression in plants and for monitoring the correlations between the 3D8 scFv expression level and plant development. The plasmids were transferred into Agrobacterium tumefaciens strain LBA4404 by electroporation and were used for leaf disc transformation of Nicotiana tabacum as described previously (Liu et al. 2012; Pires et al. 2012; Sujatha et al. 2012). In order to produce the stable transformants, the primary transformed plants (T0) were self-fertilized to generate the first generation progenies (T1) under kanamycin selection (Rudolph et al. 2003). The seeds obtained from the T1 transgenic plant lines were germinated in order to produce T2 and later progenies. The expression of 3D8 scFv in the transgenic plants was monitored by northern blot hybridization. The total cellular RNA was isolated from the leaves, stems, and roots of the wild-type and the transgenic plants using an RNeasy® plant mini kit (Qiagen Inc., Valencia, CA). The probe for the hybridization experiments was prepared by the PCR amplification of the gene encoding 3D8 scFv (700 bp) and then random labeling with [32P]-α-dCTP (Amersham Biosciences Corp., Piscataway, NJ). In order to monitor the plant growth and development based on the amounts of 3D8 scFv in the cytoplasm, two experiments were performed. Seven T2 transgenic lines that showed high 3D8 scFv expression were selected, sown, and measured for leaf and shoot lengths at 14 weeks. The second experiment for monitoring plant development was performed using tobacco leaf disc transformation following the standard protocol (Abel et al. 1986; Benvenuto et al. 1991; Yang et al. 2004) with Agrobacterium containing either pBI121-35S-3D8 scFv or pBI121-35Smini-3D8 scFv. During callus induction and shoot regeneration, the size and transformation efficiency of leaf discs were monitored in the plants with 3D8 scFv expressed under the control of either the normal or a minimal 35S promoter.
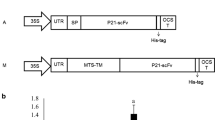
Expression of 3D8 scFv in the transgenic tobacco plants. a Schematic vector maps of pBI121-35S-3D8 scFv and pBI121-35Smini-3D8 scFv, which were used for the expression of 3D8 scFV in plants. b PCR and RT-PCR analysis of T1 transgenic lines. Seven (3, 4, 13, 14, 23, 28, and 30) out of thirty lines were selected for further T2 analysis. The line numbers for T1 in a box re-named 99-X for the T2 lines. c Northern blots for the detection of the 3D8 scFv transcripts in the leaf tissue of the WT and three T2 transgenic tobacco lines: Tx99-1, Tx99-2, and Tx99-3
Virus infection
For ss-DNA viruses [Beet curly top virus (BCTV) and Beet severe curly top virus (BSCTV)], Agrobacterium suspensions harboring each viral genome were pin-pricked onto the shoot tips (Rochester et al. 1990). Mock inoculation was performed using only the PBS buffer.
Detection of viral accumulation in plants
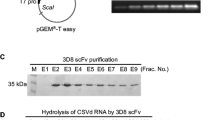
Southern blot analysis for ss-DNA viruses was used to monitor viral infection and accumulation. The CP gene of each virus, BCTV-CP and BSCTV-CP, were cloned into a pGEM®-T Easy plasmid (Promega Corp., Madison, WI). The probe for the hybridization experiments was prepared by PCR of each CP gene (500–700 bp) and then random labeling with [32P] dCTP. Southern blot analyses were performed using genomic DNA (~10 μg) and total RNAs (~20 μg) isolated from the plant leaves (Abel et al. 1986).
Nucleic acid hydrolysis activity assay of 3D8 scFv
Nucleic acid hydrolysis experiments were initiated by mixing the purified 3D8 scFv (0.1 μM) or cell lysates of plants with 2 nM of each substrate in TBS (50 mM Tris–Cl, pH7.5, 50 mM NaCl) containing 10 mM MgCl2 for DNA substrates. The DNA substrates, the supercoiled M13mp18 phagemid and ss-DNA M13mp18 phage (New England Biolabs, Ipswich, MA), were prepared as described earlier (Kim et al. 2006). After the reactions were performed at 37 °C for 1–5 h, the samples were analyzed on 1 % agarose gels by electrophoresis. Bovine serum albumin (0.1 μM) (Sigma) was used as a negative control. The intrinsic DNase activity of the 3D8 scFv was analyzed by in situ gel assays. The purified 3D8 scFv (2 μg) was applied to 12 % SDS–polyacrylamide (PAGE) gels containing 20 μg/ml of one of the following substrates: ss-DNA M13mp18 or ds-DNA M13mp18. The SDS was removed by incubating the gels with 7 M urea for 1 h at room temperature. After washing (5×) with water for 10 min, the gels were incubated overnight at 37 °C in 10 mM Tris, pH 8.0, and containing 10 mM MgCl2 and 50 mM NaCl in order to allow for the renaturation of the protein. The gels were stained with ethidium bromide in order to identify regions of substrate hydrolysis and stained with Coomassie Blue R250 to determine the position of the 3D8 scFv. The nucleic acid hydrolyzing activities of the 3D8 scFv were monitored by agarose gel electrophoresis.
Transient expression of the 3D8 scFv in tobacco leaves
Agrobacterium tumefaciens strain LBA4404 containing pBI121-β-gulucronidase (as a negative control), pBI121-3D8 scFv, or pGreen0229-3D8 scFv-Protein A was grown overnight in Luria–Bertani medium supplemented with 50 μg/ml kanamycin and 20 μM acetosyringone. pGreen0229 was obtained from the John Innes Centre (U.K.). Cells of Agrobacterium were briefly spun down and were re-suspended in MMA medium (MS salts, 10 mM MES, pH 5.6, 200 mM acetosyringone) and were incubated for at least 1 h at 28 °C. After agroinfiltration, the plants were kept in constant environmental conditions in a growth chamber at 25 °C with a 16 h photoperiod. Three days post-agroinfiltration of an Agrobacterium suspension harboring a plasmid-expressing Protein A-tagged 3D8 scFv (~34 kDa), the leaves (~0.1 g) of the transgenic plants were used to purify the fusion protein by an IgG Sepharose affinity column (Amersham Biosciences) (Kim et al. 2006). Western blot analyses were performed using rabbit IgG and anti-rabbit IgG conjugated to alkaline phosphatase (Pierce Biotechnology Inc., IL) (Kim et al. 2006). Nucleic acid hydrolyzing assays of the cell lysates of the agroinfiltrated leaves were performed as described above using the supercoiled M13mp18 phagemid as a substrate.
Intracellular distribution of GFP-fused 3D8 scFv in plants
The 3D8 scFv gene was fused in frame to the green fluorescent protein (GFP) gene in the p326smGFP vector provided by Dr. I.H. Hwang (Pohang University, Korea) in order to generate p326smGFP-scFv (Fig. 1a). The plasmids were purified using Qiagen (Valencia, CA) columns according to the manufacturer’s protocol. Protoplasts isolated from Arabidopsis thaliana were prepared according to slight modifications of the protocols of Jin et al. (2001). Plasmid DNA (total of 20–50 μg at a concentration of 2 mg/ml) was added to 300 μl of the protoplast suspension followed by 325 μl of PEG solution [400 mM mannitol, 100 mM Ca(NO3)2, and 40 % polyethylene glycol 6,000]. After incubation of the protoplasts with plasmid DNA for 30 min at room temperature, the protoplasts were recovered by centrifugation at 50×g for 5 min, resuspended in 3 ml of W5 solution, and incubated at 22 °C in the dark. Expression of the GFP-fused 3D8 scFv protein was monitored at various times after transformation and the images were captured using a Zeiss Axioplan fluorescence microscope (Jin et al. 2001). Fluorescence signals were obtained at 36 h after transformation. The data were then processed as described previously (Jin et al. 2001).
Results and discussion
Expression of 3D8 scFv in transgenic tobacco
In order to obtain transgenic tobacco plants expressing 3D8 scFv, the leaf discs of wild-type N. tabacum were transformed with A. tumefaciens containing the plasmid pBI121-3D8 scFv that was constructed in order to express 3D8 scFv constitutively in the cytosol (Benvenuto et al. 1991; Rudolph et al. 2003) (Fig. 1a). The integration of the 3D8 scFv gene into the genome was confirmed by genomic PCR of randomly chosen primary transgenic shoots (T0, 60 lines). Later, thirty lines of the first generation (T1) and then 7 lines of the second generation (T2) progenies were randomly chosen based on kanamycin selection, PCR analysis, in order to confirm the presence of the 3D8 scFv gene (Fig. 1b). Finally, three of the chosen T2 lines (Tx99-1, Tx99-2, and Tx99-3 from the T1 lines of #3, 4, and 13, respectively) showed the presence of transcripts of 3D8 scFv at the predicted size, which were not present in the wild-type plants (Fig. 1c).
Virus resistance of 3D8 scFv plants
In order to determine whether 3D8 scFv with nuclease activities confers any viral resistance to the transgenic plants, the wild-types determined by comparing to three T2 lines of transgenic tobaccos (Tx99-1, Tx99-2, and Tx99-3) at ~3–4 leaf stage were challenged with viruses of different genome types (Rochester et al. 1990; Tavladoraki et al. 1993; Jin et al. 2001). Two ss-DNA geminiviruses (BCTV and BSCTV) were tested. Wild-type plants challenged by these viruses developed typical disease symptoms, such as stunting and curling of shoot tips and leaves by the geminiviruses, in 14-28 days post-inoculation (DPI) (Fig. 2a, b). In contrast, all of the transgenic tobacco plants (Tx99-1, Tx99-2, and Tx99-3) grew normally for more than 2 months following virus inoculation, without any visible signs of viral infection. Wild-type tobacco plants exhibited an infectivity of 57 % (n = 21) for ss-DNA of BCTV and 82 % (n = 22) for ss-DNA of BSCTV monitored by disease symptoms and confirmed by southern blot hybridization. However, the transgenic tobacco plants (n = 21–28 for each line: Tx99-1, Tx99-2, and Tx99-3) did not show any accumulation of either of the ss-DNA viruses for ~1 month after virus inoculation (Table 1 and Fig. 2c). All of the bioassay experiments were conducted under the same growth chamber for both wild-type and transgenic tobacco plants and there was no difference in the growth and development between wild-type and transgenic plants.
Suppression of disease symptoms and the inhibition of virus accumulation in the T2 transgenic tobacco plants. Two ss-DNA geminiviruses (BCTV or BSCTV) were challenged on T2 transgenic tobacco expressing 3D8 scFv. a Identification of virus accumulation in the WT and Tx99-2 tobacco plants with infectious BCTV and BSCTV clones. b Representative images of the leaves of the WT and Tx99-2 tobacco plants inoculated with ss-DNA geminiviruses. c Southern blots of the geminivirus DNA accumulation in the WT and Tx99-2 tobacco plants. Samples were taken from the newly produced upper leaves at 14 DPI. Virus replication intermediates were designated as ss for single-stranded DNA, sc for supercoiled ds-DNA, and rc for relaxed circular ds-DNA. In (c), M indicates ‘mock’ for a buffer control
DNase activity of 3D8 scFv
The 3D8 scFv subcloned into the pIg20 vector was expressed and purified from bacterial culture supernatants (Kim et al. 2006). The gels were stained with ethidium bromide (Fig. 3a: lane 1) in order to identify the regions of substrate hydrolysis, then with Coomassie Blue R250 (Fig. 3a: lane 2) in order to determine the position of 3D8 scFv. In situ gel assays revealed that degradation (dark black bands) was due to the activity of the 3D8 scFv and not the activity of the contaminating enzymes from Escherichia. coli (Fig. 3a). Catalytic affinity assay was performed, which was dependent on the reaction time. According to the agarose gel electrophoresis, the hydrolysis of ss-DNA and ds-DNA was proportional to the reaction time. Therefore, the 3D8 scFv has DNase activity in vitro (Fig. 3b).
The intrinsic DNase activity of 3D8 scFv. a In situ gel assays of the nucleic acid hydrolyzing activities of 3D8 scFv. The purified 3D8 scFv(2 μg) was applied to 12 % SDS–polyacrylamide gels containing 20 μg/ml of one of the following substrates: ss-DNA M13mp18 or ds-DNA M13mp18. The dark black bands on the ethidium bromide stained gels were due to nucleic acid hydrolysis. b The nucleic acid hydrolyzing activities of the 3D8 scFv monitored by agarose gel electrophoresis. The nucleic acid hydrolyzing experiments were initiated by mixing the purified 3D8 scFv (0.1 mM) with 2 nM of DNA substrate in TBS (50 mM Tris–Cl, pH 7.5, 50 mM NaCl) containing 10 mM MgCl2 or 50 mM EDTA. The BSA was used as a negative control. The arrows indicate supercoiled DNA (sc), linear DNA (lin), and relaxed circular (rc) DNAs
It was difficult to detect the expression of 3D8 directly by western blots for the cell lysates from the T1 and T2 transgenic plants. However, the transient expression of Protein A-tagged 3D8 scFv in the leaves of wild-type N. tabacum and the subsequent affinity purification demonstrated the expression of 3D8 scFv in the tobacco plants, as monitored by SDS-PAGE (Fig. 4a, b). The purified Protein A-tagged 3D8 scFv possessed nucleic acid degrading activities in proportion to the protein concentrations, as shown by degrading the plasmid DNA (Fig. 4c), which was indicative of the expression of functional 3D8 scFv in the transgenic tobacco plants.
The nuclease activity of purified 3D8 scFv expressed in tobacco plants by agroinfiltration. a Schematic vector maps of pGreen0229-3D8 scFv-Protein A, b SDS-PAGE of the purified Protein A-tagged 3D8 scFv (designated as 3D8 scFv-Protein A), which was transiently expressed in the leaves of the wild-type N. tabacum. The position of the 3D8 scFv-Protein A is indicated by an arrow. The cell lysates from the non-transiently expressed control leaves, which were treated with the same purification procedure, were loaded for comparison (designated as WT). ‘MW’ indicates the molecular weight standard marker. c Nuclease activity assay of the purified 3D8 scFv-Protein A that was transiently expressed in the leaves of wild-type N. tabacum, as described in (b). The purified Protein A-tagged 3D8 scFv possessed nucleic acid degrading activities in proportion to the protein concentrations by degrading the M13mp18 plasmid DNA
GFP fused 3D8 scFv exhibit cytosolic localization of 3D8 scFv
In order to monitor the intracellular distribution of 3D8 scFv in tobacco plants, GFP-fused 3D8 scFv, in which the GFP gene was fused in frame to the 5′ end or 3′ end of the 3D8 scFv gene (Fig. 5a), was expressed in the protoplasts of A. thaliana (Jin et al. 2001). Green fluorescent signals from the GFP-fused 3D8 scFv were observed in the cytosol (Fig. 5b), demonstrating the cytosolic localization of the 3D8 scFv in the transgenic tobacco plants (Jin et al. 2001).
The cytosol expression of 3D8 scFv a Schematic vector maps of p326smGFP-3D8 scFv-N and p326smGFP-3D8 scFv-C. b Subcellular distribution of GFP-fused 3D8 scFv expressed in the protoplasts of Arabidopsis. Two constructs were prepared for this study by the fusion of the GFP to the 3D8 scFv protein of either the C-terminus (upper panel) or the N-terminus (lower panel) of the 3D8 scFv protein. GFP, chlorophyll, merge, and WL indicate the green fluorescent signal, the autofluorescent signals of chlorophyll, the overlap of the GFP, and the autofluorescent signals, and the white light, respectively. Bar = 5 μm. (Color figure online)
3D8 scFv has no cytotoxic-effects on the plant growth of transgenic tobacco
The stability of the 3D8 scFv cytosolic expression on plant development was tested. 3D8 scFv was fused to both CaMV 35S normal and minimal promoters and transformed into tobacco plants. The highly expressed 3D8 scFv protein, under the control of the normal 35S promoter, led to fewer and smaller regenerated leaf discs in comparison to the less expressed 3D8 scFv protein under the control of a 35S minimal promoter (Fig. 6a–c). However, after the stable transformant lines were established, there was no significant difference observed in the wild-type tobacco plants vs. Tx99-2 in terms of plant growth and development (Fig. 6d, e). Explants containing the normal 35S promoter occurred more frequently in small size clusters compared to the explants with the 35S minimal promoter (Fig. 6b). The 3D8 scFv expression in fully developed transgenic plants was barely detected by western blot analysis (data not shown). Based on these observations, we concluded that the expression of a relatively high amount of 3D8 scFv may inhibit normal plant development so that no transformant was produced, but expression of lower levels of the 3D8 scFv could protect against virus infection with no harmful effects on plant development.
3D8 scFv protein could decrease the transformation efficiency, but had little effects on the established stable transformants (a) CaMV 35S normal and CaMV 35S minimal promoters fused with the 3D8 scFv gene were used for the introduction of the 3D8 scFv gene by the Agrobacterium-mediated transformation. The normal CaMV 35S promoter harboring leaf discs produced explants smaller in size and fewer in number than the CaMV 35S minimal promoter. b Regenerated explants were classified into 5 groups based on their size. c The expression levels of the 3D8 scFv protein in the regenerated leaf discs under the control of either the normal CaMV 35S promoter or the CaMV 35S minimal promoter. Tobacco leaf discs containing the constructs 35S::3D8 scFv and 35Smini::3D8 scFv were analyzed for 3D8 scFv expression by indirect ELISA using a polyclonal antiserum to the 3D8 scFv protein. d The leaf lengths of the transgenic plants and the wild-type plants were of similar sizes at each week [2, 4, 6, and 8 weeks after germination (WAG)] and the average leaf sizes were 22–25 cm at 8 WAG. e The average stem lengths of the transgenic and wild-type plants were 12 cm at 6 WAG, but the stem length of the transgenic plants at 8 WAG was 12–32 % shorter than that of the wild-type plants
Recently, antiviral studies have been focused on recombinant antibodies that bind to and inactivate essential viral proteins, such as viral enveloped proteins, coated proteins, replication initiator proteins, and viral RNA-dependent RNA polymerase, thereby interrupting virus propagation (Fecker et al. 1997; Boonrod et al. 2004; Safarnejad et al. 2009). However, antiviral agents were generally only effective for one or two of the target viruses, but to the lack of protection against new viruses is a severe problem. In order to overcome this problem, there were several trials for virus resistant developments in order to digest the viral genome itself by using nucleases. The antiviral effects experiment using nuclease was performed using an animal virus. Montandon et al. (1982) demonstrated virus resistance to the Moloney murine leukemia virus (M-MuLV) with DNase I. In this study, endogenous unmethylated proviral MuLV DNA was digested by DNase I in NIH-3T3 cell line (Montandon et al. 1982). In the plant virus resistance research, Pac1 (RNase activity), the transgenic potato showed virus resistance against the potato spindle tuber viroid (PSTVd). It is known that the anti-viral effects of these RNases depends on their RNase activity (Sano et al. 1997).
Through collaboration with several research groups, it was determined that the 3D8 recombinant antibody (3D8 scFv) has nuclease activity and their abzyme substrates can be all types of nucleic acids including single stranded DNA, double stranded DNA, single stranded RNA and double stranded RNA. It was also determined that 3D8 scFv could hydrolyze both DNA and RNA non-sequence specifically (Kim et al. 2006). Also, porcine kidney cell lines (PK15), which expressed the 3D8 scFv proteins showed induced virus resistance against the challenging classical swine fever virus (CSFV), which was a RNA virus. The virus protection seemed to be performed by RNase activity of 3D8 scFv proteins (Jun et al. 2010).
Our results demonstrated that 3D8 scFv targeted into cytosol can reduce DNA virus accumulation by DNase activity in 3D8 scFv transgenic tobacco. We described a novel strategy that targets the viral genome itself, rather than the viral gene products, in order to generate virus-resistance. Considering that field crops are usually infected with a range of viruses (Prins 2003; Soosaar et al. 2005), viral genome targeting is a novel strategy for the complete control of a broad spectrum of viruses, including uncharacterized and synergetically recombinant viruses at the molecular level, regardless of their genome types and variations in the gene products. The potential of 3D8 scFv for conferring resistance against viruses specific to other crops may provide an interesting avenue for future research.
References
Abel PP, Nelson RS, De B, Hoffmann N, Rogers SG, Fraley RT, Beachy RN (1986) Delay of disease development in transgenic plants that express the tobacco mosaic virus coat protein gene. Science 232:738–743
Baker B, Zambryski P, Staskawicz B, Dinesh-Kumar S (1997) Signaling in plant-microbe interactions. Science 276:726–733
Baulcombe DC (1996) Mechanisms of pathogen-derived resistance to viruses in transgenic plants. Plant Cell 8:1833
Benfey PN, Chua NH (1990) The cauliflower mosaic virus 35S promoter: combinatorial regulation of transcription in plants. Science 250:959–966
Benvenuto E, Ordas RJ, Tavazza R, Ancora G, Biocca S, Cattaneo A, Galeffi P (1991) ‘Phytoantibodies’: a general vector for the expression of immunoglobulin domains in transgenic plants. Plant Mol Biol 17:865–874
Blackburn P, Wilson G, Moore S (1977) Ribonuclease inhibitor from human placenta. Purification and properties. J Biol Chem 252:5904–5910
Boonrod K, Galetzka D, Nagy PD, Conrad U, Krczal G (2004) Single-chain antibodies against a plant viral RNA-dependent RNA polymerase confer virus resistance. Nat Biotechnol 22:856–862
Briddon R, Markham P (2000) Cotton leaf curl virus disease. Virus Res 71:151–159
Chellappan P, Masona MV, Vanitharani R, Taylor NJ, Fauquet CM (2004) Broad spectrum resistance to ssDNA viruses associated with transgene-induced gene silencing in cassava. Plant Mol Biol 56:601–611
Fauquet C, Bisaro D, Briddon R, Brown J, Harrison B, Rybicki E, Stenger D, Stanley J (2003) Virology division news: revision of taxonomic criteria for species demarcation in the family Geminiviridae, and an updated list of begomovirus species. Arch Virol 148:405–421
Fecker LF, Koenig R, Obermeier C (1997) Nicotiana benthamiana plants expressing beet necrotic yellow vein virus (BNYVV) coat protein-specific scFv are partially protected against the establishment of the virus in the early stages of infection and its pathogenic effects in the late stages of infection. Arch Virol 142:1857–1863
Hong Y, Stanley J (1995) Regulation of African cassava mosaic virus complementary-sense gene expression by N-terminal sequences of the replication-associated protein AC1. J Gen Virol 76:2415–2422
Jang J, Jeong J, Jun H, Lee S, Kim J, Kim Y, Kwon M (2009) A nucleic acid-hydrolyzing antibody penetrates into cells via caveolae-mediated endocytosis, localizes in the cytosol and exhibits cytotoxicity. Cell Mol Life Sci 66:1985–1997
Jin JB, Kim YA, Kim SJ, Lee SH, Kim DH, Cheong GW, Hwang I (2001) A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13:1511–1526
Jun HR, Pham CD, Lim SI, Lee SC, Kim YS, Park S, Kwon MH (2010) An RNA-hydrolyzing recombinant antibody exhibits an antiviral activity against classical swine fever virus. Biochem Biophys Res Commun 395:484–489
Kim YR, Kim JS, Lee SH, Lee WR, Sohn JN, Chung YC, Shim HK, Lee SC, Kwon MH, Kim YS (2006) Heavy and light chain variable domains of anti-DNA binding antibody hydrolyze both double- and single-stranded DNAs without sequence specificity. J Biol Chem 281:15287–15295
Kwon MH, Lee MS, Kim KH, Park S, Shin HJ, Jang YJ, Kim HI (2002) Production and characterization of an anti-idiotypic single chain Fv that recognizes an anti-DNA antibody. Immunol Invest 31:205–218
Liu C, Liu S, Wang F, Wang Y, Liu K (2012) Expression of a rice CYP81A6 gene confers tolerance to bentazon and sulfonylurea herbicides in both Arabidopsis and tobacco. Plant Cell Tissue Organ Cult (PCTOC) 109:419–428
Lucas JA (1998) Plant pathology and plant pathogens. Blackwell Science Ltd., Oxford
Mansoor S, Zafar Y, Briddon RW (2006) Geminivirus disease complexes: the threat is spreading. Trends Plant Sci 11:209–212
Moffat AS (1999) Geminiviruses emerge as serious crop threat. Science 286:1835
Montandon PE, Montandon F, Fan H (1982) Methylation state and DNase I sensitivity of chromatin containing Moloney murine leukemia virus DNA in exogenously infected mouse cells. J Virol 44:475–486
Noris E, Accotto G, Tavazza R, Brunetti A, Crespi S, Tavazza M (1996) Resistance to tomato yellow leaf curl geminivirus in Nicotiana benthamiana plants transformed with a truncated viral C1 gene. Virology 224:130–138
Pires AS, Rosa S, Castanheira S, Fevereiro P, Abranches R (2012) Expression of a recombinant human erythropoietin in suspension cell cultures of Arabidopsis, tobacco and Medicago. Plant Cell Tissue Organ Cult (PCTOC) 110:171–181
Prins M (2003) Broad virus resistance in transgenic plants. Trends Biotechnol 21:373–375
Prins M, Laimer M, Noris E, Schubert J, Wassenegger M, Tepfer M (2008) Strategies for antiviral resistance in transgenic plants. Mol Plant Pathol 9:73–83
Rochester DE, Kositratana W, Beachy RN (1990) Systemic movement and symptom production following agroinoculation with a single DNA of tomato yellow leaf curl geminivirus (Thailand). Virology 178:520–526
Rudolph C, Schreier PH, Uhrig JF (2003) Peptide-mediated broad-spectrum plant resistance to tospoviruses. Proc Natl Acad Sci U S A 100:4429–4434
Safarnejad MR, Fischer R, Commandeur U (2009) Recombinant-antibody-mediated resistance against Tomato yellow leaf curl virus in Nicotiana benthamiana. Arch Virol 154:457–467. doi:10.1007/s00705-009-0330-z
Sano T, Nagayama A, Ogawa T, Ishida I, Okada Y (1997) Transgenic potato expressing a double-stranded RNA-specific ribonuclease is resistant to potato spindle tuber viroid. Nat Biotechnol 15:1290–1294. doi:10.1038/nbt1197-1290
Scholthof K-BG, Scholthof HB, Jackson AO (1993) Control of plant virus diseases by pathogen-derived resistance in transgenic plants. Plant Physiol 102:7–12
Soosaar JL, Burch-Smith TM, Dinesh-Kumar SP (2005) Mechanisms of plant resistance to viruses. Nat Rev Microbiol 3:789–798
Sujatha M, Vijay S, Vasavi S, Reddy PV, Rao SC (2012) Agrobacterium-mediated transformation of cotyledons of mature seeds of multiple genotypes of sunflower (Helianthus annuus L.). Plant Cell Tissue Organ Cult (PCTOC) 110:275–287
Tavladoraki P, Benvenuto E, Trinca S, De Martinis D, Cattaneo A, Galeffi P (1993) Transgenic plants expressing a functional single-chain Fv antibody are specifically protected from virus attack. Nature 366:469–472
Thresh J, Otim-Nape G, Thankappan M, Muniyappa V (1998) The mosaic diseases of cassava in Africa and India caused by whitefly-borne geminiviruses. Rev Plant Pathol 77:935–945
Vance V, Vaucheret H (2001) RNA silencing in plants–defense and counterdefense. Science 292:2277–2280
Varma A, Malathi V (2003) Emerging geminivirus problems: a serious threat to crop production. Ann Appl Biol 142:145–164
Yang SJ, Carter SA, Cole AB, Cheng NH, Nelson RS (2004) A natural variant of a host RNA-dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana. Proc Natl Acad Sci U S A 101:6297–6302
Acknowledgments
This study was supported by a grant from the Next Generation BioGreen21 program (No. PJ0079842012) from the Rural Development Administration (RDA) and the Korea Institute of Ocean Science and Technology project (No. PE99154) of Korea.
Author information
Authors and Affiliations
Corresponding author
Additional information
Gunsup Lee, Hye-Kyung Shim and Myung-Hee Kwon have contributed equally to this study.
Rights and permissions
About this article
Cite this article
Lee, G., Shim, HK., Kwon, MH. et al. A nucleic acid hydrolyzing recombinant antibody confers resistance to curtovirus infection in tobacco. Plant Cell Tiss Organ Cult 115, 179–187 (2013). https://doi.org/10.1007/s11240-013-0357-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-013-0357-4