Abstract
Chrysanthemum stunt viroid (CSVd), the smallest plant pathogen known to infect chrysanthemums, is a single-stranded circular RNA viroid that induces stunting that results in an overall height reduction of 30–50 % in mature plants. A catalytic single-chain variable antibody, 3D8 scFv, which exhibits intrinsic DNase and RNase activities, was expressed in chrysanthemums to generate transgenic plant resistance to CSVd infection. Moreover, a codon-optimized version of the 3D8 scFv gene for chrysanthemums was also transformed into plants; these codon-optimized transgenic chrysanthemums expressed twice as much 3D8 scFv and displayed 60 % more resistance to CSVd infection, compared with transgenic chrysanthemums harboring the original 3D8 scFv gene. CSVd challenge experiments with codon-optimized and original 3D8 scFv-transgenic chrysanthemums showed that CSVd in newly produced leaves of both codon-optimized and original 3D8 scFv-transgenic plants was not detected by RT-PCR. This is the first report describing the development of a CSVd-resistant chrysanthemum harboring a catalytic single-chain antibody, 3D8 scFv, which has intrinsic RNase activity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chrysanthemum stunt viroid (CSVd) was first identified in 1973 as the causal agent of chrysanthemum stunt disease (Diener and Lawson 1973; Hollings and Stone 1973). CSVd consists of a single-stranded, circular RNA molecule that is 354–356 nucleotides in length; this viroid has been shown to be transmitted both mechanically and by vegetative propagation (Gross et al. 1982). Infection by CSVd results in stunting and an approximate reduction in the height of mature chrysanthemums by 30–50 %; the extent of these symptoms depends on the cultivar and environmental conditions such as temperature and light (Handley and Horst 1988; Hollings 1960). Foliar symptoms, such as smaller and paler leaves, have also been observed but are less common (Brierley 1950). CSVd is widely distributed in all regions in which chrysanthemums are grown, such as Europe, North America, South America, Africa, and Asia. However, in many cultivars, more than 30 % of the infected chrysanthemums exhibit no apparent symptoms (OEPP/EPPO 1989). These apparently uninfected chrysanthemums are believed to be a viroid reservoir for rapid CSVd spread.
For CSVd-free propagation of chrysanthemums, biocontrol appears to be the most effective approach. Meristem-tip culture and heat treatments were also applied for CSVd-free propagation of chrysanthemums; however, these approaches failed to eliminate CSVd from infected chrysanthemum plants (Hollings and Stone 1970). For most viroids, spread occurs readily by mechanical contact, such as plant-to-plant contact, handling during cultivation, and contact with contaminated tools. To overcome the limitations of viroid-free plant propagation, disease resistance enabled by genetic engineering has recently become an important goal for many crops and vegetables, as well as horticultural plants. In the past few decades, many transgenic plants have been developed with resistance against bacteria, viruses, and viroids (Fitchen and Beachy 1993; Ishida et al. 2002; Prins et al. 2008). Specifically, transgenic plants expressing recombinant dsRNA-specific RNases display a broad spectrum resistance to a variety of plant RNA viruses and viroids (Fitchen and Beachy 1993; Prins et al. 2008; Sano et al. 1997).
Some companies have developed transgenic plants tolerant to herbicide and harmful insect (Prado et al. 2014). In several countries, genetically modified bean and corn were allowed to be sold commercially (James 2013). Even though transgenic plants have been many questioned, they are thought to be one of the best solutions to improve the productivity of crops easily, which could contribute to poverty reduction in the developing countries (Qaim 2010).
In general, substantial transformation research has thus far been documented for horizontal gene transfer among different species. However, a remaining challenge is to maximize the expression of the introduced gene in the genome of a heterologous organism. First, codon usage for protein expression is known to depend on the genome of the host species (Burgess-Brown et al. 2008; Menzella 2011). Many of the introduced genes show low expression levels in different genomes, because these genes contain codons that are rarely used in the desired host genome (Burgess-Brown et al. 2008). These unusual codons reduce the translation rate and induce translation errors during protein synthesis. Therefore, transgenes have been redesigned to match the pattern of codon usage in the heterologous organism by systematic altering the codons in the transgenes to enhance the yields of the expressed proteins (Fiedler et al. 1997; Menzella 2011; Owens and Hammond 2009).
The 3D8 single-chain variable fragment (3D8 scFv) is an antinucleic acid antibody that can bind and hydrolyze nucleic acid in a sequence-independent manner (Kim et al. 2012). Recently, 3D8 scFv molecules have been shown to induce resistance to both DNA and RNA viruses in tobacco. The progenies of 3D8 scFv-transgenic tobacco plants exhibited complete resistance against two single-stranded (ss) DNA geminiviruses, Beet curly top virus, and Beet severe curly top virus, without viral accumulation or the manifestation of any disease symptoms (Lee et al. 2013a). Progenies of these transgenic tobacco plants also acquired complete resistance against four ss-RNA tobamoviruses and one cucumovirus, without any viral accumulation, and with a delayed onset of disease symptoms (Lee et al. 2013b). Moreover, the progenies of 3D8 scFv-transgenic tobacco plants did not show noticeable adverse effects with respect to either growth or reproduction. In addition, Turnip mosaic virus (TuMV) resistance was developed in most Chinese cabbage (Brassica rapa) lines transformed with the 3D8 scFv gene (Jung et al. 2009).
In the present study, the original and codon-optimized versions of the 3D8 scFv gene for chrysanthemum were introduced into a horticultural plant species, chrysanthemum, and the anti-CSVd effects were investigated in the resultant transgenic chrysanthemum plants. Our results demonstrate a novel approach for chrysanthemum protection against CSVd infection using a nucleic acid-hydrolyzing recombinant 3D8 scFv antibody.
Materials and Methods
Plant Materials and Preparation of Viroids
Florist’s chrysanthemum (Chrysanthemum × morifolium) plants (Cultivar “Vivid Scarlet”) were provided from the Rural Development Administration (RDA), Korea, and grown in an inoculation box containing charcoal MS medium lacking hormones (Naing et al. 2014). Newly produced chrysanthemum plants were subcultured on new MS medium every 2 months by asexual propagation. After obtaining CSVd inoculum kindly provided by Dr. Bong Nam Chung (Rural Development Administration, NCBI GenBank number AF394452), healthy chrysanthemum plants were inoculated with CSVd inoculum and maintained in a growth chamber under a 16:8-h light/dark cycle at 24 °C.
CSVd RNA Transcript Preparation and Infection of Transgenic Chrysanthemums
Full-length CSVd-cDNA was synthesized from total RNA isolated from CSVd-infected leaf tissue and subsequently cloned into pGEM®-T Easy, a vector that harbors the T7 promoter (Promega, Madison, WI, USA) (Fig. 1a, b). CSVd RNA was synthesized from the cloned cDNA using an in vitro transcription kit (HiScribe T7 In Vitro Transcription; New England Biolabs, Ipswich, MA, USA). CSVd transcripts were gently applied with a cotton swab onto the surface of the chrysanthemum leaves for infection.
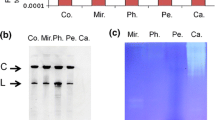
CSVd transcript preparation and analysis of hydrolysis activity of 3D8 scFv. a CSVd cDNA was subcloned into pGEM®-T Easy, a vector that contains a T7 promoter and a ScaI restriction enzyme site. b Verification of ScaI-mediated linearization of CSVd cDNA by electrophoresis. In vitro transcription was performed to generate capped CSVd RNA transcripts from the linearized cDNA. c Expression and purification of recombinant 3D8 scFv. Protein induction was performed in E. coli BL21(DE3)pLysE cells; purified proteins were resolved by SDS-PAGE and stained with Coomassie blue. E1-9, eluate fractions. d Hydrolysis activity assay of 3D8 scFv. CSVd RNA transcripts (3 μg) were incubated with purified 3D8 scFv (2 μg) in TBS with MgCl2 (0.1 mM) at 37 °C for different lengths of time. Reaction products were analyzed by electrophoresis
Expression and Purification of 3D8 scFv
To test its nucleic acid-hydrolyzing activity, 3D8 scFv was expressed recombinantly in bacteria and purified by IgG-Sepharose affinity chromatography as previously described (Kim et al. 2006). Protein concentrations (mg ml−1 cm−1) were determined using an extinction coefficient of 1.995 at 280 nm, which was calculated from the amino acid sequence of the 3D8 scFv protein. Eluted proteins were resolved by SDS-PAGE and stained with Coomassie blue (Fig. 1c).
Codon Optimization of the 3D8 scFv Gene
Variations in codon usage frequency between animals and plants can cause differences in translation efficiency for protein synthesis. In this study, the 3D8 scFv gene used was originally derived from the autoimmune-prone MRL-lpr/lpr mouse. The codon sequence of the original 3D8 scFv gene was optimized to maximize its protein expression level in chrysanthemums. A codon-optimized version of the 3D8 scFv gene was designed based on the preferential codon usage of chrysanthemums, using the back-translation program (Fig. 2). Codon bias patterns were measured by the codon adaptation index (CAI) in the CAI Application by comparing the codons used in the original 3D8 scFv gene and the codon-optimized version of 3D8 scFv with typical chrysanthemum codon usage patterns (Nakamura et al. 2000). The optimized 3D8 scFv gene was synthesized by Noble Bioscience, Inc., USA.
Comparison of original and codon-optimized 3D8 scFv sequences. The codon-optimized 3D8 scFv sequence contained 250 codons; the original 3D8 scFv sequence contained 249 codons. The similarities and differences between the codon-optimized and the original 3D8 scFv codon sequences are shown. Both DNA sequences encode the same amino acid sequence; however, their codon sequences exhibited many differences. For instance, only 87 out of 250 codons were identical in both the original and codon-optimized 3D8 scFv sequences
Construction of the Recombinant Expression Vector
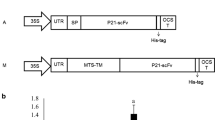
Initially, an Agrobacterium transformant (strain LBA4404) harboring the pBI121 vector was cultured in this study. The pBI121 vector (harboring the nptII kanamycin resistance gene) also contained the coding sequence of the original 3D8 scFv, which was used as the template to synthesize the codon-optimized 3D8 scFv gene. To subclone the codon-optimized 3D8 scFv fragment into pGEM®-T Easy, the codon-optimized sequence was PCR-amplified and ligated with the gene encoding protein A. The resultant fragment was subsequently subcloned into pGEM®-T Easy. Finally, the recombinant expression vector was constructed by subcloning the coding regions into pCAMBIA 1304 (harboring the hptII hygromycin resistance gene) for the expression of codon-optimized 3D8 scFv (Fig. 3a, b).
Generation of expression vectors and cloning strategy for the codon-optimized 3D8 scFv construct. a Construction of a pCAMBIA 1304-based vector for the expression of codon-optimized 3D8 scFv, compared with that of a pBI121-based vector for the expression of nonoptimized 3D8 scFv. b Cloning strategy for the codon-optimized 3D8 scFv gene. After subcloning into pGEM-T® Easy, the codon-optimized 3D8 scFv gene was subcloned into pCAMBIA1304 using the SpeI and BstEII sites. For comparison, the pBI121 recombinant vector containing the original 3D8 scFv sequence is also shown
Agrobacterium-Mediated Transformation of Chrysanthemums
The recombinant expression vectors were introduced into the GV3101 strain of Agrobacterium tumefaciens for the expression of codon-optimized 3D8. The LBA4404 strain was used for the expression of the original gene. Agrobacteria were cultured in YEP medium with kanamycin (50 μg/ml). Cells were harvested by centrifugation (3000 rpm for 15 min) and resuspended in Murashige and Skoog (MS) medium (Murashige and Skoog 1962). Chrysanthemum leaf disks were inoculated with agrobacteria and cultured on antibiotic-free MS medium supplemented with indole-3-acetic acid (IAA, 0.5 mg/l) and 6-benzyladenine (6-BA, 1.0 mg/l) for 2 days (Fig. 4a). Independent experiments were performed for transformation of the nonoptimized 3D8 scFv gene (four replicates) and the codon-optimized gene (five replicates). To rescue nonoptimized 3D8 scFv-transgenic plants, calli were transferred to MS medium supplemented with indole-3-acetic acid (IAA, 0.5 mg/l), 6-benzyladenine (6-BA, 1.0 mg/l), cefotaxime (100 mg/l), and kanamycin (50 mg/l) for shoot regeneration and grown for 3 weeks (Fig. 4b). Subsequently, the shoots were cultured for 3 weeks on MS medium containing the same concentrations of antibiotics (cefotaxime and kanamycin) as the shooting medium, with half the amount of IAA (0.25 mg/l), for rooting (Fig. 4c). The transgenic chrysanthemum plants were maintained in a sterile transparent plastic box containing MS medium supplemented with cefotaxime (100 mg/l), kanamycin (50 mg/l), and charcoal (1.0 mg/l) (Fig. 4d), and finally acclimatized to a pot (Fig. 4e). The same selection scheme was used for the cell culture and regeneration of codon-optimized 3D8 scFv-transgenic plants, except that hygromycin selection was used.
Agrobacterium-mediated transformation in tissue culture. a Leaf disks were dipped in Agrobacterium solution and then placed on MS medium. b Shoots induced from a callus, 3 weeks after inoculation into shooting medium. c Rooting was induced from plantlets after transfer to rooting and selection medium. d Healthy chrysanthemum plants with full roots were transferred to MS medium supplemented with charcoal and antibiotics (cefotaxime 100 mg/l and kanamycin 20 mg/l). e After confirmation by RT-PCR, plants were transferred to soil for viroid resistance tests
Analysis of 3D8 scFv RNA Transcript and Protein Levels in Transgenic Chrysanthemums
Bona fide transgenic lines were selected from the putative 3D8 scFv transformants and the putative codon-optimized 3D8 scFv transformants by RT-PCR. The transgenes and the selectable marker genes were detected in the codon-optimized 3D8 scFv (hptII)-transgenic plants and the original 3D8 scFv (nptII)-transgenic plants using specific primer sets (Table 1). Total RNA was isolated from transgenic plants using RNeasy Plant Mini Kit (Qiagen, Valencia, CA), and cDNA was synthesized from total RNA using an oligo(dT)18 primer and M-MLV reverse transcriptase (Bioneer, Daejeon, Korea). Amplification was performed with AccuPower PCR PreMix (Bioneer). The pBI121 expression vector, harboring the original 3D8 scFv gene, was used as a positive control. Protein levels were quantified by performing enzyme-linked immunosorbent assays (ELISAs) on extracts from transgenic plants. Levels of the original (unmodified) and the codon-optimized 3D8 scFv protein were assessed in the transgenic chrysanthemum plants to evaluate the effects of 3D8 scFv codon optimization for the chrysanthemum genome on protein translation efficiency. Total protein extracts were prepared from young leaf tissue of chrysanthemum plants and diluted to 20 μg/ml in coating buffer (1.59 g sodium carbonate, 2.93 g sodium bicarbonate, and 0.2 g sodium azide dissolved in 1000 ml distilled water). All subsequent reactions were performed at room temperature for 2 h. Diluted extracts were aliquoted directly into a 96-well polyvinyl chloride microtiter plate and incubated at 4 °C overnight to allow protein binding. Next, the wells were washed twice with phosphate-buffered saline (PBS) and then blocked with BSA in PBS (5 % w/w). The wells were then washed three times with PBS and incubated with anti-3D8 scFv polyclonal antibodies [1:1000 dilution in dilution buffer; BSA (1 % w/v) in PBS]. Anti-3D8 scFv antibody was kindly provided by Dr. Myung-Hee Kwon (Ajou Medical School, Korea). Wells were washed four times with PBS-T (PBS with 0.05 % Tween-20) and then incubated with a 1:5000 dilution of alkaline phosphatase-conjugated goat anti-rabbit IgG antibodies. After a final four washes with PBS-T, 4-nitrophenyl phosphate disodium salt hexahydrate substrate was added to the wells. The substrate was diluted in ELISA PNP buffer (0.1 g magnesium chloride hexahydrate, 0.2 g sodium azide, and 97 ml diethanolamine), with the pH adjusted to 9.8 with hydrochloric acid, and its final volume adjusted to 1000 ml with distilled water. The absorbances of the wells at 405 nm were measured with an ELISA plate reader (TECAN Sunrise). Protein quantifications were carried out independently in triplicate. Protein levels were compared with those of BSA (an internal control) and a nontransgenic plant.
CSVd Resistance Test and Analysis of Disease Symptoms in Transgenic Chrysanthemums
Representative codon-optimized and original 3D8 scFv-transgenic plants were selected to monitor the manifestation of disease symptoms and the development of CSVd resistance at 10-week postinoculation. For observations of disease symptoms, noninfected plants (mock) were included as an internal control. CSVd was detected in the inoculated leaves and young leaves of transgenic plants and noninfected plants by RT-PCR. CSVd-specific primers were designed based on the sequences of CSVd (Table 1).
Results
In Vitro Hydrolysis of CSVd Transcripts by Purified 3D8 scFv Protein
To verify that 3D8 scFv could hydrolyze the CSVd genome, CSVd cDNA was subcloned into pGEM®-T Easy (Fig. 1a), and genomic CSVd RNA was synthesized using an in vitro transcription kit (New England BioLabs) (Fig. 1b). 3D8 scFv protein was expressed in E. coli BL21(DE3) pLysE cells transformed with pIg20-3D8 scFv and purified via its protein A tag on IgG-Sepharose resin (Fig. 1c). The ability of purified 3D8 scFv to hydrolyze genomic CSVd RNA over time was then assessed in in vitro assays (Fig. 1d). After 30 min of incubation, degradation of genomic CSVd RNA was observed; this degradation was nearly complete after 180 min of incubation. This result indicates that 3D8 scFv can hydrolyze genomic CSVd RNA, in addition to the other viral nucleic acids it is also known to hydrolyze (Jun et al. 2010; Lee et al. 2013b).
Codon Optimization of 3D8 scFv Gene Sequence for Chrysanthemums
The 3D8 scFv coding sequence (GenBank Accession Numbers: 3D8VH, AF232220 and 3D8VL, AF232221) was originally obtained from an autoimmune-prone MRL-lpr/lpr mouse (Kwon et al. 2002). In previous studies using Nicotiana tabacuum, the original 3D8 scFv gene was introduced into tobacco by the leaf disk-mediated agrobacterium transformation method. However, in this study, the intended purpose for expressing the 3D8 scFv gene was to protect chrysanthemums from CSVd infection rather than from viral infection. According to the codon usage table for chrysanthemums in the database (Tables 2 and 3), the original 3D8 scFv sequence contains rare codons that are used at low frequency in chrysanthemums. Based on the codon usage database, the rare codons in the original 3D8 scFv sequence include 2 CUC and 10 CUG codons, encoding leucine; 1 CGC codon for arginine; and 8 UCC and 6 AGC codons for serine (Table 3). It is likely that the presence of these rare codons in the original 3D8 scFv gene sequence impairs the expression of 3D8 scFv in chrysanthemums. By analyzing the codon usage in chrysanthemums (http://www.kazusa.or.jp/codon/), the 3D8 scFv sequence was codon-optimized for chrysanthemums by changing the low-frequency codons to high-frequency codons and optimizing the GC content from 52 to 43 %. This codon optimization was hypothesized to improve the expression of 3D8 scFv in chrysanthemums. Initially, the 3D8 scFv coding sequence was optimized using the most frequent codons found in chrysanthemums. However, the GC content of the new gene sequence was 34 % (data not shown), which was an unacceptable level for chrysanthemums, which normally exhibit a GC content of 43 % (Table 3). Therefore, we substituted some codons with the second-most frequent codons, thus increasing the final GC content to 43 %. After codon optimization, the optimized 3D8 scFv gene sequence contained 87 codons that were the same as in the original sequence (black codons), and 163 codons that were different from those in the original sequence (red codons) (Fig. 2). Altogether, the codon-optimized 3D8 scFv gene contained a total of 250 codons, encoding 19 different amino acids. The codon-optimized version contained 196 most frequently used codons and 54 second-most frequently used codons in the Chrysanthemum × morifolium genome; no rare codons were present in optimized 3D8 scFv gene (Table 2)
Genetic Transformation of Chrysanthemum Plants
The transformation efficiencies of the leaf disks and plantlets were determined at each step of the five-step transformation procedure. The transformation efficiency was first calculated as the number of confirmed plantlets divided by the number of selected rooting plantlets. The transformation efficiencies for the original 3D8 scFv and the codon-optimized version of 3D8 scFv were calculated as the average transformation efficiency divided by the time of replication. Overall, the transformation efficiency for 3D8 scFv (18.2 %) was nearly equal to the transformation efficiency of the codon-optimized version of 3D8 scFv (19.7 %) (Table 4).
Enhanced Expression of 3D8 scFv in Codon-Optimized 3D8 scFv-transgenic Plants
The expression levels of the original 3D8 scFv and the codon-optimized version of 3D8 scFv were analyzed on both the RNA transcript level and the protein level. RT-PCR was employed to detect 3D8 scFv transcripts in a population of transgenic plants harboring both the original and codon-optimized versions of 3D8 scFv. The transgenic plants expressed RNA transcripts of the original 3D8 scFv gene (Fig. 5a (b)) as well as the codon-optimized 3D8 scFv gene (Fig. 5a (a)). Additional RT-PCR analysis was performed to detect two selectable marker genes, the hygromycin resistance gene (hptII) (for the optimized 3D8 scFv transgene, Fig. 5a (a)) and the kanamycin resistance gene (nptII) (for the original 3D8 scFv transgene, Fig. 5a (b)), in the transformed plants. Therefore, the plants were confirmed as transgenic for the target gene and also for the selectable marker gene. RT-PCR analysis showed that all transgenic plants harboring the original (5 lines) and optimized version (12 lines) of 3D8 scFv genes expressed 3D8 scFv. The levels of scFv protein in the same transgenic lines (5 lines for original and 12 lines for optimized version) were quantified by ELISA using anti-3D8 scFv antibodies. Generally, transgenic plants expressing the optimized 3D8 scFv gene showed more 3D8 scFv protein than the original 3D8 scFv transgenic plants (Fig. 5b (a, b)). Only two transgenic plants (line Ori-5-2 and Ori-6), which expressed the original 3D8 scFv gene, produced scFv protein at a detectable level with the anti-3D8 scFv antibodies. Taken together, compared with the original 3D8 scFv-transgenic plants (Fig. 5b (b)), the codon-optimized transgenic plants (lines Opt-14, Opt-16-2, Opt-17, and Opt-24) produced more than twice as much 3D8 scFv protein (Fig. 5b (a)).
Transformation analysis and comparison of protein expression levels. a Confirmation of putative 3D8 scFv-transgenic and optimized 3D8 scFv-transgenic chrysanthemum plants on the RNA level. RT-PCR was performed with primers specific for the original 3D8 scFv sequence and the hygromycin resistance gene (hptII) in the codon-optimized 3D8 scFv-transgenic plants (a), or with primers specific for the original 3D8 scFv sequence and the kanamycin resistance gene (nptII) for the codon-optimized 3D8 scFv-transgenic plants (b). Controls: cDNA synthesized from a wild-type chrysanthemum (N) or a plasmid harboring either the original or the codon-optimized 3D8 scFv sequence (P). b ELISA analysis of protein extracts from 5 original 3D8 scFv-transgenic plants and 12 codon-optimized 3D8 scFv-transgenic plants that were previously confirmed by RT-PCR. Controls: bovine serum albumin (BSA) or protein from a wild-type chrysanthemum plant (none)
Antiviroid Effects of the 3D8 scFv Transgene Against CSVd in Chrysanthemum Plants
Transgenic plant lines harboring either the codon-optimized or the original version of the 3D8 scFv gene were inoculated with in vitro-synthesized CSVd transcripts (Fig. 6). Infected plants were then examined for symptoms of viroid infection and the development of CSVd resistance at 10-week postinoculation. The development of disease symptoms was clearly different between wild-type and 3D8 scFv-transgenic chrysanthemum plants; in all transgenic plants expressing either the original or the codon-optimized version of 3D8 scFv, no symptoms of viroid infection were detected, indicating that CSVd resistance had developed in these plants. However, the wild-type plants exhibited severe symptoms such as leaf curling and plant stunting, as shown in Fig. 6a. Viroid resistance was confirmed at the molecular level in all 3D8 scFv-transgenic plants that did not exhibit any symptom of infection. To this end, RT-PCR was used to assess the presence of CSVd in the leaves that had been initially inoculated, as well as in the newly sprouted young leaves. While CSVd RNA was detected in inoculated leaves of both transgenic lines by RT-PCR, almost no CSVd RNA was detected in the young leaves of the transgenic lines, with two exceptions: one codon-optimized 3D8 scFv line (Opt-4) and one original 3D8 scFv transgenic line (Ori-5-2) (Fig. 6b).
Analysis of viroid resistances in original 3D8 scFv-transgenic and codon-optimized 3D8 scFv-transgenic chrysanthemum plants and representative disease symptoms observed in chrysanthemum plants carrying Chrysanthemum stunt viroid (CSVd). a Comparison of different symptoms between wild-type and 3D8 scFv-transgenic plants infected with CSVd. The wild-type plant had a reduced height compared with the transgenic plants. b Detection of CSVd in inoculated plants by RT-PCR. RT-PCR was used to detect CSVd RNA in both original 3D8 scFv-transgenic and optimized 3D8 scFv-transgenic plants using RNA isolated from young leaves. RT-PCR was used to detect CSVd RNA in both original 3D8 scFv-transgenic and optimized 3D8 scFv-transgenic plants using RNA isolated from inoculated leaves. Although CSVd RNA was detected in all inoculated leaves by RT-PCR, CSVd RNA was not detected in the young leaves from some of the lines. These lines were Opt-4, derived from the optimized 3D8 scFv-transgenic plants, and Ori-5-2, derived from the original 3D8 scFv-transgenic plants
Discussion
In our previous studies, 3D8 scFv was shown to exhibit antiviral activity against various plant viruses (Beet curly top virus (BCTV), Beet severe curly top virus (BSCTV), Pepper mild mottle virus (PMMoV), Tobacco mild green mosaic virus (TMGMV), Tomato mosaic virus (ToMV), Tobacco mosaic virus (TMV), and Cucumber mosaic virus (CMV)), and animal viruses (Pseudorabies virus (PRV), Herpes simplex virus (HSV), Classical swine fever virus (CSFV), and H9N2 influenza virus) (Jun et al. 2010; Kim et al. 2012; Lee et al. 2013a, b, 2014), even at low 3D8 scFv protein concentrations. In the transgenic cells or mouse system which we developed, 3D8 scFv could only be detected by ELISA, but not by Western blot analysis of protein lysates (Jun et al. 2010; Lee et al. 2014). In our previous works, the stability of 3D8 scFv cytosolic expression on plant development was tested because of intrinsic nuclease activity of 3D8 scFv proteins (Lee et al. 2013a). We observed that the highly expressed 3D8 scFv protein produced relatively fewer and smaller leaf disks compared to the less expressed 3D8 scFv protein. However, once the stable transformants were established, they did not show significant difference in wild-type versus transgenic plants. 3D8 scFv expression in fully developed transgenic plants was hardly detected by Western blot analysis even though they showed the complete resistance against virus infection. This finding was also observed on transgenic animal cell lines and mouse system (Lee et al. 2014). Taken together, we concluded that expression of low levels of 3D8 scFv could be good enough to protect against virus infection with no harmful effects on plant development.
Based on our previous results, in this study, we tried to express more 3D8 scFv protein on chrysanthemums by codon optimization of this protein for these plants. 3D8 scFv in transgenic chrysanthemums expressing either the original or codon-optimized version were detected by ELISA, but not by Western blot analysis (Fig. 5). However, codon optimization of the 3D8 scFv sequence for chrysanthemums, in which rare codons were substituted for high-frequency codons, enabled higher expression of 3D8 scFv in transgenic chrysanthemum. ELISAs showed that the level of codon-optimized 3D8 scFv protein was almost two times higher than that of the original 3D8 scFv protein (Fig. 5). Moreover, a positive correlation was observed between the level of expression of 3D8 scFv and resistance to CSVd infection (Fig. 6).
The antiviral effect experiment by certain type of nuclease was introduced in animal virus work (Montandon et al. 1982), and Pac1 (dsRNA-specific ribonuclease) was first applied for induced viroid resistance against potato spindle tuber viroid (PSTVd) (Sano et al. 1997). Compared to Pac1, 3D8 scFv can digest all types of nucleic acids such as single-stranded and double-stranded DNA/RNA. Our results demonstrated that 3D8 scFv can reduce viroid RNA accumulation by intrinsic RNase activity in 3D8 scFv transgenic chrysanthemum. This 3D8 scFv, which can presumably degrade a broad spectrum of viroid RNAs, represents a major advantage in field crops because they are usually infected with a range of viroid and viruses (Prins 2003; Soosaar et al. 2005).
Resistance to CSVd was analyzed in both the originally inoculated leaves and in the newly produced young leaves (Fig. 6). At 10-week postinoculation, CSVd RNA was detected in all of the inoculated leaves by RT-PCR, indicating that both transgenic and wild-type chrysanthemum plants were infected with CSVd. However, CSVd was not detected in the newly produced young leaves of transgenic plants expressing the original 3D8 scFv and the codon-optimized version of 3D8 scFv. Out of the 12 transgenic lines expressing codon-optimized 3D8 scFv, CSVd RNA was only detected in the young leaves of one (Opt-4) of these lines. In the original 3D8 scFv transgenic plants, one line (Ori-5-2) out of five transgenic lines was infected by CSVd. But, CSVd RNA accumulations of both original and optimized 3D8 scFv transgenic plants were much reduced compared to wild-type chrysanthemum plants. Codon-optimized 3D8 scFv-transgenic plants did show higher resistance to CSVd infection compared with the original 3D8 scFv-transgenic plants, could be due to the higher expression level of 3D8 scFv in the codon-optimized 3D8 scFv-transgenic plants compared with the original 3D8 scFv-transgenic plants. In both transgenic plants, 3D8 scFv was capable of protecting chrysanthemum from CSVd infection. In our previous studies, in which tobacco plants were challenged with two DNA and five RNA viruses, 3D8 scFv was not detected by Western blot analysis in transgenic tobacco plants, although all viruses tested failed to infect them (Lee et al. 2013a; Lee et al. 2013b). Thus, we infer that the transgenic plants were expressing a small, but sufficiently protective, amount of 3D8 scFv, since 3D8 scFv was shown to digest various viral genomes.
In conclusion, the present study demonstrates that animal-derived 3D8 scFv antibodies can confer viroid resistance to plants. Specifically, the results presented here indicate that systemic invasion of CSVd is prevented by 3D8 scFv. When CSVd infects wild-type chrysanthemums, it successfully replicates and disseminates. However, in 3D8 scFv-transgenic chrysanthemums, CSVd RNA was detected in both infected leaves of transgenic plants by (1) possible replication of inoculum CSVd RNAs on infected leaves or (2) just RNA amplification of inoculum RNA without CSVd replication process. To understand the resistance mechanism shown in this study, further investigation is needed. Taken together, CSVd RNA in 3D8 scFv-transgenic chrysanthemums seems to be digested by the intrinsic nuclease activity of 3D8 scFv present in leaves or stems. A subsequent viroid infection study is currently ongoing for 3D8 scFv-transgenic chrysanthemum plants focusing on another chrysanthemum-specific viroid, CChMVd. These and other follow-up studies will potentially support a role for 3D8 scFv in conferring protection to plants against a broad variety of viroids.
References
Brierley P (1950) Some host plants of Chrysanthemum stunt virus. Phytopathology 40:869
Burgess-Brown NA, Sharma S, Sobott F, Loenarz C, Oppermann U, Gileadi O (2008) Codon optimization can improve expression of human genes in Escherichia coli: A multi-gene study. Protein Expr Purif 59:94–102
Diener T, Lawson R (1973) Chrysanthemum stunt: a viroid disease. Virology 51:94–101
Fiedler U, Phillips J, Artsaenko O, Conrad U (1997) Optimization of scFv antibody production in transgenic plants. Immunotechnol: Int J Immunol Eng 3:205–216
Fitchen JH, Beachy RN (1993) Genetically engineered protection against viruses in transgenic plants. Annu Rev Microbiol 47:739–763
Gross HJ, Krupp G, Domdey H, Raba M, Jank P, Lossow C, Alberty H, Sänger HL, Ramm K (1982) Nucleotide sequence and secondary structure of citrus exocortis and chrysanthemum stunt viroid. Eur J Biochem 121:249–257
Handley M, and Horst R (1988) The effect of temperature and light on chrysanthemum stunt viroid infection of florists chrysanthemum. Paper presented at: VII International Symposium on Virus Diseases of Ornamental Plants 234
Hollings M (1960) American stunt virus in English chrysanthemum stocks. Ann Rep Glasshouse Crops Res Inst 1959
Hollings M, Stone OM (1970) Attempts to eliminate chrysanthemum stunt from chrysanthemum by meristem‐tip culture after heat‐treatment. Ann Appl Biol 65:311–315
Hollings M, Stone OM (1973) Some properties of chrysanthemum stunt, a virus with the characteristics of an uncoated ribonucleic acid. Ann Appl Biol 74:333–348
Ishida I, Tukahara M, Yoshioka M, Ogawa T, Kakitani M, Toguri T (2002) Production of anti-virus, viroid plants by genetic manipulations. Pest Manag Sci 58:1132–1136
James C (2013) Global status of commercialized biotech/GM crops: 2012. International Service for the Acquisition of Agri-Biotech Applications SEAsia Center (ISAAA), Ithaca
Jun HR, Pham CD, Lim SI, Lee SC, Kim YS, Park S, Kwon MH (2010) An RNA-hydrolyzing recombinant antibody exhibits an antiviral activity against classical swine fever virus. Biochem Biophys Res Commun 395:484–489
Jung Y, Rhee Y, Auh C-K, Shim H, Choi J-J, Kwon S-T, Yang J-S, Kim D, Kwon M-H, Kim Y-S (2009) Production of recombinant single chain antibodies (scFv) in vegetatively reproductive Kalanchoe pinnata by in planta transformation. Plant Cell Rep 28:1593–1602
Kim YR, Kim JS, Lee SH, Lee WR, Sohn JN, Chung YC, Shim HK, Lee SC, Kwon MH, Kim YS (2006) Heavy and light chain variable single domains of an anti-DNA binding antibody hydrolyze both double- and single-stranded DNAs without sequence specificity. J Biol Chem 281:15287–15295
Kim A, Lee JY, Byun SJ, Kwon MH, Kim YS (2012) Viral genome RNA degradation by sequence-selective, nucleic-acid hydrolyzing antibody inhibits the replication of influenza H9N2 virus without significant cytotoxicity to host cells. Antivir Res 94:157–167
Kwon MH, Lee MS, Kim KH, Park S, Shin HJ, Jang YJ, Kim HI (2002) Production and characterization of an anti-idiotypic single chain Fv that recognizes an anti-DNA antibody. Immunol Investig 31:205–218
Lee G, Shim H-K, Kwon M-H, Son S-H, Kim K-Y, Park E-Y, Lee T-K, Lee W-R, Auh C-K, Kim D et al (2013a) A nucleic acid hydrolyzing recombinant antibody confers resistance to curtovirus infection in tobacco. Plant Cell Tissue Organ Cult 115:179–187
Lee G, Shim H-K, Kwon M-H, Son S-H, Kim K-Y, Park E-Y, Yang J-K, Lee T-K, Auh C-K, Kim D et al (2013b) RNA virus accumulation is inhibited by ribonuclease activity of 3D8 scFv in transgenic Nicotiana tabacum. Plant Cell Tissue Organ Cult 115:189–197
Lee G, Yu J, Cho S, Byun SJ, Kim DH, Lee TK, Kwon MH, Lee S (2014) A nucleic-acid hydrolyzing single chain antibody confers resistance to DNA virus infection in hela cells and C57BL/6 mice. PLoS Pathog 10, e1004208
Menzella HG (2011) Comparison of two codon optimization strategies to enhance recombinant protein production in Escherichia coli. Microb Cell Factories 10:15
Montandon PE, Montandon F, Fan H (1982) Methylation state and DNase I sensitivity of chromatin containing Moloney murine leukemia virus DNA in exogenously infected mouse cells. J Virol 44:475–486
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–497
Naing AH, Jeon SM, Han J-S, Lim SH, Lim KB, and Kim CK (2014) Factors influencing in vitro shoot regeneration from leaf segments of Chrysanthemum. Comptes Rendus Biologies
Nakamura Y, Gojobori T, Ikemura T (2000) Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res 28:292
Owens RA, Hammond RW (2009) Viroid pathogenicity: one process, many faces. Viruses 1:298–316
Prado JR, Segers G, Voelker T, Carson D, Dobert R, Phillips J, Cook K, Cornejo C, Monken J, Grapes L (2014) Genetically engineered crops: From idea to product. Annu Rev Plant Biol 65:769–790
Prins M (2003) Broad virus resistance in transgenic plants. Trends Biotechnol 21:373–375
Prins M, Laimer M, Noris E, Schubert J, Wassenegger M, Tepfer M (2008) Strategies for antiviral resistance in transgenic plants. Mol Plant Pathol 9:73–83
Qaim M (2010) Benefits of genetically modified crops for the poor: household income, nutrition, and health. New Biotechnol 27:552–557
Sano T, Nagayama A, Ogawa T, Ishida I, Okada Y (1997) Transgenic potato expressing a double-stranded RNA-specific ribonuclease is resistant to potato spindle tuber viroid. Nat Biotechnol 15:1290–1294
Soosaar JL, Burch-Smith TM, Dinesh-Kumar SP (2005) Mechanisms of plant resistance to viruses. Nat Rev Microbiol 3:789–798
Acknowledgments
This study was supported by a grant from the Next Generation BioGreen 21 program (No. PJ007984012014; Rural Development Administration, Korea) and a grant from the Korea Institute of Ocean Science and Technology (No. PE99154).
Author contributions
D.T. and S.L. conceived the study. D.T., Y.R., and P.H. designed and performed experiments and analyzed data. E. K. and T.L. gave conceptual advice and technical support for the clinical analysis. D.T., S.C., and P.H. performed experiments. J.K. provided conceptual advice for codon optimization of 3D8 scFv. K.L. and Y.R. performed statistical analysis. D.T., S.C., Y.R., and S.L. supervised the study and wrote the paper.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Tran, D.T., Cho, S., Hoang, P.M. et al. A Codon-Optimized Nucleic Acid Hydrolyzing Single-Chain Antibody Confers Resistance to Chrysanthemums Against Chrysanthemum Stunt Viroid Infection. Plant Mol Biol Rep 34, 221–232 (2016). https://doi.org/10.1007/s11105-015-0915-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-015-0915-5