Abstract
A critical step in the development of a reproducible Agrobacterium tumefaciens mediated transformation system for a recalcitrant species, such as pearl millet, is the establishment of optimal conditions for efficient T-DNA delivery into target tissue from which plants can be regenerated. A multiple shoot regeneration system, without any intervening callus phase, was developed and used as a tissue culture system for Agrobacterium-mediated transformation. Agrobacterium super virulent strain EHA105 harboring the binary vector pCAMBIA 1301 which contains a T-DNA incorporating the hygromycin phosphotransferase (hpt II) and β-glucuronidase (GUS) genes was used to investigate and optimize T-DNA delivery into shoot apices of pearl millet. A number of factors produced significant differences in T-DNA delivery; these included optical density, inoculation duration, co-cultivation time, acetosyringone concentration in co-cultivation medium and vacuum infiltration assisted inoculation. The highest transformation frequency of 5.79% was obtained when the shoot apex explants were infected for 30 min with Agrobacterium O.D.600 = 1.2 under a negative pressure of 0.5 × 105 Pa and co-cultivated for 3 days in medium containing 400 μM acetosyringone. Histochemical GUS assay and polymerase chain reaction (PCR) analysis confirmed the presence of the GUS gene in putative transgenic plants, while stable integration of the GUS gene into the plant genome was confirmed by Southern analysis. This is the first report showing reproducible, rapid and efficient Agrobacterium-mediated transformation of shoot apices and the subsequent regeneration of transgenic plants in pearl millet. The developed protocol will facilitate the insertion of desirable genes of useful traits into pearl millet.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pearl millet (Pennisetum glaucum (L.) R. Br.) is a robust, cross pollinating, diploid (2n = 2x = 14) annually grown major cereal crop. It is a high yielding, drought tolerant summer crop and can be grown in low rainfall areas where other crops such as maize and sorghum are not profitable (FAO 2004). Nutritionally, it is comparable to rice and is an excellent forage crop because of its low hydrocyanic acid content (Chowdari et al. 1998). Increased tillering to compensate for stress induced loss of yield, rapid deep root penetration, roots with specialized cell walls to prevent desiccation and an efficient C4 mechanism with potential growth rates make pearl millet an ideal crop for arid and semi arid tropics.
The development of an efficient method of genetic transformation is a pre-requisite for the application of bio-molecular techniques to the improvement of a given crop species. Cereals including millet crops have been primary targets for improvement by genetic transformation (Vasil 2005; Ceasar and Ignacimuthu 2009). In comparison to other major cereals, there have been only a few reports on successful genetic transformation of pearl millet (Taylor and Vasil 1991; Taylor et al. 1993; Lambe et al. 1995, 2000; Girgi et al. 2002, 2006; Goldman et al. 2003; O’Kennedy et al. 2004; Latha et al. 2006), and these were limited to biolistic methods. Although microprojectile bombardment has revolutionized the field of genetic transformation of cereals there were considerable variation seen in stability, integration and expression of the introduced transgene (Kohli et al. 1999). Moreover, this technique is expensive and requires specialized instruments such as biolistic/DNA gun for bombardment. Hence, there is a requirement for alternate methods such as Agrobacterium tumefaciens mediated genetic transformation, which appears more effective at regenerating transgenic plants with low transgene copy number. These transgenic plants were more stable over generations and have reduced gene silencing associated with integration of T-DNA into euchromatic regions (Barakat et al. 1997; Shou et al. 2004).
The two critical steps to be optimized for genetic transformation of plants are transfer of foreign DNA into the plant cells and regeneration of plants from transformed cells (Yookongkaew et al. 2007). In many species, transgenic plant recovery was difficult, because the cells transformed may not regenerate owing to several factors such as cells accessible for gene transfer are not suitable for plant regeneration (Komari et al. 1998). Therefore, development of a simple and effective approach for gene transfer is of major interest. We have developed an efficient in vitro plant regeneration protocol through somatic embryogenesis (from seeds, shoot apices and immature inflorescences explants) and direct shoot organogenesis (from shoot apex explant) for pearl millet (Jha et al. 2009). It has been observed that genetic mutations (methylation, albinism) and somaclonal variations were low in plants regenerated directly from shoot meristematic cultures as compared to plants regenerated from embryogenic calli (Bregitzer et al. 2002). Probable reason for this low frequency of mutation may be the absence of tissue dedifferentiation steps that are common in the initiation of callus and somatic embryo culture (Hirochika 1993). Thus, direct shoot organogenesis could be the preferred method for genetic transformation of pearl millet in order to minimise somaclonal variation and genotype dependence observed during callus-mediated regeneration. Transformation of shoot apices was first reported in 1988 (McCabe et al. 1988). Shoot apex explant can either be transformed using Agrobacterium or through biolistic methods (Gould and Magallanes-Cedeno 1998; Zapata et al. 1999; Cho et al. 2003; Goldman et al. 2003; Yookongkaew et al. 2007).
Two methods were used to obtain transgenic plants by transfer of DNA into the shoot apical meristem. In the first method transgenic progeny are directly produced from the shoot apex explants, or meristem cells, followed by development of transgenic plants having partially transformed reproductive organs. T0 plants produced by this way will always be chimeric. In the second method transformed shoot apical meristem cells are multiplied by treatment with growth regulators, then reprogrammed into the developmental stage under in vitro conditions (Zhong et al. 1996). Thus, manipulation of transgenic meristem cells by treatment with growth regulator to induce multiple shoot regeneration could result in more stably transformed plants (Yookongkaew et al. 2007). We used the second method (Multiple shoot induction) to optimize transformation protocol.
To the best of our knowledge, there are no reports on Agrobacterium-mediated transformation of pearl millet. Herein we report a reproducible, rapid and efficient Agrobacterium-mediated transformation protocol using Agrobacterium strain EHA 105 harboring the binary vector pCAMBIA 1301. This Agrobacterium strain EHA 105 is one of the popular strains (with vector pCAMBIA 1301) used for plant transformation (Yookongkaew et al. 2007; Li et al. 2010, 2011). Binary vector pCAMBIA 1301 contain hygromycin phosphotransferase (hpt II) and β-Glucuronidase (GUS) genes in its T-DNA. hpt II gene is used as the selectable marker gene and GUS gene is used as reporter in order to optimize parameters for high frequency transformation and subsequent regeneration of transgenic pearl millet plants.
Materials and methods
Multiple shoot induction (shoot organogenesis) from shoot apical meristem cultures
Mature seeds of pearl millet inbred genotype 843B (obtained from ICRISAT, India) were surface disinfected with 70% ethanol for 1 min and then with 0.1% mercuric chloride for 5 min, followed by five washes in sterile distilled water. Surface disinfected seeds were grown on Murashige and Skoog (MS) medium (Murashige and Skoog 1962) containing 30 g/l sucrose and 8 g/l agar (Bactoagar, Qualigens). The pH of medium (used in all the shoot organogenesis experiments) was adjusted to 5.8 before agar was added. The media were autoclaved at 121°C for 15 min and 25 ml of media was dispensed into each 100 ml autoclaved conical flask (Borosil) and closed by sterile cotton plugs.
Emerging shoot apices consisting of shoot apical meristem and a part of mesocotyl were excised from 3 to 4 days old seedlings and cultured on MS medium + 17.6 μM BAP + 30 g/l sucrose + 8 g/l agar (pH 5.8) for multiple shoot induction (Jha et al. 2009). Cultures were maintained under 16 h light provided by fluorescent lamps with the light intensity of 50 μmol m−2 s−1 at 25 ± 2°C. Shoot tips were subcultured after every 20 days in the same induction medium. Multiple shoots produced from the shoot tips were carefully separated. For shoot elongation, small shoots were cultured on MS medium.
The shoot apices of 5–6 mm length, obtained from 3 to 4 day old aseptically grown seedlings, were used as explants for Agrobacterium-mediated transformation. These were precultured for 12 h on MS + 17.6 μM BAP + 30 g/l sucrose + 8 g/l agar (pH 5.8) 1 day before co-cultivation with Agrobacterium.
Sensitivity of shoot apices of pearl millet to hygromycin
Prior to genetic transformation, the amount of hygromycin required to inhibit shoot growth was tested. The non-transformed shoot apices were placed on MS + 17.6 μM BAP + 30 g/l sucrose + 8 g/l agar (pH 5.8) supplemented with 0, 5, 10, 15, 20, 25, 30, 35 and 50 mg/l hygromycin and subcultured 2 times at 15 days interval. The survival rates of the explants were evaluated. The concentration of hygromycin that killed most of the explant was used in subsequent transformation experiments.
Agrobacterium strain, binary vector and preparation of Agrobacterium suspension
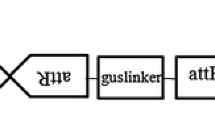
Agrobacterium tumefaciens supervirulent strain EHA 105, harboring the binary vector pCAMBIA 1301 was used in the present study. The T-DNA of pCAMBIA 1301 (Fig. 1) contains CaMV 35S promoter driven GUS gene interrupted by a modified castor bean catalase intron and CaMV 35S promoter driven hpt II gene conferring resistance to hygromycin. The intron within the GUS gene prevents its expression by Agrobacterium and allows the detection of eukaryotic expression only (Vergne et al. 2010).
A single bacterial colony was grown overnight in liquid YENB (0.75% yeast extract, 0.8% nutrient broth [pH 7.5]) medium supplemented with 50 mg/l kanamycin at 25°C with an orbital shaking of 150 rpm. Next morning the culture was resuspended in the same medium such that O.D.600 nm was approximately 0.1. It was then grown for 4–5 h under same conditions until an O.D.600 nm of 0.8–1.0 was reached. The suspension was pelleted down at 5,000 rpm for 5 min. The pellet was subsequently resuspended in MS-inf medium [MS + 68.5 g/l sucrose + 36.04 g/l glucose + 1 g/l casamino acid + 200 μM acetosyringone (pH 5.2) (modified from Ishida et al. 1996)] to be used for Agrobacterium-infection.
Transformation of shoot apices with Agrobacterium
Precultured explants (shoot apices) were immersed in MS-inf medium for different time durations (10–60 min). The O.D.600 nm of Agrobacterium in MS-inf medium was adjusted from 0.4 to 3.0 before Agrobacterium-infection. To evaluate the effect of negative pressure (0.5 × 105 Pa), a vacuum pump was used during Agrobacterium-infection (for 30 min). Following Agrobacterium-infection, shoot apex explants were blotted dry on sterile filter paper to remove excess Agrobacterium and then transferred to fresh co-cultivation medium [MS + 17.6 μM BA + 20 g/l sucrose + 10 g/l glucose + 0.7 g/l proline + 0.5 g/l MES + 0, 100, 200 or 400 μM acetosyringone + 0.5 g/l casamino acid + 8 g/l agar (pH 5.8) (modified from Ishida et al. 1996)] for 1–6 days. Co-cultivation was carried out at 25°C in 16 h light. Different concentrations of acetosyringone (Acs) (0, 100, 200 and 400 μM) were tested in co-cultivation medium. The co-cultivated shoot apices were first washed with cefotaxime solution (250 mg/l) for 5 min, then washed four times with sterile distilled water and finally blot dried and transferred onto recovery medium [MS + 17.6 μM BA + 30 g/l sucrose + 8 g/l agar + 500 mg/l cefotaxime (pH 5.8) (modified from Jha et al. 2009)] for 7 days to inhibit the growth of Agrobacterium. After recovery phase, shoot apices were transferred on to the selection medium [MS + 17.6 μM BA + 30 g/l sucrose + 8 g/l agar + 250 mg/l cefotaxime + 30 mg/l hygromycin (pH 5.8) (modified from Jha et al. 2009)] for 30 days, with 15 days subculture intervals, to stimulate the production of transgenic shoots. Surviving hygromycin-resistant shoots were transferred onto pre-regeneration medium [MS + 17.6 μM BA + 30 g/l sucrose + 8 g/l agar + 250 mg/l cefotaxime (pH 5.8) (modified from Jha et al. 2009)] for 4–6 weeks. Surviving shoots were regenerated on regeneration medium [MS + 30 g/l sucrose + 8 g/l agar + 30 mg/l hygromycin (pH 5.8) (modified from Jha et al. 2009)] for 2–3 weeks and were rooted on rooting medium [½ MS + 0.4% w/v activated charcoal + 15 g/l sucrose + 8 g/l agar + 30 mg/l hygromycin (pH 5.8) (modified from Jha et al. 2009)].
Analysis of reporter gene expression
Transformed tissues (shoot apices after co-cultivation and leaves after selection) were analyzed for β-glucuronidase expression by using X-Gluc as the substrate (Jefferson 1987). The histochemical reaction was allowed to proceed at 37°C overnight. Subsequently, the tissues were cleared in 70% ethanol. To serve as control against any background GUS staining, untransformed tissues were included at all staining occasions.
PCR and Southern blot analyses
Genomic DNA was isolated from leaves of untransformed and putatively transformed plants using Qiagen DNeasy Plant Medi Kit. PCR analysis was carried out in a 50 μl reaction mixture containing DNA (500 ng), 200 μM of each dNTP, 1 μl of each primer (10 μM), 1 unit of Taq DNA polymerase, 2.0 mM MgCl2 and 1 × Taq Buffer. PCR conditions used were 94°C for 5 min for initial melting followed by 30 cycles of amplification with each cycle consisting of the following steps: 94°C for 30 s, 60°C for 1 min and 72°C for 1 min with a final extension at 72°C for 7 min. GUS gene specific primers were used for detection of GUS gene. These primers amplify a product of 1,029 bp from pCAMBIA 1301 plasmid. The sequences of primers used were 5′-GCC ATT TGA AGC CGA TGT CAC GCC-3′ (forward primer), 5′-GTA TCG GTG TGA GCG TCG CAG AAC-3′ (reverse primer). Primers used for amplification of hpt II gene were 5′-CTATCGGCGAGTACTTCTACAC-3′ (forward primer), 5′-GTGTCACGTTGCAAGACCTA-3′ (reverse primer). These primers amplify an internal hpt II sequence of 694 bp.
About 10 μg of genomic DNA isolated from independently transformed plants was digested with NcoI and Eco72I to release ~2 kb GUS gene from the T-DNA region of pCAMBIA 1301. The digested DNA was electrophoresed on 1% agarose gel and transferred to Hybond-N+ membrane (Amersham Pharamacia Biotech, UK) using standard protocols (Sambrook et al. 1989). These DNAs were hybridized against a random primed radiolabelled probe of 1,029 bp fragment obtained by PCR amplification of pCAMBIA1301 with GUS specific primers (forward primer 5′-GCC ATT TGA AGC CGA TGT CAC GCC-3′, reverse primer 5′-GTA TCG GTG TGA GCG TCG CAG AAC-3′). Scanning and recording of images were performed in phosphorimager (FUGIFILM, FLA 5000).
Data analysis
Explants showing GUS blue spots were recorded as positive. Each experiment was replicated 3 times and each set with a replicate consisted of at least 55 explants. The transformation frequency was calculated as the total number of transgenic plantlets produced relative to the total number of explants infected by Agrobacterium. The data was analyzed using SPSS software version 10 (www.spss.en.softonic.com). Analysis of variance (ANOVA) was used to test the statistical significance, and the significance of differences among means was carried out using LSD (Least significant difference) at a significance of P = 0.05.
Results and discussion
The T-DNA transfer from Agrobacterium into host plant genome is known to be influenced by several factors (reviewed by Cheng et al. 2004; Jones et al. 2005). As Agrobacterium-mediated transformation is a multi factor, complex interaction process (Liu and Pijut 2010), it is very difficult to analyze that which factor (such as inoculation, co-culture, co-cultivation, selection, acetosyringone etc.) contributes the most to the transformation efficiency. Step by step optimization of such factors proved to be of considerable importance for the establishment of successful transformation systems in monocotyledonous crops. Since there has been no report of successful transformation of pearl millet using Agrobacterium tumefaciens, it was considered important to investigate the effects of some of the factors on T-DNA delivery. In the present study, various factors affecting gene delivery were determined by assaying the activity of GUS gene in leaf tissues of putatively transformed plants by histochemical assays after selection. Most of the studies of Agrobacterium-mediated transformation of cereals use tissues consisting of actively dividing meristematic cells, such as immature embryos and callus induced from scutella. There are only a few reports on Agrobacterium-mediated cereal transformation using shoot apices or shoot apical meristem as explant (Sairam et al. 2003 in maize; Park et al. 1996; Arockiasamy and Ignacimuthu 2007; Yookongkaew et al. 2007 in rice; Zhang et al. (2010) in Kentucky blue grass). In pearl millet, direct shoot organogenesis without visible intervening callus phase was developed by us (Jha et al. 2009) using shoot apices as explants, which was a rapid and reproducible regeneration protocol. In the present study, pearl millet shoot apices were transformed with Agrobacterium and transformed shoot apices were propagated by multiple shoot formation.
Shoot organogenesis
For shoot organogenesis without any intervening callus phase (i.e. multiple shoot induction), aseptically grown shoot apices were cultured on MS medium containing 17.6 μM BA and subcultured every 20 days onto the same induction medium. Multiple shoots (Fig. 2a) produced within 4–6 weeks were carefully separated for shoot elongation. Small shoots were cultured for 2–3 weeks on MS medium for shoot elongation. As reported previously, a maximum of 22 shoots formed in 9 weeks on MS + 17.6 μM BA (Jha et al. 2009). Shoots were rooted on ½ strength MS containing 0.4% (w/v) activated charcoal.
Agrobacterium-mediated transformation and plant regeneration in P. glaucum. a Multiple shoot induction from shoot apex explants on MS medium containing 17.6 μM BA (scale bar 10 mm). b, c Transient GUS expression in shoot apex explants co-cultivated with A. tumefaciens strain EHA105 harbouring the plasmid pCAMBIA1301 (scale bar 5 mm). d Shoots growing on hygromycin containing selection medium (scale bar 10 mm). e GUS expression in transformed and control leaves (scale bar 5 mm). f Regeneration of transformed plants on regeneration medium (scale bar 10 mm). g, h Rooting and hardening of transgenic plants (scale bar 10 mm). i Transgenic plants 6 weeks after transfer to pots
This kind of meristematic shoot culture resulting in the formation of multiple shoots by differentiating axillary and adventitious buds by the use of BA have been described for maize (Zhong et al. 1992), oat (Zhang et al. 1996), barley (Zhang et al. 1998), sorghum (Zhong et al. 1998), pearl millet (Devi et al. 2000; Jha et al. 2009), finger millet (Kumar et al. 2001) and wheat (Ahmad et al. 2002). All cereal crops tested have shown a similarity of response to medium containing different concentrations of BA. The medium containing lower concentrations of BA (2.2 or 4.4 μM) promoted more axillary buds than adventitious buds even after 3 months of cultures. The medium with 8.8 μM or 17.6 μM BA mostly stimulated shoot apical domes to enlarge, followed by differentiation of both axillary and adventitious buds from the leaf axils and enlarged apical domes within the first 2–3 months of incubation (reviewed by Sticklen and Oraby 2005). Kinetin was also reported to induce multiple shoots in finger millet (Kumar et al. 2001) and pearl millet (Jha et al. 2009).
Sensitivity of shoot apex explants to different concentrations of hygromycin
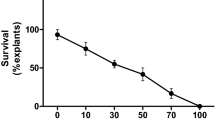
The effects of various concentrations of hygromycin were evaluated on shoot apex explants to determine the appropriate selection dose. Our analysis revealed that fewer than 5% of the shoot apices were able to survive in the presence of 30 mg/l hygromycin and that no shoots survived in the presence of higher concentrations of hygromycin (Fig. 3). Therefore, 30 mg/l hygromycin was used in subsequent transformation experiments.
Influence of Agrobacterium cell density, inoculation period, co-cultivation period, acetosyringone concentration and vacuum condition on transformation frequency
Cell density
A critical factor in the shoot apices transformation system is the density of the Agrobacterium inoculum in the inoculation medium (Li et al. 2007). We obtained the best transformation frequency (3.34%) using Agrobacterium inoculum at O.D.600 1.2 (Table 1). A reduction in the mean transformation frequency was observed following inoculation with higher densities of Agrobacterium cells, possibly because of increased production of toxins to the receptor cells (Gu et al. 2008). Several workers reported the optimum cell density for Agrobacterium-infection in O.D. < 1 [Zhao et al. (2000) in sorghum; Kumria et al. (2001) in rice; Sarker and Biswas (2002) in wheat; Gasparis et al. (2008) in oat] while several reported O.D. = 1 as best suited for Agrobacterium-infection [Kumar et al. (2005a) in rice; Shrawat et al. (2007) in barley; Yookongkaew et al. (2007) in rice]. Our results are in accord with that of Amoah et al. (2001), where they reported O.D. between 1 and 1.5 as best suited for producing highest GUS expression frequency in wheat callus. Determining the optimal inoculation density was important because at high O.D. levels, the explant tissues were almost wholly colonized by the bacteria, elimination of which becomes more difficult following the co-cultivation and, subsequently, during selection and regeneration stages. Usually this would necessitate the use of antibiotics at higher levels which, in itself, has detrimental effects on plant tissue development.
Inoculation period
The duration of the exposure time of explant to Agrobacterium cells also influences the transformation frequency. Pearl millet shoot apex explants incubated for 30 min with Agrobacterium cells at O.D.600 = 1.2 showed significantly increased frequency of transformation compared to those transformed for 10 and 20 min, while exposure to Agrobacterium for more than 45 min resulted in a decline in transformation frequency (Table 2). Prolonged inoculation time adversely affected the explant because of overgrowth of Agrobacterium. Different inoculation periods were reported by different workers. Zhao et al. (2000); Howe et al. (2006) in sorghum and Ishida et al. (2007) in maize, reported 5 min of inoculation period as optimal, while Sarker and Biswas (2002) reported 50 min of inoculation period as optimal along with 5 min vacuum treatment in rice. Our results are in accord with Lee et al. (2006) in orchardgrass and Zhao et al. (2011) in chinese upland rice, where they recommended an inoculation period of 30 min.
Co-cultivation period
The co-cultivation duration is a crucial factor influencing Agrobacterium-mediated gene transfer in pearl millet. Using the optimal conditions described above (O.D.600 = 1.2 and inoculation period of 30 min), the effect of varying the length of co-cultivation period was investigated. Co-cultivation duration for 2–3 days was generally considered suitable for Agrobacterium-mediated transformation as reported for many plant species such as wheat (Amoah et al. 2001; Mitic et al. 2004), rice (Al-Forkan et al. 2004; Hoque et al. 2005; Yookongkaew et al. 2007); buffel grass (Kumar et al. 2005b) and switchgrass (Somleva et al. 2002). Lee et al. (2006) and Ma et al. (2010) conducted co-cultivation for 1–7 days on orchardgrass callus and bast fiber plant ramie callus, respectively. Lee et al. (2006) found co-cultivation of 3 days as optimal, while Ma et al. (2010) reported 2 days of co-cultivation as optimal. In the present study, co-cultivation was carried out in the 16 h light from 0 to 6 days. Table 3 shows that the transformation frequency increased from 0.57 ± 0.13% at 1 day to 4.07 ± 0.63% at 3 days. Extension of co-cultivation period from 1 to 3 days significantly enhanced the frequency of transformation. Although co-cultivation for more than 2 days resulted in slight Agrobacterium growth around the explant, GUS expression was more intense when compared to shoot apices cultured for 2 days in transient GUS expression studies (Fig 2b, c). Extending the co-cultivation to longer than 3 days resulted in an abundant proliferation of Agrobacterium, tissue necrosis and subsequent cell death. Our results are in agreement with the observation of Wu et al. (2003) and Shrawat et al. (2007) where they reported 3 days co-cultivation as optimal for barley callus. Yang et al. (2010) optimized several factors such as Agrobacterium cell density, inoculation period and co- cultivation period for transformation of boston ivy and reported that co-cultivation period was critical for transformation.
Acetosyringone
Plant specific phenolic compounds that induce the expression of Agrobacterium vir genes are important for gene transfer (Stachel et al. 1985). In monocots, where such compounds are not synthesized, addition of phenolic compounds such as acetosyringone during plant and bacterial interaction supports the gene transfer (Stachel et al. 1985; Hiei et al. 1994). Using the optimal conditions described above (O.D.600 = 1.2, inoculation period of 30 min and co-cultivation period of 3 days), the effect of varying concentration of acetosyringone (0–400 μM) in co-cultivation medium was investigated. No GUS expression was observed when acetosyringone was excluded from the co-cultivation medium (Table 4). It was found that increasing the concentration of acetosyringone from 100 to 400 μM significantly enhanced the transformation frequency, the highest frequency 4.66 ± 0.64% being obtained on medium containing 400 μM acetosyringone. These results suggest that inclusion of acetosyringone in co-cultivation medium significantly influences the transformation frequency and therefore is essential for successful transformation of pearl millet. This observation supports earlier reports in a number of species showing that the addition of acetosyringone during co-cultivation increases the number of transformed cells in the target tissue in rice (Hiei et al. 1994), wheat (Wu et al. 2003) and barley (Shrawat et al. 2007). Although acetosyringone has not been found to be essential for Agrobacterium-mediated transformation of barley (Tingay et al. 1997; Fang et al. 2002) it has been reported to be a key component to successful transformation of rice (Hiei et al. 1997), maize (Ishida et al. 1996) and wheat (Cheng et al. 2003; Wu et al. 2003). The difference in the requirement of acetosyringone for successful transformation of cereals may be because of the differences in the inoculation and co-cultivation duration and also in the competence of target tissues (Shrawat et al. 2007).
Vacuum infiltration of shoot apices with Agrobacterium
Vacuum treatment to infiltrate tissues with Agrobacterium has been successfully used in transformation of cereals (Dong et al. 2001; Amoah et al. 2001). In our system, a negative pressure of 0.5 × 105 Pa, created by the vacuum pump in the vacuum desiccator, resulted in increased transformation frequency (5.79 ± 0.43%) in comparison to that obtained at atmospheric pressure (Fig. 4). It has been suggested that a vacuum pump creates a negative pressure environment that results in an increase in effective Agrobacterium volatilization, a condition conducive to the transfer of a foreign gene into plant cells (Gu et al. 2008). Vacuum treatment of shoot apices with Agrobacterium for more than 30 min resulted in explant tissues being completely colonized by the Agrobacterium, making it more difficult to eliminate it in the recovery and subsequent stages resulting in loss of shoot apices and subsequent growth (data not shown). In the present study, when combined with optimized parameters, vacuum infiltration assisted transformation of shoot apices of pearl millet significantly enhances the transformation frequency. Our results are in agreement with Shrawat et al. (2007) who reported that vacuum infiltration (along with sonication) assisted in Agrobacterium-mediated transformation of barley.
Effect of vacuum infiltration treatment on transformation frequency in pearl millet. Shoot apex explants were inoculated with A. tumefaciens strain EHA 105 harbouring pCAMBIA 1301 at O.D.600 = 1.2 with and without negative pressure of 0.5 × 105 Pa created by vacuum pump for infection time of 30 min and co-cultivated for 3 days on co-cultivation medium containing 400 μM acetosyringone. Transformation frequency was calculated as number of transgenic plants/total number of explants inoculated ×100%
Regeneration of stably transformed plants
Using the optimal transformation procedure, the shoot apices immersed in Agrobacterium suspension (MS-inf medium) of O.D.600 = 1.2 for 30 min under vacuum treatment, and then co-cultivated for 3 days on co-cultivation medium gave the maximum transformation frequency. If the shoot apices were cultured on the selection medium immediately following co-cultivation these shoot apices appeared to suffer from the combined stress of Agrobacterium and selection. To alleviate this problem, the explants were transferred to recovery medium (as described in materials and methods). After the recovery phase, shoot apices were transferred to selection medium containing hygromycin for 30 days, with 15 days subculture intervals, to allow the growth of transformed shoots. The shoot apices that survived on selection medium (Fig. 2d) were transferred onto pre-regeneration medium for 4–6 weeks to stimulate the production of transgenic multiple shoots. These transformed plants showed GUS expression in leaves (Fig. 2e). Transgenic shoots were then separated individually and regenerated on regeneration medium for 2–3 weeks (Fig. 2f). The well grown shoots with leaves were rooted on rooting medium (Fig. 2g, h) and then hardened in pots containing autoclaved agropeat. The transgenic plants were covered with polythene bags to retain moisture and nutrients were provided by Hoagland’s medium (Hoagland and Arnon 1950) (Fig. 2i).
Establishment of high-efficiency Agrobacterium-mediated transformation has greatly facilitated the widespread application of transformation in cereals (Repellin et al. 2001; Cheng et al. 2004). This technique has been widely used to introduce genes of interest into cereal genomes and also as a common means of testing gene function by enhancing or inhibiting expression of target genes (Kobayashi et al. 2001; Nagasaki et al. 2001; Mori et al. 2002). There are no reports of Agrobacterium-mediated transformation in pearl millet. All the reports have used biolistics for transformation of pearl millet (Taylor and Vasil 1991; Taylor et al. 1993; Lambe et al. 1995, 2000; Girgi et al. 2002, 2006; Goldman et al. 2003; O’Kennedy et al. 2004; Latha et al. 2006) and have used precultured immature zygotic embryos and embryogenic callus as a transformation target. Transgenic plants regenerated by a more or less long term callus phase are at enhanced risk of somaclonal variations and problems in transgene inheritance and stability of transgene expression (Bregitzer and Tonks 2003). In our study, a system in which shoot apices having shoot apical meristems was used as the material for gene transfer in pearl millet inbred genotype 843B. Transgenic plants regenerated directly from shoot apical meristems will always be chimeric. However, a feasible technique for avoiding this phenomenon was to multiply transgenic shoot apical meristem cells by treatment with plant growth regulator (Zhong et al. 1996). We used BA to multiply the transformed shoot apices containing shoot apical meristems and produced multiple shoots from single transformed shoot tips. These shoots were regenerated after selection and used for transgenic analysis. In recent years, several reports on use of multiple-shoot apical meristem as (Agrobacterium-mediated) transformation targets are available (Zhang et al. 2010 in Kentucky blue grass; Zhang et al. 2005 and Sairam et al. 2003 in corn; Yookongkaew et al. 2007 in rice, Gu et al. 2008 in winter jujube), indicating great potential of this technique.
Genomic DNA was extracted from the leaves of untransformed control plants and transformed plantlets obtained after selection and regeneration. PCR amplification of the GUS region was performed to detect the presence of the transgene in the genome of T0 generation transformants (named as (T01–T012).The GUS gene (1,029 bp) was amplified by using genomic DNA from transformant lines tested (Fig. 5a). Out of ten GUS primer positive transgenic lines, six lines (T02, T03, T04, T05, T08 and T010) were again cross checked by hpt II gene specific primers and showed amplification of a 694 bp (Fig. 5b). Stable integration of the transgene in PCR positive plants (T02, T03, T04, T05, T08 and T010) was confirmed by Southern analysis. Genomic DNA digested with NcoI and Eco72I released ~2 kb GUS gene internal to the T-DNA region and hybridized to a 1,029 bp radiolabelled probe specific for the GUS gene from pCAMBIA 1301. Out of six, five lines were shown to contain the GUS transgene, thus confirming stable transgenics (Fig. 5c).
PCR analysis and Southern blot analyses of transgenic plants of Pennisetum glaucum. a PCR amplification of 1,029 bp GUS fragment gene in hygromycin resistant plants after selection (lane 1 1 kb marker, lane 2 positive control plasmid pCAMBIA1301, lanes 3–14 (named as T01–T012): Putative GUS positive transgenic plants of pearl millet, lane 15 untransformed control plant). b PCR amplification of 694 bp hpt II gene in 6 transformed plants (T02, T03, T04, T05, T08 and T010) (lane 1 1 kb marker, lane 2 positive control plasmid pCAMBIA1301, lane 3 untransformed control plant, lanes 4–9: hpt II gene positive transgenic plants). c Southern blot analysis of transgenic plants (T02, T03, T04, T05, T08 and T010). Genomic DNA was digested with NcoI and Eco72I and hybridized with alpha p32 dCTP labeled GUS probe. (lane 1 positive control plasmid pCAMBIA 1301, lane 2 untransformed control plant, lanes 3–8 transgenic plants analysed)
In conclusion, we have effectively accomplished multiple shoot regeneration from shoot apex explant containing shoot apical meristems in pearl millet, which served as an explant source for Agrobacterium-mediated transformation. Establishment of an easy, rapid, and widely applicable transformation system for pearl millet is very important for crop improvement and for study of gene function. Our results showed that optimization of optical density, inoculation duration, co-cultivation period, acetosyringone concentration and vacuum infiltration assisted inoculation improved the probability of T-DNA delivery. By using this procedure, multiple shoots resistant to hygromycin were obtained and transgenic plants have been regenerated. It was shown by PCR and Southern blot hybridization analysis that the GUS gene was integrated into the genome of the T0 generation transgenic plants. This is a pioneering report on Agrobacterium-mediated transformation of pearl millet inbred genotype 843B using shoot apices as the target tissue. Successful transformation requires a balance between the different factors affecting the transformation frequency, in order to achieve maximum number of T-DNA transfer events to occur in the target tissue, while maintaining the regenerability of recipient plant cells.
Abbreviations
- CaMV 35S:
-
Cauliflower mosaic virus promoter
- GUS:
-
β-Glucuronidase
- Hpt:
-
Hygromycin phosphotransferase
- BA:
-
Benzyladenine
- X-Gluc:
-
5-Bromo-4-chloro-3-indolyl- β-d-glucuronide
- Acs:
-
Acetosyringone
- PCR:
-
Polymerase chain reaction
- Min:
-
Minutes
References
Ahmad A, Zhong H, Wang W, Sticklen MB (2002) Shoot apical meristem: in vitro regeneration and morphogenesis in wheat (Triticum aestivum L.). In Vitro Cell Dev Biol plant 38:163–167
Al-Forkan M, Brain Power J, Anthony P, Lowe KC, Davey MR (2004) Agrobacterium-mediated transformation of Bangladeshi indica rice. Cell Mol Biol Lett 9:287–300
Amoah BK, Wu H, Sparks C, Jones HD (2001) Factors influencing Agrobacterium-mediated transient expression of uidA in wheat inflorescence tissue. J Exp Bot 52:1135–1142
Arockiasamy S, Ignacimuthu ES (2007) Regeneration of transgenic plants from two indica rice (Oryza sativa L.) cultivars using shoot apex explants. Plant Cell Rep 26:1745–1753
Barakat A, Carels N, Bernardi G (1997) The distribution of genes in the genome of Gramineae. Proc Natl Acad Sci USA 94:6857–6861
Bregitzer P, Tonks D (2003) Inheritance and expression of transgenes in barley. Crop Sci 43:4–12
Bregitzer P, Zhang S, Cho M-J, Lemaux PG (2002) Reduced somaclonal variation in barley is associated with culturing highly differentiated, meristematic tissues. Crop Sci 42:1303–1308
Ceasar SA, Ignacimuthu S (2009) Genetic engineering of millets: current status and future prospects. Biotechnol Lett 31:779–788
Cheng M, Hu T, Layton J, Liu CN, Fry JE (2003) Desiccation of plant tissues post Agrobacterium infection enhances T-DNA delivery and increases stable transformation efficiency in wheat. In Vitro Cell Dev Biol plant 39:595–604
Cheng M, Lowe BA, Spencer TM, Ye X, Armstrong CL (2004) Factors influencing Agrobacterium-mediated transformation of monocotyledonous species. In Vitro Cell Dev Biol plant 40:31–45
Cho MJ, Choi HW, Okamoto D, Zhang S, Lemaux PG (2003) Expression of green fluorescent protein and its inheritance in transgenic oat plants generated from shoot meristematic cultures. Plant Cell Rep 21:467–474
Chowdari KV, Davierwala AP, Gupta VS, Ranjekar PK, Govila OP (1998) Genotype identification and assessment of genetic relationships in pearl millet [Pennisetum glaucum (L.) R. Br.] using microsatellites and RAPDs. Theor Appl Genet 97:154–162
Devi P, Zhong H, Sticklen MB (2000) In vitro morphogenesis of pearl millet [Pennisetum glaucum. (L.) R. Br]: efficient production of multiple shoots and inflorescences from shoot apices. Plant Cell Rep 19:546–550
Dong J, Kharb P, Teng W, Hall TC (2001) Characterization of rice transformed via an Agrobacterium-mediated inflorescence approach. Mol Breed 7:187–194
Fang YD, Akula C, Altpeter F (2002) Agrobacterium-mediated barley (Hordeum vulgare L.) transformation using green fluorescent protein as a visual marker and sequence analysis of the T-DNA: barley genomic DNA junctions. J Plant Physiol 159:1131–1138
FAO (2004) Food and agriculture organization of the United Nations. Pennesitum americanum (L.) Leeke: species description. Last accessed October 2004. http://www.fao.org/ag/AGP/AGPC/doc/Gbase/DATA/Pf000297.htm
Gasparis S, Bregier C, Orczyk W, Nadolska-Orczyk A (2008) Agrobacterium-mediated transformation of oat (Avena sativa L.) cultivars via immature embryo and leaf explants. Plant Cell Rep 27:1721–1729
Girgi M, O’Kennedy MM, Morgenstern A, Mayer G, Lorz H, Oldach KH (2002) Transgenic and herbicide resistant pearl millet (Pennisetum glaucum L.) R. Br. via microprojectile bombardment of scutellar tissue. Mol Breed 10:243–252
Girgi M, Breese WA, Lorz H, Oldach KH (2006) Rust and downy mildew resistance in pearl millet (Pennisetum glaucum) mediated by heterologous expression of the afp gene from Aspergillus giganteus. Transgenic Res 15:313–324
Goldman JJ, Hanna WW, Fleming G, Ozias-Akins P (2003) Fertile transgenic pearl millet [Pennisetum glaucum (L.) R. Br.] plants recovered through microprojectile bombardment and phosphinothricin selection of apical meristem-, inflorescence-, and immature embryo-derived embryogenic tissues. Plant Cell Rep 21:999–1009
Gould JH, Magallanes-Cedeno M (1998) Adaptation of cotton shoot apex culture to Agrobacterium-mediated transformation. Plant Mol Biol Rep 16:283
Gu XF, Meng H, Qi G, Zhang JR (2008) Agrobacterium-mediated transformation of the winter jujube (Zizyphus jujuba Mill.). Plant Cell Tissue Organ Cult 94:23–32
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282
Hiei Y, Komari T, Kubo T (1997) Transformation of rice by Agrobacterium tumefaciens. Plant Mol Biol 35:205–218
Hirochika H (1993) Activation of tobacco retrotransposons during tissue culture. EMBO J 12:2521–2528
Hoagland M, Arnon DI (1950) The water culture method to grow plants without soil. Calif Agric Expt Station Cir 347. Berkeley California
Hoque ME, Mansfield JW, Bennett MH (2005) Agrobacterium-mediated transformation of indica rice genotypes: an assessment of factors affecting the transformation efficiency. Plant Cell Tissue Organ Cult 82:45–55
Howe A, Sato S, Dweikat I, Fromm M, Clemente T (2006) Rapid and reproducible Agrobacterium-mediated transformation of sorghum. Plant Cell Rep 25:784–791
Ishida Y, Saito H, Ohta S, Hiei Y, Komari T, Kumashiro T (1996) High efficiency transformation of maize (Zea mays L.) mediated by Agrobacterium tumefaciens. Nat Biotechnol 14:745–750
Ishida Y, Hiei Y, Komari T (2007) Agrobacterium-mediated transformation of maize. Nat Protoc 2(7):1614–1621
Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5:387–405
Jha P, Yadav CB, Anjaiah V, Bhat V (2009) In vitro plant regeneration through somatic embryogenesis and direct shoot organogenesis in Pennisetum glaucum (L.) R. Br. In Vitro Cell Dev Biol plant 45:145–154
Jones HD, Doherty A, Wu H (2005) Review of methodologies and a protocol for the Agrobacterium-mediated transformation of wheat. Plant Methods 1:5
Kobayashi T, Nakanishi H, Takahashi M, Kawasaki S, Nishizawa NK, Mori S (2001) In vivo evidence that Ids3 from Hordeum vulgare encodes a dioxygenase that converts 2-deoxymugineic acid to mugineic acid in transgenic rice. Planta 212:864–871
Kohli A, Gahakwa D, Vain P, Laurie DA, Christou P (1999) Transgene expression in rice engineered through particle bombardment: molecular factors controlling stable expression and transgene silencing. Planta 208:88–97
Komari T, Hiei Y, Ishida Y, Kumashiro T, Kubo T (1998) Advances in cereal gene transfer. Curr Opin Plant Biol 1:161–165
Kumar S, Agarwal K, Kothari SL (2001) In vitro induction and enlargement of apical domes and formation of multiple shoots in finger millet, Eleusine coracana (L.) Gaertn and crowfoot grass, Eleusine indica (L.) Gaertn. Curr Sci 81:1482–1485
Kumar J, Shukl SM, Bhat V, Gupta S, Gupta MG (2005a) In vitro plant regeneration and genetic transformation of Dichantium annulatum. DNA Cell Biol 24:670–679
Kumar KK, Maruthasalam S, Loganathan M, Sudhakar D, Balasubramanian P (2005b) An improved Agrobacterium-mediated transformation protocol for recalcitrant elite indica rice cultivars. Plant Mol Biol Rep 23:67–73
Kumria R, Waie B, Rajam MV (2001) Plant regeneration from transformed embryogenic callus of an elite indica rice via Agrobacterium. Plant Cell Tissue Organ Cult 67:63–71
Lambe P, Dinant M, Matagne RF (1995) Differential long-term expression and methylation of the hygromycin phosphotransferase (hph) and b-glucuronidase (GUS) genes in transgenic pearl millet (Pennisetum americanum) callus. Plant Sci 108:51–62
Lambe P, Dinant M, Deltour R (2000) Transgenic pearl millet (Pennisetum glaucum). In: Bajaj YPS (ed) Biotechnology in agriculture and forestry, transgenic crops I, vol 46. Springer, Berlin, pp 84–108
Latha AM, Rao KV, Reddy TP, Reddy VD (2006) Development of transgenic pearl millet (Pennisetum glaucum (L.) R. Br.) plants resistant to downy mildew. Plant Cell Rep 25:927–935
Lee SH, Lee DG, Woo HS, Lee KW, Kim DH, Kwak SS, Kim JS, Kim H, Ahsan N, Choi MS, Yang JK, Lee BH (2006) Production of transgenic orchardgrass via Agrobacterium-mediated transformation of seed-derived callus tissues. Plant Sci 171:408–414
Li ZN, Fang F, Liu GF, Bao MZ (2007) Stable Agrobacterium-mediated genetic transformation of London plane tree (Platanus acerifolia Willd.). Plant Cell Rep 26:641–650
Li M, Li H, Hu X, Pan X, Wu G (2010) An Agrobacterium tumefaciens-mediated transformation system using callus of Zoysia tenuifolia Willd. ex Trin. Plant Cell Tissue Organ Cult 102:321–327
Li M, Li H, Hu X, Pan X, Wu G (2011) Genetic transformation and overexpression of a rice Hd3a induces early flowering in Saussurea involucrata Kar. et Kir. ex Maxim. Plant Cell Tissue Organ Cult. doi:10.1007/s11240-011-9927-5
Liu X, Pijut PM (2010) Agrobacterium-mediated transformation of mature Prunus serotina (black cherry) and regeneration of transgenic shoots. Plant Cell Tissue Organ Cult 101:49–57
Ma X, Yu C, Tang S, Guo S, Zhang R, Wang Y, Zhu A, Zhu S, Xiong H (2010) Genetic transformation of the bast fiber plant ramie (Boehmeria nivea Gaud.) via Agrobacterium tumefaciens. Plant Cell Tissue Organ Cult 100:165–174
McCabe DE, Swain WF, Martinell BJ, Christou P (1988) Stable transformation of soybean (Glycine max) by particle acceleration. Nat Biotechnol 6:923–925
Mitić N, Nikolić R, Ninković S, Miljuš-Djukić J, Nešković M (2004) Agrobacterium-mediated transformation and plant regeneration of Triticum aestivum L. Biol Plant 48(2):179–184
Mori M, Nomura T, Ooka H, Ishizaka M, Yokota T, Sugimoto K, Okabe K, Kajiwara H, Satoh K, Yamamoto K, Hirochika H, Kikuchi S (2002) Isolation and characterization of a rice dwarf mutant with a defect in brassinosteroid biosynthesis. Plant Physiol 130:1152–1161
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Nagasaki H, Sakamoto T, Sato Y, Matsuoka M (2001) Functional analysis of the conserved domains of a rice KNOX homeodomain protein, OSH15. Plant Cell 13:2085–2098
O’Kennedy MM, Burger JT, Botha FC (2004) Pearl millet transformation system using the positive selectable marker gene phosphomannose isomerase. Plant Cell Rep 22:684–690
Park SH, Pinson SRM, Smith RH (1996) T-DNA integration into genomic DNA of rice following Agrobacterium inoculation of isolated shoot apices. Plant Mol Biol 32:1135–1148
Repellin A, Baga M, Jauhar PP, Chibbar RN (2001) Genetic enrichment of cereal crops via alien gene transfer: new challenges. Plant Cell Tissue Organ Cult 64:159–183
Sairam RV, Parani M, Franklin G, Lifeng Z, Smith B, MacDougall J, Wilber C, Sheikhi H, Kashikar N, Meeker K, Al-Abed D, Berry K, Vierling R, Goldman SL (2003) Shoot meristem: an ideal explant for Zea mays L. transformation. Genome 46:323–329
Sambrook J, Maniatis T, Fritsch E (1989) Molecular cloning: a laboratory manual, 2nd edn. Colspring Harbor Laboratory Press, Cold Spring Harbor, NY
Sarker RH, Biswas A (2002) In vitro plantlet regeneration and Agrobacterium-mediated genetic transformation of wheat (Triticum aestivum L.). Plant Tissue Cult 12(2):155–165
Shou H, Frame BR, Whitham SA, Wang K (2004) Assessment of transgenic maize events produced by particle bombardment or Agrobacterium-mediated transformation. Mol Breed 13:201–208
Shrawat AK, Becker D, Lo¨rz H (2007) Agrobacterium tumefaciens-mediated genetic transformation of barley (Hordeum vulgare L.). Plant Sci 172:281–290
Somleva MN, Tomaszewski Z, Conger BV (2002) Agrobacterium-mediated genetic transformation of Switchgrass. Crop Sci 42:2080–2087
Stachel SE, Messens E, Van Montagu M, Zambryski P (1985) Identification of the signal molecules produced by wounded plant cells that activate T-DNA transfer in Agrobacterium tumefaciens. Nature 318:624–629
Sticklen MB, Oraby HF (2005) Shoot apical meristem: a sustainable explant for genetic transformation of cereal crops. In Vitro Cell Dev Biol plant 41:187–200
Taylor MG, Vasil IK (1991) Histology of, and physical factors affecting, transient GUS expression in pearl millet (Pennisetum glaucum (L.) R. Br.) embryos following microprojectile bombardment. Plant Cell Rep 10:120–125
Taylor MG, Vasil V, Vasil IK (1993) Enhanced GUS gene expression in cereal/grass cell suspensions and immature embryos using the maize ubiquitin-based plasmid pAHC25. Plant Cell Rep 12:491–495
Tingay S, McElroy D, Kalla R, Fieg S, Wang M, Thornton S, Brettell R (1997) Agrobacterium tumefaciens-mediated barley transformation. Plant J 11:1369–1376
Vasil IK (2005) The story of transgenic cereals: the challenge, the debate, and the solution—a historical perspective. In Vitro Cell Dev Biol plant 41:577–583
Vergne P, Maene M, Gabant G, Chauvet A, Debner T, Bendahmane M (2010) Somatic embryogenesis and transformation of the diploid Rosa chinensis cv Old Blush. Plant Cell Tissue Organ Cult 100:73–81
Wu H, Sparks C, Amoah B, Jones HD (2003) Factors influencing successful Agrobacterium-mediated genetic transformation of wheat. Plant Cell Rep 21:659–668
Yang Y, Bao M, Liu G (2010) Factors affecting Agrobacterium-mediated genetic transformation of embryogenic callus of Parthenocissus tricuspidata Planch. Plant Cell Tissue Organ Cult 102:373–380
Yookongkaew N, Srivatanakul M, Narangajavana J (2007) Development of genotype-independent regeneration system for transformation of rice (Oryza sativa ssp. indica). J Plant Res 120:237–245
Zapata C, Srivatanakul M, Park S-H, Lee B-M, Salas MG, Smith RH (1999) Improvements in shoot apex regeneration of two fiber crops: cotton and kenaf. Plant Cell Tissue Organ Cult 56:185–191
Zhang S, Zhong H, Sticklen MB (1996) Production of multiple shoots from shoot apical meristems of oat (Avena sativa L.). J Plant Physiol 148:667–671
Zhang S, Williams-Carrier R, Jackson D, Lemaux PG (1998) Expression of CDC2Zm and KNOTTED1 during in vitro axillary shoot meristem proliferation and adventitious shoot meristem formation in maize (Zea mays L.) and barley (Hordeum vulgare L.). Planta 204:542–549
Zhang JR, Li XH, Quan RD, Zhang QW, Shang M, Yang AF, Li GS (2005) Method for creating transgenic receptor system of corn and application. China Patent 00110842.5, 29 October 2003
Zhang K, Wang J, Hu X, Yang A, Zhang J (2010) Agrobacterium-mediated transformation of shoot apices of Kentucky bluegrass (Poa pratensis L.) and production of transgenic plants carrying a betA gene. Plant Cell Tissue Organ Cult 102:135–143
Zhao ZY, Cai T, Tagliani L, Miller M, Wang N, Pang H, Rudert M, Schroeder S, Hondred D, Seltzer J, Pierce D (2000) Agrobacterium-mediated sorghum transformation. Plant Mol Biol 44:789–798
Zhao W, Zheng S, Ling H-Q (2011) An efficient regeneration system and Agrobacterium-mediated transformation of Chinese upland rice cultivar Handao 297. Plant Cell Tissue Organ Cult. doi:10.1007/s11240-011-9946-2
Zhong H, Srinivasan C, Sticklen M (1992) Morphogenesis of corn (Zea mays L.) in vitro II. Transdifferentiation of shoots, tassels, and ear primordia from corn shoots. Planta 187:483–489
Zhong H, Sun B, Warkentin D, Zhang S, Wu R, Wu T, Sticklen MB (1996) The competence of maize shoot meristems for integrative transformation and inherited expression of transgenes. Plant Physiol 110:1097–1107
Zhong H, Wang W, Sticklen M (1998) In vitro morphogenesis of Sorghum bicolor (L.) Moench: efficient plant regeneration from shoot apices. J Plant Physiol 153:719–726
Acknowledgments
Pooja Jha wishes to thank Council for Scientific and Industrial Research, India for financial assistance in the form of J.R.F and S.R.F. The funding by Board of Research in Nuclear Sciences (BRNS), Department of Atomic Energy (DAE), Government of India, for carrying out this research work is gratefully acknowledged. Authors wish to thank Professor Neera Bhalla Sarin, School of Life Sciences, Jawaharlal Nehru University, New Delhi for her help towards Southern analysis. Authors are grateful to Dr. S. Eapen and Dr. T. R. Ganapathi (Bhabha Atomic Research Center, Mumbai, India) for kindly going through the manuscript and for their critical suggestions. We gratefully acknowledge the research grant from Delhi University to cover part of this work. We wish to duly acknowledge Dr. K.N. Rai, (ICRISAT), Patancheru, A.P. India, for providing the seeds of pearl millet used in this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jha, P., Shashi, Rustagi, A. et al. Efficient Agrobacterium-mediated transformation of Pennisetum glaucum (L.) R. Br. using shoot apices as explant source. Plant Cell Tiss Organ Cult 107, 501–512 (2011). https://doi.org/10.1007/s11240-011-0001-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-011-0001-0