Abstract
This review summarizes progress on the genetic transformation of millets and discusses the future prospects for the development of improved varieties. Only a limited number of studies have been carried out on genetic improvement of millets despite their nutritional importance in supplying minerals, calories and protein. Most genetic transformation studies of millets have been restricted to pearl millet and bahiagrass and most studies have been limited to the assessment of reporter and marker gene expression. Biolistic-mediated gene delivery has been frequently used for the transformation of millets but Agrobacterium-mediated transformation is still lagging. Improved transformation of millets, allied to relevant gene targets which may offer, for example, improved nutritional quality, resistance to abiotic and biotic stresses, and resistance to fungal infection will play important roles in millet improvement.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant biotechnology is founded on the demonstrated totipotency of plant cells, combined with the delivery, stable integration, and expression of transgene in plant cells, the regeneration of transformed plants, and the Mendelian transmission of transgenes to the progeny (Vasil 2008). Transgenic technology has been utilized in the past decade for the improvement of many crop plants including major cereals such as maize, wheat and rice. Many of these transgenic cereals have already reached the field for large scale cultivation. But a genetic improvement program for millets has been initiated only in recent years and has been paid less attention despite their nutritional importance.
Millet crops [pearl millet, Pennisetum glaucm (L.) R.Br; finger millet, Eleusine coracana (L) Gaertn; kodo millet, Paspalum scorbiculatum Linn; bahiagrass, Paspalum notatum Flugge; foxtail millet, Setaria italica (L.) P. Beauv; little millet, Panicum miliare Lam; proso millet, Panicum miliaceum (L.); barnyard millet, Echinochola crusgalli (Linn.) P. Beauv; guinea grass, Panicum maximum Jacq; elephant grass, Pennisetum purpurium Schum] are grasses that belong to the Family Poaceae of the monocotolydon group. Millets are staple foods that supply a major proportion of calories and protein to large segments of populations in the semi-arid tropical regions of Africa and Asia (O’Kennedy et al. 2006). The semi-arid tropics are characterized by unpredictable weather, limited and erratic rainfall, nutrient-poor soils and suffer from a host of agricultural constraints (Sharma and Ortiz 2000). The projected food demand for 2025 will require the yield of cereals, including millets, to rise from 2.5 to 4.5 t ha−1 (Borlaug 2002). Production of millets, like finger millet, is also constrained by fungal diseases; so genetic engineering of millets is essential for conferring high level of resistance to improve the yield (Ceasar and Ignacimuthu 2008). Genetic engineering technology can also be harnessed for improving the nutritional quality of millets and conferring resistance to abiotic stresses (drought and saline).
Among the available reports of millet transformation, the biolistic method has been mostly used for gene delivery. Agrobacterium-mediated transformation of millets lags behind although it is a versatile method of gene delivery for many cereals which also belong to the Poaceae. In contrast, Agrobacterium-mediated transformation of other cereals is effective with rice (Ignacimuthu et al. 2000; Bajaj and Mohanty 2005; Shrawat and Lörz 2006), wheat (Jones et al. 2005), barley and sorghum (Shrawat and Lörz 2006; O’Kennedy et al. 2006). We have also reported the Agrobacterium-mediated transformation system for Indian rice cultivars (Ignacimuthu and Arockiasamy 2006; Arockiasamy and Ignacimuthu 2007). Efforts made for the Agrobacterium-mediated transformation of cereals need to be extended to millet crops in the near future for their genetic improvement to meet the growing demand for food.
Millets are capable of surviving in some of the most inhospitable ecosystems of the world providing food and fodder to millions where other quality cereals fail to grow. Hence with an ultimate aim of supplementing conventional breeding efforts the genetic transformation protocols for millets need to be developed so that important quality traits may be incorporated across the barriers of incompatibility (Gupta et al. 2001). Two reviews published on millets describe the widespread role of biotechnology for the improvement of millets in general (Kothari et al. 2005) and pearl millet in particular (O’Kennedy et al. 2006). The scope of the current review is narrow and specifically describes the recent advances in genetic engineering and transgenic aspects of millets; we review the plasmid, promoter, marker and reporter genes and different methods of gene delivery used for the transformation of millets; we also discuss the future prospects for the Agrobacterium-mediated transformation of millets.
In vitro culture of millets
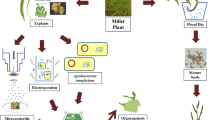
Establishment of an efficient in vitro regeneration system is a vital prerequisite for the successful transformation and recovery of transgenic millet crops. Many reports are available for the in vitro culture of millets and different methods of in vitro culture have been reported in millets (reviewed by Kothari et al. 2005). Development of in vitro regeneration protocol through somatic embryogenesis seems to be more helpful for the efficient transformation and recovery of transgenic plant (Veluthambi et al. 2003). Somatic embryogenesis and plant regeneration protocols have been reported for pearl millet, finger millet, kodo millet and foxtail millet (Kothari et al. 2005). Recently we have also developed this protocol for finger millet (Ceasar and Ignacimuthu 2008). Similar protocols need to be developed in future for the rest of the millets to produce transgenic millets more efficiently, particularly for the Agrobacterium-mediated approaches.
Genetic engineering of millets
Pearl millet
Pearl millet (Pennisetum glaucm) has been given the highest priority among millets in transformation studies; in all the available reports, the pearl millet was transformed by biolistic method of gene delivery (Table 1). The first pearl millet transformation was made by Taylor et al. (1991) through microprojectile bombardment (biolistic); immature embryos were used as the target explants. Plasmid pMON 8678 was used for transformation; it contained β-glucuronidase (GUS or uidA) gene under the control of maize alcohol dehydrogenase gene (adh1) promoter. Transformation was confirmed by GUS histochemical assay. Later the pearl millet was transformed in the same laboratory using plasmids pBARGUS and pAHC25 (Taylor et al. 1993); they observed that expression of the uidA gene in plasmid pAHC25 was superior to pBARGUS. Following this, Lambe et al. (1995) transformed pearl millet by the biolistic method with two plasmids (p35SGUS and pROB5). The p35SGUS contained the GUS gene and pROB5 contained the hygromycin phosphotransferase gene (hpt) conferring resistance to hygromycin; expression of both genes was under the control of separate CaMV35S promoters. 1–2 μm tungsten particles coated with plasmids were used for bombardment and a distance of 8 cm was maintained between the target material (embryonic calli or embryonic cell suspension) and the stopping plate of macroprojectile. They also confirmed the integration of transgene by Southern blot analysis (Lambe et al. 1995).
Another approach for pearl millet transformation was made by Girgi et al. (2002) who delivered genes to scutellar tissue by two alternative transformation protocols, particle gun 1,000/He (PDS) or particle inflow gun (PIG). Plasmid pAHC25 was used for PIG transformation; it harbored the GUS reporter gene and bar (phospinothricin acetyltransferase) marker gene; both the GUS and bar genes were put under the control of uq1 (ubiquitin from maize) promoter. Plasmid p35SAcS and pubi.gus were used in co-transformation experiments with PDS; p35SAcS contained the bar selection marker gene under the control of CaMV35S promoter and pubi.gus had the GUS gene (uidA) driven by the uq1 promoter. However, in this approach Girgi et al. (2002) obtained very low (0.18%) frequency of pearl millet transformation in spite of employing two different gene delivery methods (PIG and PDS).
Genotype-independent pearl millet transformation system was developed by Devi and Sticklen (2002) using shoot apical meristem as target tissue for gene delivery. Plasmid pAct1-F containing uidA gene driven by rice Act1 (rice actin) promoter was used for transformation; they used tungsten particles of two different sizes (0.9 and 1.2 μm). The important parameters influencing the transformation like size of the tungsten particles, density of the particles, pressure of the helium gas, distance from the target and osmoticum of the culture medium were optimized (Devi and Sticklen 2002).
The transformation frequency of pearl millet was greatly increased by Goldman et al. (2003) using three different explants (embryogenic tissue, inflorescences and apical meristems); the transformation frequency ranged from 5 to 85%. They used plasmids pAHC25 and p524EGFP; plasmid pAHC25 contained the selectable bar gene and the reporter gene uidA, both under the control of separate uq1 promoter. Plasmid p524EGFP.1 contained the double cauliflower mosaic virus 35S promoter sequence followed by the alfalfa mosaic virus enhancer sequence controlling expression of an enhanced green fluorescent protein-encoding (gfp) gene. They coated the DNA on 0.6 or 0.75 μm gold particles; the distance of 6 cm was maintained between the target and stopping screen.
The transformation protocol for pearl millet was also optimized by O’Kennedy et al. (2004) using a phosphomannose isomerase (manA) transgene driven under the control of the uq1 promoter placed in the plasmid pNOV3604ubi. Transgenic manA expressing cells acquired the ability to convert mannose 6-phosphate to fructose 6-phosphate while the non-transgenic cells accumulated mannose 6-phosphate and could not survive (positive selection). The manA gene has been shown to be superior to antibiotic or herbicide (pat or bar) resistant selectable marker genes for transformation of many plants including maize and wheat (Reed et al. 2001). This gene could be utilized in future for the transformation of other millets.
The first transgenic pearl millet expressing functionally active foreign gene conferring resistance to fungal disease (downy mildew) has only recently been produced by Girgi et al. (2006). They have used the antifungal protein (afp) gene isolated from the ascomycete, Aspergillus giganteus; immature zygotic embryos have been used as target for biolistic transformation. Transformation of pearl millet was confirmed by molecular analysis; the disease resistance was also increased up to 90% when compared to non transformed control plants.
Latha et al. (2006) have also developed a transgenic pearl millet conferring resistance to downy mildew disease by inserting a chemically synthesized prawn antifungal protein encoding gene (pin); embryogenic calli were used as target explants for bombardment. The transformed plants were selected by incorporating phosphinothricin (PPT) in the selection medium and were subsequently confirmed by PCR, Southern blot and northern blot analysis. They have also confirmed the Mendelian inheritance pattern of par and pin genes in T1 progenies. The transformed plants showed high level of resistance to fungal pathogen when compared to untransformed control after challenging with fungal spores.
Even though the transformation system for pearl millet was developed earlier (Taylor et al. 1991) only two fertile transgenic pearl millets expressing functional foreign genes have been reported so far (Girgi et al. 2006; Latha et al. 2006). Since downy mildew is a major constraint in pearl millet production (Rachie and Majumdar 1980) genes like chitinase and glucanase could be transferred to pearl millet in the near future in order to offer the prospect of conferring high levels of resistance against fungal pathogens and thereby improving pearl millet production. These genes (chitinase and glucanase) seem to be promising in the fight against fungal pathogens (Kumar et al. 2003; Kalpana et al. 2006; Maruthasalam et al. 2007). In rice these genes were introduced by Agrobacterium-mediated transformation but there is no Agrobacterium-mediated transformation yet reported in pearl millet.
Bahiagrass
Bahiagrass (Paspalum notatum) is a major subtropical forage species that is extensively grown in the southeastern United States and from Central Mexico to Argentina. The transformation system for this important forage grass was first established by Smith et al. (2002) using the biolistic method. Transformed plants were selected on phosphinothricin and later confirmed by PCR and Southern blot analysis. Gondo et al. (2003) conducted preliminary experiments for particle inflow gun-mediated transformation of diploid bahiagrass. Later the same laboratory transformed diploid bahiagrass with efficient plant recovery system employing modified callus induction and plant regeneration protocols; 22 transgenic plants were recovered out of 360 explants bombarded (Gondo et al. 2005).
Altpeter and James (2005) have developed a transformation protocol for bahiagrass using the nptll (neomycin phosphotransferase) marker gene expressed under the control of uql promoter using biolistic method. Later the same laboratory also developed a transgenic bahiagrass with improved turf quality (Hangning et al. 2007). Over-expression of Arabidopsis ATHB16 transcription factor in bahiagrass produced significantly more vegetative and fewer reproductive tillers (Hangning et al. 2007). ATH1 is an Arabidopsis homeobox transcription factor-encoding gene; in ryegrass the expression of this gene also produced late-heading or non-flowering plants with more vegetative parts (Van der Valk et al. 2004). They utilized the same protocol developed by Altpeter and James (2005) for the delivery of the ATHB16 transcription factor-encoding gene. Transformed plants were selected on 50 mg paromomycin sulfate l−1 and later confirmed by molecular (PCR and Southern blot) analysis.
Recently James et al. (2008) developed a transgenic bahiagrass resistant to abiotic stress by expressing the stress-inducible gene HsDREB1A; transformed plants survived under severe salt and repeated cycles of dehydration stresses. HsDREB1A encodes a transcription factor; it was isolated from barley and subcloned under the control of the stress-inducible barley HVAls promoter. Tolerance of transgenic plants to drought stress and salt stress was confirmed by exposing them to severe drought and salt stresses (James et al. 2008). This study can be of great use in the development of abiotic (drought and salt) stress resistant millets in the future. Since millets are the native of semi-arid tropics where unpredicted drought is most prevalent, transferring the genes like HsDREB1A to important millets like pearl millet and finger millet will help to improve the productivity of these millets and offer food security in this region.
Finger millet
Finger millet (Eleusine coracana) is the primary food source for millions of people in tropical dry land regions. Finger millet also has nutritional qualities superior to that of rice and is on par with that of wheat (Latha et al. 2005). One of the major biotic constraints in the finger millet crop is the high seed yield loss (>50%) due to leaf blast disease caused by the pathogenic fungus, Pyricularia grisea; this demands genetic improvement of finger millet by transferring fungal resistance genes (Ceasar and Ignacimuthu 2008). Only a little work has been done for the improvement of this millet in spite of its nutritional importance.
Preliminary work on finger millet transformation was performed by Gupta et al. (2001) who compared the efficiency of five gene promoters (CaMV35, Actl, uql, rice ribulose 1, 5-biphosphate carboxylase small subunit (RbcS) gene promoter and Flaveria trinervia (Ft) gene promoter) for the expression of the GUS reporter gene. They confirmed that promoters Actl and uql gave best response for the GUS gene expression; RbcS and CaMV35S promoters produced medium response and Ft promoter was found to be ineffective for GUS expression (Gupta et al. 2001). This study may be more helpful to choose the type of promoter for the high level expression of valuable genes in finger millet in future.
A transgenic finger millet resistant to fungal blast disease was developed by Latha et al. (2005). Antifungal protein (PIN) of prawn was chemically synthesized and cloned in the plasmid pPin35S and the bar reporter gene was cloned in the plasmid pBar35S; both the genes were put under the control of CaMV35S promoter. The transformed plants were selected on PPT-supplemented medium. Stable integration and expression of the PIN gene were confirmed by Southern and northern blot analyses. This is the first and only report available for transgenic finger millet exhibiting high-level of resistance to leaf blast disease. The genetic improvement of finger millet to improve the grain yield by conferring resistance to fungal disease and other stresses is a priority.
Foxtail millet
Foxtail millet (Setaria italica) is an important food crop grown in India, China and Japan on salinity-prone lands as well as during adverse conditions such as severe drought. It is also planted in Australia, North Africa and South America for hay and silage (Sreenivasulu et al. 2004). Only two reports are available so far for the transformation of foxtail millet; in both cases Agrobacterium-mediated transformation was used. Agrobacterium-mediated transformation system for foxtail millet was first developed by Liu et al. (2005). The transformed explants were selected on 50 mg kanamycin l−1 and assayed for GUS gene expression and later confirmed by Southern blot analysis; they obtained 6.6% transformation frequency through this approach. Afterwards the same protocol was utilized by Fang et al. (2007) for the transformation of foxtail millet with pollen specific gene Si401 expressed under the control of pollen specific promoter of Zea mays (Zm13) cloned into the plasmid pBIZm13Si401; the vector also contained nptII gene for kanamycin selection. They reported that Si401 expression in foxtail millet resulted in multiple abnormalities during the late stages of anther development including premature degeneration of the tapetum, pre-deposition of Wbrous bands in endothecium cells, and aborted pollen grains. These interesting findings may be harnessed in the near future for the development of agronomically important male sterile cereal crops.
Since these two studies on foxtail millet have confirmed the potential of Agrobacterium-mediated transformation of millet, efforts need to be taken to extend this protocol to many other important millet crops for the transfer of essential genes and production of improved traits using Agrobacterium-mediated approaches.
Other millets
A limited number of studies have been reported on other millets including barnyard millet, elephant grass and guinea grass for transformation studies; preliminary experiments alone were conducted but the transgenic crops expressing functionally active foreign genes are yet to be produced in these millets. Elephant grass and guinea grass were transformed by Hauptmann et al. (1987, 1988). Protoplasts were used as the target explant and DNA was delivered by electroporation. They used three plasmids, viz. pMON145, pMON273 and pMON41, containing marker genes chlorophenicol acetyl transferase (cat), nptII and hpt, respectively; all three genes were put under the control of separate CaMV35S promoters. Transformed protoplasts were selected on the antibiotics and later confirmed by Southern hybridization; they obtained very low (10−5 to 2 × 10−6) frequency of transformation. Taylor et al. (1993) also conducted transformation studies using biolistic method in these two grasses (elephant grass and guinea grass) along with pearl millet; details of this transformation was discussed in the pearl millet section. Only one report is available for the transformation of barnyard millet; this millet was also used along with finger millet for assessment of the efficiency of five gene promoters as discussed in finger millet transformation part of this review (Gupta et al. 2001). In barnyard millet, only the uq1 promoter was effective for the expression of GUS gene; other four promoters (CaMV35S, Act1, RbcS and Ft) were ineffective based on the histochemical analysis.
Future prospects for Agrobacterium-mediated transformation of millets
The biolistic method of gene delivery offered a genotype-independent transformation system for many cereals and made a great breakthrough in the transformation of monocots which were earlier thought to be inaccessible for Agrobacterium-mediated system. But a major drawback in the biolistic system is the considerable variation seen in stability, integration and expression of the introduced transgene (Kohli et al. 1999). The Agrobacterium-mediated system on the other hand offers the precise integration of a small number of gene copies into the plant genome and shows a greater degree of stability for the transgene (Dai et al. 2001). One of the major barriers in the use of Agrobacterium to transform cereals is the absence of wound responses and the associated activation of virulence genes. These problems were overcome with the use of actively dividing embryogenic cells such as immature embryos and calli induced from scutella which were co-cultivated with Agrobacterium in the presence of acetosyringone, a potent inducer of virulence genes (Vasil 2005). After a detailed study on the molecular events occurring during T-DNA transfer many laboratories have transformed cereals using Agrobacterium by manipulating transformation conditions and successfully obtained transgenic cereals. The natural ability of Agrobacterium to deliver a discrete segment of DNA into the recipient genome has been exploited in Agrobacterium-mediated transformation of cereals (Cheng et al. 2004; Shrawat and Lörz 2006). The possibility of utilizing Agrobacterium-mediated system for millet transformation in the future is presented in the following section of this review.
Several factors are shown to be involved in the Agrobacterium-mediated transformation of cereals; these include the screening of most responsive genotype and explant, Agrobacterium strain, binary vector, selectable marker gene and promoter, inoculation and co-culture conditions and tissue culture medium (Shrawat and Lörz 2006). Screening of genotypes for tissue culture response is an important aspect for subsequent transformation by Agrobacterium; genotypes responding well in tissue culture are readily amenable for transformation in wheat, maize, barley and rice (Cheng et al. 2004; Shrawat and Lörz 2006). Genotype response in tissue culture studies were reported in a few millets (Kothari et al. 2005); based on these reports, elite genotypes can be selected which may be readily amenable for Agrobacterium-mediated transformation. These model genotypes need to be isolated from the rest of the millets which give good response on tissue culture and can be utilized for the subsequent transgenic plant production. In wheat, maize, barley and sugarcane model genotypes have worked well for Agrobacterium-mediated transformation. These model genotypes include maize cv. A188 or its hybrids, wheat cvs. Bobwhite and Fielder, barley cvs. Golden Promise and Igri and sugarcane cv. Ja 60-5. (Cheng et al. 2004).
Somatic embryo is the more suitable explant for co-cultivation with Agrobacterium and efficient transformation in cereals, like rice, due to its active cell division (Kumar et al. 2003). This approach may be more helpful for the efficient Agrobacterium-mediated transformation of millets. Shoot apical meristems have also been successfully employed as starting material to recover stably transformed maize, wheat, rice, oat, barley, sorghum and millet (Sticklena and Orabya 2005).
The type of Agrobacterium strain also played a key role for the efficient cereal transformation. Strains LBA4404 and EHA101 and their derivatives (EHA105 from EHA101, AGL0 and AGL1 from EHA101) were most frequently used for transformation of cereals (Cheng et al. 2004). In millets the strain LBA4404 alone was used to deliver the gene into millet and only one millet (foxtail millet) has been transformed so far (Liu et al. 2005; Fang et al. 2007); other millets are still not tested for the transformation efficiency of any Agrobacterium strains. So screening of different Agrobacterium strains with various superbinary vectors can be of great use in the successful transformation of millets.
Recently Ding et al. (2007) have optimized the Agrobacterium-mediated transformation protocol for wheat using mature embryos as explants; mature embryos pre-cultured for 14 days before co-cultivation gave better response to transformation. This could also be attempted in millets in the future; since millet seeds can be stored for long periods without any insect damage, transformation experiments can be performed at any period of the year using mature embryos. Desiccation of explants prior to or post Agrobacterium infection has proved to be an important factor in increasing the efficiency of transformation in cereals (reviewed by Cheng et al. 2004). Though the molecular events involved in desiccation are still not clear it is believed that desiccation treatment had suppressed the over-growth of Agrobacterium and subsequently facilitated better cell recovery after co-culture when compared to the control without desiccation (Shrawat and Lörz 2006).
Other factors that influence the Agrobacterium-mediated transformation of cereals are addition of a vir gene-inducing compound (acetosyringone) in the co-culture medium, osmotic treatment, antinecrotic treatment, Agrobacterium density, addition of surfactants (e.g. silwet L77; pluronic acid F68) and temperature maintenance during co-culture (Cheng et al. 2004). Testing of these factors in millets could be helpful in Agrobacterium-mediated transformation. Agrobacterium infection using techniques such as desiccation, antinecrotic mixture for pre-induction as well as plant growth regulation treatment are emerging. Understanding the molecular basis of several factors such as desiccation and antinecrotic treatments affecting both T-DNA delivery and stable transformation may facilitate application of these treatments to other species including millets in the near future (Opabode 2006).
In conclusion, the millet transformation is dominated by physical methods of gene delivery (biolistic; electroporation). Many reports of millet transformation are also restricted to the analysis of marker or reporter gene expression. Production of transgenic millets expressing functional foreign genes has been initiated only in recent years. Efforts made in the Agrobacterium-mediated transformation of other cereals need to be extended to millets in the near future to produce transgenic millets expressing agronomically important foreign genes. This will greatly help to improve millet production by conferring resistance to biotic and abiotic stresses.
References
Altpeter F, James VA (2005) Genetic transformation of turftype bahiagrass (Paspalum notatum Flugge) by biolistic gene transfer. Int Turfgrass Soc Res J 10:1–5
Arockiasamy S, Ignacimuthu S (2007) Regeneration of transgenic plants from two indica rice (Oryza sativa L.) cultivars using shoot apex explants. Plant Cell Rep 26:1745–1753
Bajaj S, Mohanty A (2005) Recent advances in rice biotechnology-towards genetically superior transgenic rice. Plant Biotechnol J 3:275–307
Borlaug NE (2002) Feeding a world of 10 billion people: the miracle ahead. In Vitro Cell Dev Biol Plant 8:221–228
Ceasar SA, Ignacimuthu S (2008) Efficient somatic embryogenesis and plant regeneration from shoot apex explants of different Indian genotypes of finger millet (Eleusine coracana (L.) Gaertn.). In Vitro Cell Dev Biol Plant 44:427–435
Cheng M, Lowe BA, Spencer TM, Ye X, Armstrong CL (2004) Factors influencing Agrobacterium-mediated transformation of monocotyledonous species. In Vitro Cell Dev Biol Plant 40:31–45
Dai S, Zheng P, Marmey P, Zhang S, Tian W, Chen S, Beachy RN, Fauquet C (2001) Comparative analysis of transgenic rice plants obtained by Agrobacterium-mediated transformation and particle bombardment. Mol Breed 7:25–33
Devi P, Sticklen M (2002) Culturing shoot-tip clumps of pearl millet [Pennisetum glaucum (L.) R. Br.] and optimal microprojectile bombardment parameters for transient expression. Euphytica 125:45–50
Ding L, Li S, Gao J, Wang Y, Yang G, He G (2007) Optimization of Agrobacterium-mediated transformation conditions in mature embryos of elite wheat. Mol Biol Rep. doi:10.1007/s11033-007-9148-5
Fang FQ, Qian Z, Guang MA, Jing JY (2007) Co-suppression of Si401, a maize pollen speciWc Zm401 homologous gene, results in aberrant anther development in foxtail millet. Euphytica 163:103–111
Girgi M, O’Kennedy MM, Morgenstern A, Smith G, Lorz H, Oldach KH (2002) Transgenic and herbicide resistant pearl millet (Pennisetum glaucum L.) R.Br. via microprojectile bombardment of scutellar tissue. Mol Breed 10:243–252
Girgi M, Breese WA, Lorz H, Oldach KH (2006) Rust and downy mildew resistance in pearl millet (Pennisetum glaucum) mediated by heterologous expression of the afp gene from Aspergillus giganteus. Transgenic Res 15:313–324
Goldman JJ, Hanna WW, Fleming G, Ozias-Akins P (2003) Fertile transgenic pearl millet [Pennisetum glaucum (L.) R. Br.] plants recovered through microprojectile bombardment and phosphinothricin selection of apical meristem-, inflorescence-, and immature embryo-derived embryogenic tissues. Plant Cell Rep 21:999–1009
Gondo T, Ishii Y, Akashi R, Kawamura O (2003) Efficient induction of embryogenic callus from mature seeds of bahiagrass (Paspalum notatum Flügge) and conditions for genetic transformation by particle bombardment. Grassl Sci 49:33–37
Gondo T, Shin-ichi T, Ryo A, Osamu K, Franz H (2005) Green, herbicide-resistant plants by particle inflow gun-mediated gene transfer to diploid bahiagrass (Paspalum notatum). J Plant Physiol 16:1367–1375
Grando MF, Franklin CI, Shatters JRG (2002) Optimizing embryogenic callus production and plant regeneration from ‘Tifton 9’ bahiagrass seed explants for genetic manipulation. Plant Cell Tissue Organ Cult 71:213–222
Gupta P, Raghuvanshi S, Tyagi AK (2001) Assessment of the efficiency of various gene promoters via biolistics in leaf and regenerating seed callus of millets, Eleusine coracana and Echinochloa crusgalli. Plant Biotechnol 18:275–282
Hangning Z, Paula L, Fredy A (2007) Improved turf quality of transgenic bahiagrass (Paspalum notatum Flugge) constitutively expressing the ATHB16 gene, a repressor of cell expansion. Mol Breed 20:415–423
Hauptmann RM, Ozias-Akins P, Vasil V, Tabaeizadeh Z, Rogers SG, Horsch RB, Vasil I, Fraley RT (1987) Transient expression of eletroporated DNA in monocotyledonous and dicotyledonous species. Plant Cell Rep 6:265–270
Hauptmann RM, Vasil V, Ozias-Akins P, Tabaezadh Z, Rogers SG, Fraley RT, Horsch RB, Vasil IK (1988) Evaluation of selectable markers for obtaining stable transformants in the Gramineae. Plant Physiol 86:602–606
Ignacimuthu S, Arockiasamy S (2006) Agrobacterium-mediated transformation of an elite indica rice for insect resistance. Curr Sci 90:829–835
Ignacimuthu S, Arockiasamy S, Terada R (2000) Genetic transformation of rice: current status and future prospects. Curr Sci 79:186–195
James VA, Neibaur JI, Altpeter F (2008) Stress inducible expression of the DREB1A transcription factor from xeric, Hordeum spontaneum L. in turf and forage grass (Paspalum notatum Flugge) enhances abiotic stress tolerance. Transgenic Res 17:93–104
Jones HD, Doherty A, Wu H (2005) Review of methodologies and a protocol for the Agrobacterium-mediated transformation of wheat. Plant Methods 1:5–13
Kalpana K, Maruthasalam S, Rajesh T, Poovannan K, Kumar KK, Kokiladevi E, Raja JAJ, Sudhakar D, Velazhahan R, Samiyappan R, Balasubramanian P (2006) Engineering sheath blight resistance in elite indica rice cultivars using genes encoding defense proteins. Plant Sci 170:203–215
Kohli A, Gahakwa D, Vain P, Laurie DA, Christou P (1999) Transgene expression in rice engineered through particle bombardment: molecular factors controlling stable expression and transgene silencing. Planta 208:88–97
Kothari SL, Kumar S, Vishnoi RK, Kothari SL, Watanabe KN (2005) Applications of biotechnology for improvement of millet crops: review of progress and future prospects. Plant Biotechnol 22:81–88
Kumar KK, Poovannan K, Nandakumar R, Thamilarasi K, Geetha C, Jayashree N, Kokiladevi E, Raja JAJ, Samiyappan R, Sudhakar D, Balasubramanian P (2003) A high throughput functional expression assay system for a defence gene conferring transgenic resistance on rice against the sheath blight pathogen, Rhizoctonia solani. Plant Sci 165:969–976
Kumar KK, Maruthasalam S, Loganathan MD, Sudhakar D, Balasubramanian P (2005) An improved Agrobacterium-mediated transformation protocol for recalcitrant elite indica rice cultivars. Plant Mol Biol Rep 23:67–73
Lambe P, Dinant M, Matagne RF (1995) Differential long-term expression and methylation of the hygromycin phosphotransferase (hph) and b-glucuronidase (GUS) genes in transgenic pearl millet (Pennisetum americanum) callus. Plant Sci 108:51–62
Lambe P, Dinant M, Deltour R (2000) Transgenic pearl millet (Pennisetum glaucum). In: Bajaj YPS (ed) Biotechnology in agriculture and forestry, transgenic crops I, vol 46. Springer, Berlin, pp 84–108
Latha MA, Venkateswara Rao K, Dashavantha Reddy V (2005) Production of transgenic plants resistant to leaf blast disease in finger millet (Eleusine coracana (L.) Gaertn.). Plant Sci 169:657–667
Latha MA, Rao KV, Reddy TP, Reddy VD (2006) Development of transgenic pearl millet (Pennisetum glaucum (L.) R. Br.) plants resistant to downy mildew. Plant Cell Rep 25:927–935
Liu YH, Yu JJ, Zhao Q, Ao GM (2005) Genetic transformation of millet (Setaria italica) by Agrobacterium-mediated. Agric Biotechnol J 13:32–37
Maruthasalam S, Kalpana K, Kumar KK, Loganathan M, Poovannan K, Raja JAT, Kokiladevi E, Samiyappan R, Sudhakar D, Balasubramanian P (2007) Pyramiding transgenic resistance in elite indica rice cultivars against the sheath blight and bacterial blight. Plant Cell Rep 26:791–804
McCormac AC, Wu H, Bao M, Wang Y, Xu R, Elliott MCD, Chen DF (1998) The use of visual marker genes as cellspecific reporters of Agrobacterium-mediated T-DNA delivery to wheat (Triticum aestivum L.) and barley (Hordeum vulgare L.). Euphytica 99:17–25
O’Kennedy MM, Burger JT, Botha FC (2004) Pearl millet transformation system using the positive selectable marker gene phosphomannose isomerase. Plant Cell Rep 22:684–690
O’Kennedy MM, Grootboom A, Shewry PR (2006) Harnessing sorghum and millet biotechnology for food and health. J Cereal Sci 44:224–235
Opabode JT (2006) Agrobacterium-mediated transformation of plants: emerging factors that influence efficiency. Biotechnol Mol Biol Rev 1:12–20
Rachie KO, Majmudar JV (1980) Pearl millet. Pennsylvania State University Press, University Park, p 307
Reed J, Privalle L, Powell ML, Meghji M, Dawson J, Dunder E, Suttie J, Wenck A, Launis K, Kramer C, Chang YF, Hansen G, Wright M (2001) Phosphomannose isomerase: an efficient selectable marker for plant transformation. In Vitro Cell Dev Biol Plant 37:127–132
Sharma KK, Ortiz R (2000) Program for the application of genetic transformation for crop improvement in the semi-arid tropics. In Vitro Cell Dev Biol Plant 36:83–92
Shrawat AK, Lörz H (2006) Agrobacterium-mediated transformation of cereals: a promising approach crossing barriers. Plant Biotechnol J 4:575–603
Smith RL, Grando MF, Li YY, Seib JC, Shatters RG (2002) Transformation of bahiagrass (Paspalum notatum Flugge). Plant Cell Rep 20:1017–1021
Sreenivasulu N, Miranda M, Prakash HS, Wobus U, Weschke W (2004) Transcriptome changes in foxtail millet genotypes at high salinity: identification and characterization of a PHGPX gene specifically upregulated by NaCl in a salt-tolerant line. J Plant Physiol 161:467–477
Sticklena MB, Orabya HF (2005) Shoot apical meristem: a sustainable explant for genetic transformation of cereal crops. In vitro Cell Dev Biol Plant 41:187–200
Taylor MG, Vasil V, Vasil IK (1991) Histology of, and physical factors affecting, transient GUS expression in pearl millet (Pennisetum glaucum (L.) R. Br.) embryos following microprojectile bombardment. Plant Cell Rep 10:120–125
Taylor MG, Vasil V, Vasil IK (1993) Enhanced GUS gene expression in cereal/grass cell suspensions and immature embryos using the maize ubiquitin-based plasmid pAHC25. Plant Cell Rep 12:491–495
Van der Valk P, Proveniers MCG, Pertijs JH, Lamers J, Van Dun CMP, Smeekens JCM (2004) Late heading of perennial ryegrass caused by introducing an Arabidopsis homeobox gene. Plant Breed 123:531–535
Vasil IK (2005) The story of transgenic cereals: the challenge, the debate, and the solution. In Vitro Cell Dev Biol Plant 41:577–583
Vasil IK (2008) A short history of plant biotechnology. Phytochem Rev 7:387–394
Veluthambi K, Gupta K, Sharma A (2003) The current status of plant transformation technologies. Curr Sci 84:368–380
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ceasar, S.A., Ignacimuthu, S. Genetic engineering of millets: current status and future prospects. Biotechnol Lett 31, 779–788 (2009). https://doi.org/10.1007/s10529-009-9933-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-009-9933-4