Abstract
DNA barcoding (molecular characterisation) is a useful tool for describing the taxonomy and systematics of organisms. Over 250 species of avian haemosporidian parasites have been described using morphological characters, yet molecular techniques based on polymerase chain reaction (PCR) suggest this diversity is underestimated. Moreover, molecular techniques are particularly useful for the detection of chronic infections and tissue stages of these parasites. Species delimitation is problematic among haemosporidians, and many questions about the mechanisms and patterns of speciation, host specificity and pathogenicity are still unresolved. Accumulation of additional genetic and morphological information is needed to approach these questions. Here, we combine microscopic examination with PCR-based methods to develop molecular characterisation of Haemoproteus (Parahaemoproteus) manwelli Bennett, 1978 and Haemoproteus (Parahaemoproteus) gavrilovi Valkiūnas & Iezhova, 1990, both of which parasitise the bee-eater Merops apiaster L. We also describe a new species, Haemoproteus (Parahaemoproteus) palloris n. sp., from the blood of the willow warbler Phylloscopus trochilus (L.). We performed phylogenetic analyses with a set of mitochondrial cytochrome b (cyt b) gene lineages, which have been linked to parasite morphospecies and are available in the MalAvi database. Our findings show that morphological characters, which have been traditionally used in the description of haemosporidians, exhibit phylogenetic congruence. This study contributes to a better understanding of avian haemosporidian diversity and provides new molecular markers (cyt b and apicoplast gene sequences) for the diagnostics of inadequately investigated haemosporidian infections.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Recently, molecular characterisation or DNA barcoding has become a benchmark in parasite species identification and epidemiology research for many vector-transmitted diseases (Ondrejicka et al., 2014), particularly haemosporidians (Haemosporida, Apicomplexa) (Clark et al., 2014). These protozoan parasites, which infect humans and other animal hosts worldwide, are transmitted by blood-sucking dipteran insects. Haemosporidian parasites include agents of malaria (Plasmodium spp.), which are still responsible for approximately 600,000 human deaths every year (WHO, 2014). Haemosporidian parasites are also diverse and widespread in birds, which are competent hosts for over 250 species of the genera Haemoproteus Kruse, 1890, Plasmodium Marchiafava & Celli, 1885 and Leucocytozoon Berestneff, 1904 (see Valkūinas, 2005). Information based on the traditional light microscopic examination of Giemsa-stained thin blood films as well as some characters of life-history traits is the gold standard for species identification, taxonomy and systematics of haemosporidian parasites. However, accurate identification of parasite species is often not possible in wild birds or vectors. The main obstacles to haemosporidian identification include: (i) natural infections are predominantly light (i.e. low intensity); (ii) co-infections of two or more parasite species in the same individual host are common; (iii) the synchronised development of infection (in some Plasmodium spp.) can result in the absence of all blood stages necessary for identification; and (iv) the limitations of morphological characters that sometimes overlap between different parasite species. In addition, haemosporidians have complex life-cycles, including different tissue stages in the vertebrate hosts and sporogonic stages in dipteran vectors that often lack diagnostic morphological characters (Valkūinas, 2005). These difficulties in haemosporidian species identification can be partly overcome by the application of molecular diagnostic methods to identify DNA barcodes (Valkiūnas et al., 2014).
Numerous molecular assays based on polymerase chain reaction (PCR) have been developed in the past twenty years to better understand the diversity and phylogenetic relationships among haemosporidian parasites. Molecular markers targeting different genes of the parasite genome have been developed. These include (i) nuclear-encoded genes such as 18S small subunit (SSU) of the ribosomal RNA (rRNA) (Feldman et al., 1995; Li et al., 1995), dihydrofolate reductase-thymidylate synthase (DHFR-TS) (Bensch et al., 2004) and adenylosuccinate lyase, (asl) (Martinsen et al., 2008); (ii) mitochondrial encoded genes such as cytochrome b (cyt b) gene (Bensch et al., 2000; Perkins & Schall, 2002; Ricklefs & Fallon, 2002; Richard et al., 2002; Beadell et al., 2004; Waldenström et al., 2004), cytochrome c oxidase subunit 1 (cox1) (Martinsen et al., 2008), and regions of the mitochondrial small and large subunit rRNA genes (Fallon et al., 2003); and (iii) apicoplast encoded genes such as caseinolytic protease C gene (clpc) (Martinsen et al., 2008). However, the most widely used protocols are based on the mitochondrial cyt b gene (Bensch et al., 2009). This reliance on cyt b is due to the high sensitivity of screening methods (Richard et al., 2002; Hellgren et al., 2004) the high levels of polymorphism between lineages, and the strong congruence between phylogenetic and traditional classifications of haemosporidian morphospecies (Hellgren et al., 2007; Iezhova et al., 2011; Outlaw & Ricklefs, 2014). Moreover, a well-organised online database now exists to store and access information about the diversity and distributions of cyt b lineages (Bensch et al., 2009, see MalAvi: http://mbio-serv2.mbioekol.lu.se/Malavi/index.html). Finally, phylogenies based on partial cyt b gene can clearly predict vector groups of haemosporidians (Martinsen et al., 2008; Bukauskaitė et al., 2015), a pattern that can be helpful for research on haemosporidian epidemiology.
Sequencing entire genomes of avian haemosporidians seems a logical next step for answering many questions regarding phylogenetic relationships and evolution of haemosporidian parasites. However, obtaining good quality haemosporidian DNA templates for genome sequencing remains difficult, primarily since these parasites inhabit nucleated erythrocytes and the host DNA predominates in the blood samples. Hence, the application of full genome sequencing in avian haemosporidian research is still in its infancy (Palinauskas et al., 2013b; Hellgren et al., 2013; Lauron et al., 2014; Coral et al., 2015). As such, the majority of recent studies using avian haemosporidians as a model system are based on short sequences of mitochondrial cyt b gene (of approximately 500 nucleotides) obtained using general primers (Ferraguti et al., 2013; Clark et al., 2014; Drovetski et al., 2014; Ricklefs et al., 2014). Even for studies using the cyt b gene, the accurate estimation of global diversity patterns of avian haemosporidians is limited due to a bias towards sampling passerine birds from Europe and North America, and the lack of microscopic examination of samples in the majority of molecular studies (Clark et al., 2014). To overcome these issues, additional efforts have to be made to more broadly sample understudied avian host groups and to implement traditional microscopy in parallel with PCR-based techniques.
The limited number of haemosporidian morphological studies is likely due to a lack of experience in both the preparation of good quality blood smears and the proper microscopic techniques used for examination and taxonomy (Valkiūnas et al., 2008a). Understanding this problem has provoked the collaboration between molecular biologists and traditional taxonomists for interdisciplinary studies (Hellgren et al., 2007; Iezhova et al., 2011). Currently, molecular methods are largely available and applicable for diagnostics of avian haemosporidian parasites in laboratories around the world. The number of collaborative studies has increased lately, with more research groups implementing both traditional taxonomy and molecular biology to describe new parasite species and to develop molecular characterisations for previously described species (Mantilla et al., 2013; Silveira et al., 2013; Matta et al., 2014; Clark et al., 2015).
This study uses both molecular and microscopic techniques to identify a previously undescribed Haemoproteus sp. (Haemoproteidae) that parasitises the willow warbler Phylloscopus trochilus (L.). In addition, we identify new DNA diagnostic markers for two Haemoproteus spp. infecting European bee-eaters Merops apiaster L. Morphological descriptions and identifications are based on microscopic examinations of Giemsa-stained thin blood smears with single parasite infections. Phylogenetic analyses performed with partial sequences of the mitochondrial cyt b gene are used to reconstruct the relationships between our new DNA lineages and previously characterised haemosporidian morphospecies.
Materials and methods
Study sites and collection of blood samples
We collected blood samples from 286 willow warblers caught with mist nets and song playback at Kalimok field station (44º00′N, 26º26′E), Bulgaria during April–October 2010; 95% of the birds were sampled between August and October. Additional material came from 48 European bee-eaters caught by mist nets in Burgas region (42º26′N, 27°24′E) and Kavarna region (43°25′N, 28°24′E), east Bulgaria, during June–July 2013. Birds were identified, ringed and measured using a standard protocol for ringing (Svensson, 1992; Bairlein, 1995). Approximately 30 µl of blood was withdrawn from each bird using a heparinised capillary tube after puncturing the brachial vein with a sterile syringe needle. Several drops were used immediately for preparation of blood smears on three glass slides, and the remaining blood was stored in SET buffer (0.05M tris, 0.15 M NaCl, 0.5 M EDTA, pH 8.0) for later molecular diagnostics. Blood smears were air-dried, fixed with absolute methanol, and stained with Giemsa, as described by Valkiūnas et al. (2008a). Preparations of good quality and sufficient parasite intensity from single species infections were used for morphological characterisation of Haemoproteus spp.
Examination of blood films and parasite morphology
Olympus BX41 and Zeiss Axio Imager M2 light microscopes equipped with PixeLINK and ProgRes c10 plus digital cameras and imaging software Megapixel FireWire Camera Release 3.2 and ProgRes CapturePro v2.8.0 were used to examine the blood films and prepare the illustrations. Measurements were taken from the images using the calibrated Image-Pro Plus software. The slides were examined for 15–20 min at low magnification (×400), and then at least 100 fields were studied at high magnification (×1,000). Parasite identification follows the guidelines of Valkiūnas (2005). Reported gametocytes of Haemoproteus gavrilovi were compared with the type-specimens deposited at Nature Research Centre, Vilnius, Lithuania (hapantotype accession number 2399.87 Az, see Fig. 1Q–T). A voucher specimen of Haemoproteus majoris (Laveran, 1902) deposited at Nature Research Centre, Vilnius, Lithuania (accession number 48893NS collected by Vaidas Palinauskas) isolated from a willow warbler was used to compare with the new Haemoproteus sp. described in this study. All measurements are in micrometres.
Gametocytes of Haemoproteus manwelli (A–H) and H. gavrilovi (I–T; images Q–T, are from the hapantotype) from the blood of the European bee-eater Merops apiaster: A, Young gametocyte; B–E, I–L, Q, R, Macrogametocytes; F–H, M–P, S, T, Microgametocytes. Short simple arrows indicate nucleolus of gametocyte nucleus; long simple arrows indicate gametocyte nucleus; short triangle arrows indicate volutin granules; long triangle arrows indicate single vacuoles in macrogametocytes of H. gavrilovi; triangle arrowheads indicate pigment granules. Giemsa-stained thin blood films. Scale-bar: 10 µm
Intensity of infection was estimated as a percentage by counting the number of parasites per 1,000 red blood cells or per 10,000 red blood cells in light infections (i.e. <0.1%), as recommended by Godfrey et al. (1987).
DNA extraction, PCR and sequencing
Total DNA was extracted using a standard ammonium acetate method (Richardson et al., 2001) and quantified by NanoDrop (IMPLEN Nanophotometer P330). Diluted to approximately 25 ng/µL, the total DNA was used as a template in PCR assays for detection of parasites. We used published protocols to amplify portions of the parasite mitochondrial cyt b gene (Hellgren et al., 2004) and the plastid caseinolytic protease C (clpc) gene (Martinsen et al., 2008). In all, three samples were selected from 10 available positive samples for the amplification of H. manwelli lineage hMEAPI1, 1 sample for H. gavrilovi lineage hMEAPI2 and 53 samples for the new Haemoproteus sp., lineage hWW1. All these samples contained single infections of Haemoproteus parasites, as confirmed by both the PCR testing and microscopic examination. We performed PCR amplification in 25 µL total volumes including 50 ng of total genomic DNA template (2 µL), 12.5 µL of DreamTag Master Mix, 8.5 μL nuclease-free water (Fermentas, Lithuania) and 1 μL of each primer (10 µM concentration). One negative control was used in every run of eight samples. Positive results were visualised by electrophoresing 2 µL of the final PCR product on a 2% agarose gel. Amplicons of proper (of approximately 500 bp) length were precipitated and sequenced from both ends using dye terminator cycling sequencing (BigDye) kit and loaded on an ABI PRISM ™ 3100 sequencing robot (Applied Biosystems, Florida, USA). Sequences were edited and aligned using the BioEdit programme (Hall, 1999). The presence of double peaks in sequence chromatograms was considered a co-infection.
Phylogenetic analyses
A Bayesian phylogeny of parasite lineages was reconstructed based on alignment of 45 cyt b lineages (33 of Haemoproteus spp. and 11 of Plasmodium spp.) using MrBayes version 3.1.2 (Ronquist & Huelsenbeck, 2003). One lineage of Leucocytozoon sp. (lineage SISKIN2) was used as the outgroup. We used the General Time Reversible model including invariable sites and variation among sites (GTR + I + Γ) selected by the software jModelTest 2 (Darriba et al., 2012) as the best-fit model under the Bayesian Information Criterion. Gaps and missing data in the alignment were discarded prior to analyses. Two simultaneous runs were conducted with a sample frequency of every 100th generation over 10 million generations. We discarded 25% of the trees as “burn-in” period. The remaining trees were used to construct a majority rule consensus tree. The phylogenies were visualised using Tree View 1.6.6. (Software available from http://evolution.genetics.washington.edu/phylip/software.html). The codes of cyt b lineages are given according to MalAvi database, with a letter “h” starting codes of Haemoproteus spp. lineages and a letter “p” starting codes of Plasmodium spp. lineages.
The sequence divergence between different lineages was calculated using Jukes-Cantor model of substitution, with all substitutions weighted equally (uniform rates), implemented in the programme MEGA6 (Tamura et al., 2013).
Morphological and molecular examination of blood samples
Samples with infections of Haemoproteus spp. or/and Plasmodium spp. from the bee-eaters, as determined by microscopic examination, were screened by PCR to obtain sequence data. One individual European bee-eater was co-infected with Haemoproteus manwelli and H. gavrilovi, with overall parasitaemia of 2.03%. We did not obtain a good quality sequence from this sample, but gel electrophoresis of amplified products from this sample were positive, and we observed these two parasite species on the complementary blood smears.
Haemoproteus ( Parahaemoproteus ) manwelli Bennett, 1978
Avian host and distribution: The lineage hMEAPI1 has been recorded in the bee-eater Merops apiaster L. in Bulgaria; it is a new cyt b lineage which was recorded in 10 of 48 adult bird individuals examined. Seven of these birds were caught in Burgas region, eastern Bulgaria (42°26′15.59″N, 27°24′2.30″E) and three birds were sampled in Kavarna region, north-eastern Bulgaria (43°25′21.13″N, 28°24′17.77″E). The most closely related lineage hCYAPIC01 (genetic difference in cyt b gene is 3.4%) has been reported in the azure-winged magpie Cyanopica cyanus (Pallas, 1776) in China (Zehtindjiev et al., 2013).
Site of infection: Mature erythrocytes; no other data.
Representative blood films: Voucher specimens (accession numbers 48833NS, G465770, Protozoa-2015-0002) were deposited in the Institute of Ecology, Nature Research Centre, Vilnius, Lithuania, the Queensland Museum, Brisbane, Australia, and the Institute of Biodiversity and Ecosystem Research, Sofia, Bulgaria, respectively. The label data for all these slides are: M. apiaster, 25.vii.2013, collected by K. Bedev.
DNA sequences: Mitochondrial cyt b lineage hMEAPI1 (479 bp, GenBank accession number KP462687) and apicoplast caseinolytic protease C (clpc) lineage hMEAPI1 (506 bp, GenBank accession number KT932745).
Description (Fig. 1A–H)
Young gametocytes
Young gametocytes (Fig. 1A) were rarely seen in the voucher preparations. Growing gametocytes adhere to erythrocyte nuclei and extend longitudinally along nuclei; outline even.
Macrogametocytes
Macrogametocytes (Fig. 1B–E) grow along erythrocyte nuclei, slightly enclosing them with ends. Growing gametocyte usually adheres to erythrocyte nucleus not touching erythrocyte envelope (Fig. 1B). Fully-grown gametocyte closely appressed both to erythrocyte nucleus and envelope, markedly displacing nucleus laterally (Fig. 1E, F), but does not enucleate the host cell. Mature gametocytes only slightly enclose erythrocyte nuclei and never encircle them (Fig. 1E). Cytoplasm granular in appearance, frequently containing several small vacuoles (Fig. 1C). Outline usually even. Parasite nucleus variable in shape, often roundish or oval, median or submedian in position, sometimes possesses visible nucleolus (Fig. 1B, D). Pigment granules roundish or oval, of medium (0.5–1.0) size, randomly scattered throughout the cytoplasm, 5–9 (mean ± SD, 6.8 ± 1.2) in number.
Microgametocytes
General configuration of microgametocytes (Fig. 1F–H) and other features as for macrogametocytes, with the usual haemosporidian sexual dimorphic characters (Valkiūnas, 2005). Outline of growing gametocytes sometimes slightly amoeboid (30% of microgametocytes observed in the voucher preparation) (Fig. 1F). Pigment granules 5–9 (mean 6.2 ± 1.4) in number.
Haemoproteus ( Parahaemoproteus ) gavrilovi Valkiūnas & Iezhova, 1990
Avian hosts and distribution: The lineage hMEAPI2 has been recorded in the bee-eater M. apiaster L. in Bulgaria. This is a new cyt b lineage which was recorded in one of 48 examined adult individuals. The parasite was found in a female bird caught on breeding grounds in eastern Bulgaria on the north shore of the Mandra Lake, Burgas district (42°26′15.59″N, 27°24′2.30″E). The most closely related cyt b lineage hMEBRE01 (genetic difference in cyt b gene is of 0.6 %) has been reported in the black-headed bee-eater Merops breweri (Cassin, 1859) in Gabon (Beadell et al., 2009).
Site of infection: Mature erythrocytes; no other data.
Representative blood films: Voucher specimens (accession numbers 48834NS, G465771, Protozoa-2015-0003) were deposited in the Institute of Ecology, Nature Research Centre, Vilnius, Lithuania, the Queensland Museum, Brisbane, Australia, and the Institute of Biodiversity and Ecosystem Research, Sofia, Bulgaria, respectively. Label data for all slides are: M. apiaster, 14.vii.2013, collected by K. Bedev.
DNA sequences: Mitochondrial cyt b lineage hMEAPI2 (479 bp, GenBank accession number KP462688) and apicoplast clpc lineage hMEAPI2 (506 bp, GenBank accession number KT932746).
Description (Fig. 1I–P)
Macrogametocytes
Macrogametocytes (Fig. 1I–L, type-material: Q, R) grow along erythrocyte nuclei, slightly enclosing them with ends. As gametocyte grows, displaces nucleus of infected erythrocyte first laterally (Fig. 1K, type-material: Q) and then towards one of the poles of host cell (Fig. 1L, type-material: R). Such forms were only occasionally seen in voucher preparations, but common in hapantotype. Enucleated host cells not observed. Cytoplasm granular in appearance, always contains volutin granules usually located at gametocyte ends and obscuring pigment granules. A small to medium size clear vacuole (0.6±0.2 μm in diameter), usually present in cytoplasm (Fig. 1K, type-material: R). Gametocytes adhere to nuclei and envelope of erythrocytes; contact of gametocyte edges with erythrocyte envelope varies in parasites with amoeboid outline (Fig. 1K). Outline of gametocytes varies from even (Fig. 1I, J) to slightly ameboid (Fig. 1K, type-material: Q). Parasite nucleus compact, variable in form and position. Pigment granules roundish and oval, usually small (<0.5 μm), sometimes medium-sized (0.5–1.0 μm), randomly scattered throughout cytoplasm, between 7 and 12 (on average 9 ± 1.3) in number.
Microgametocytes
General configuration of microgametocytes (Fig. 1M–P, S, T) and other features as for macrogametocytes, with the usual haemosporidian sexual dimorphic characters (Valkiūnas, 2005). Large vacuoles were not visible in the cytoplasm. Pigment granules 6–10 (mean 7.8 ± 1.4).
Haemoproteus ( Parahaemoproteus ) palloris n. sp.
Type-host: the willow warbler Phylloscopus trochilus (L.) (Passeriformes, Phylloscopidae).
Type-locality: Planted forest of black locust (Robinia pseudoacacia) and ash trees (Fraxinus spp.) at the Kalimok field station, Silistra District, Bulgaria (44º00′04′′N, 26º26′14′′E, 18 m above sea level)
Site of infection: Mature erythrocytes; no other data.
Prevalence: 18.5% (53 of 286 examined willow warblers were infected).
Type-specimens: Hapantotype (accession number 48832NS, male adult bird Phylloscopus trochilus; parasitaemia intensity 4.5%, 12.ix.2010, Kalimok field station, collected by C. Sjöholm) was deposited in the Institute of Ecology of Nature Research Centre, Vilnius, Lithuania. Parahapantotypes (accession numbers G465769 and Protozoa-2015-0001) were deposited in the Queensland Museum, Brisbane, Australia and Institute of Biodiversity and Ecosystem Research, Sofia, Bulgaria, respectively).
Distribution: According to MalAvi database, the lineage hWW1 has been recorded in 10 bird species belonging to 7 families in Europe, Africa and Asia. This lineage might be a widespread host generalist, though it remains unclear if these birds are competent hosts in which gametocytes can fully develop.
DNA sequences: Mitochondrial cyt b lineage hWW1 (478 bp, GenBank accession number AF254971) and apicoplast clpc lineage hWW1 (510 bp, GenBank accession number KT932747).
Etymology: The species name is derived from the Latin word “pallor” (in English: pallor – unusual or extreme paleness) and reflects the pale-staining of the cytoplasm of macrogametocytes (compare Fig. 2E–H with M, N).
Gametocyte of Haemoproteus palloris n. sp. (A–L) and H. majoris (M–P) from the blood of the willow warbler Phylloscopus trochilus: A, Young gametocyte; B–H, M, N, macrogametocytes; I–L, O, P, Microgametocytes. Long simple arrows indicate gametocyte nucleus; short triangle arrows indicate unfilled space between gametocyte and erythrocyte nucleus; long triangle arrows indicate vacuole-like spaces in macrogametocytes; triangle arrowheads indicate pigment granules. Giemsa-stained thin blood films. Note pale-staining of the cytoplasm in macrogametocytes of H. palloris in comparison to the relatively dark staining in macrogametocytes of H. majoris. Scale-bar: 10 µm
Description (Fig. 2A–L)
Young gametocytes
Single young gametocyte found in the type-material roundish in shape, with even outline, seen in mature erythrocyte (Fig. 2A).
Macrogametocytes
[Metrical data in Table 1.] Macrogametocytes develop in mature erythrocytes (Fig. 2B–H). Cytoplasm pale-blue, heterogeneous in appearance, volutin granules absent. Outline even (Fig. 2B) or slightly irregular at terminal gametocyte edges (Fig. 2C). Several vacuoles or vacuole-like spaces of variable size present in the majority of gametocytes (80%) (Fig. 2B, C–F). Gametocyte grows along nucleus of infected erythrocyte, encloses nucleus with ends, but does not encircle it completely. Thin cleft-like unfilled space can often be seen between growing gametocyte and erythrocyte nucleus (Fig. 2D, E). As parasite grows, gametocyte adheres to erythrocyte envelope, and unfilled space between gametocyte and erythrocyte nucleus start to close gradually. As parasite matures, gametocyte fills up erythrocyte poles and slightly displaces the nucleus of infected cell laterally (Fig. 2G, H). Parasite nucleus relatively small (Table 1), of variable form and position, usually sub-central (Fig. 2B, C, G), but also often sub-terminal (Fig. 2D) or sometimes even terminal (Fig. 2E, F). Nucleolus not seen. Pigment granules roundish or oval, small (0.5 µm) and medium-sized (0.5–1.0 µm), usually randomly scattered throughout the cytoplasm. Average number of pigment granules 12.8 (± 1.5). Influence of gametocyte on infected erythrocyte not pronounced (Table 1).
Microgametocytes
[Metrical data in Table 1.] General configuration (Fig. 2I–L) as for macrogametocytes with the usual haemosporidian sexual dimorphic characters (Valkiūnas, 2005). Cytoplasm vacuolisation not as prominent as for macrogametocytes.
Remarks
Pale-staining of the cytoplasm is a diagnostic character for several species of avian haemoproteids parasitising passerine birds. Some of them were recently described in birds of the families Picnonotidae, Sylviidae and Fringillidae (see Valkiūnas et al., 2008b; Križanauskienė et al., 2010; Dimitrov et al., 2014). It seems that this character is more common than previously thought (Križanauskienė et al., 2010). Taken separately, it is difficult to use this character for species identification of avian haemoproteids because gametocyte staining depends on the staining procedures (Valkiūnas, 2005). However, this character is clearly distinguishable when two species of haemoproteids are present in the same blood film (Križanauskienė et al., 2010; Dimitrov et al., 2014; compare Fig. 2E, F and M, N). The pale-staining of the cytoplasm depends on the density of cellular structures in gametocytes, and it is phylogenetically informative based on analyses of the parasite mitochondrial cyt b gene (Križanauskienė et al., 2010; Dimitrov et al., 2014; Fig. 3).
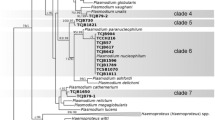
Bayesian inference phylogeny for 45 haemosposridian cytochrome b (cyt b) lineages. Cyt b lineage of Leucocytozoon sp. (SISKIN2) was used as the outgroup. Species described in this study are given in bold. Codes of the lineages (according MalAvi database, http://mbio-serv2.mbioekol.lu.se/Malavi/index.html) are given before GenBank accession numbers and species names. Grey boxes indicate groups of closely related species of haemoproteid: A, four species of Haemoproteus closely related to Haemoproteus (Parahaemoproteus) gavrilovi; B, pale-stained: parasites closely related to Haemoproteus (Parahaemoproteus) palloris; C, species of subgenus Parahaemoproteus infecting none-passerine hosts
Among the haemoproteids of passerine birds, H. palloris n. sp. is most similar to Haemoproteus pallidus Valkiūnas & Iezhova, 1991 and Haemoproteus pallidulus Križanauskienė, Pérez-Tris, Palinauskas, Hellgren, Bensch & Valkiūnas, 2010. Haemoproteus palloris n. sp. can be readily distinguished from these parasites due to the presence of vacuoles or vacuole-like spaces in the majority (80%) of the advanced macrogametocytes (Fig. 2B–F). Additional distinctive characters are the pattern of growth of parasite gametocytes. In H. palloris n. sp., an unfilled space is usually present between the gametocyte and the erythrocyte nucleus (Fig. 2D, E). This is not characteristic for H. pallidus or for H. pallidulus (see Valkiūnas, 2005; Križanauskienė et al., 2010). From this point of view, the pattern of growth of gametocytes in H. palloris n. sp. is similar to that of Haemoproteus concavocentralis Dimitrov, Zehtindjiev, Bensch, Ilieva, Iezhova & Valkiūnas, 2014, but in the latter parasite this space is more pronounced and gives the gametocyte a concave shape (Dimitrov et al., 2014), which is not the case in gametocytes of the new species.
Based on vacuolisation of the cytoplasm, H. palloris n. sp. is especially similar to Haemoproteus vacuolatus Valkiūnas, Iezhova, Loiseau, Chasar, Smith & Sehgal, 2008. The existence of one clear circular vacuole, which is present in the cytoplasm of each advanced macrogametocyte and usually adheres to the parasite nucleus, is a characteristic feature of H. vacuolatus. Similar vacuoles were also seen in H. palloris n. sp., but usually several vacuoles are present in this parasite, and they are of different size, position and shape (Fig. 2B, C, F), which is not characteristic of H. vacuolatus (Valkiūnas et al., 2008b). Additionally, the number of pigment granules is greater in macrogametocytes of H. vacuolatus (on average 20.3) in comparison to H. palloris n. sp. (on average 12.8).
Another commonly observed haemoproteid in our samples from willow warblers was H. majoris. This haemoproteid parasite is difficult to identify and can be distinguished from similar species only on the basis of a detailed analysis of a set of characters (Valkiūnas, 2005). Phylogenetic analysis showed that cyt b lineage hWW2 of H. majoris is significantly divergent from the lineage hWW1 of H. palloris n. sp. (4.7% difference, Fig. 3). Gametocytes of H. palloris can be readily distinguished from those of H. majoris due to the pale-staining of their cytoplasm and the presence of prominent vacuoles (compare Fig. 2A–H with M, N). Additionally, growing gametocytes of H. palloris do not assume a dumbbell-like shape and often do not adhere to the nuclei of erythrocytes (Fig. 2D, E), which are characteristic features of H. majoris (Valkiūnas, 2005; Fig. 2M).
Phylogenetic analysis
Phylogenetic analysis indicated that morphological characters, which have been traditionally used in taxonomy of avian haemosporidian parasites, have a phylogenetic value. The lineage hMEAPI1 of H. manwelli clustered with the lineage hCYAPIC01 of Haemoproteus sp. recorded in the azure-winged magpie Cyanopica cyanus in China, but with a relatively large genetic difference in the cyt b gene between these lineages (3.4%). Phylogenetic analyses showed that the lineage hMEAPI2 of H. gavriovi is closely related to hMEBRE1 (genetic difference of 0.6%), a lineage reported in black-headed bee-eater Merops breweri in Gabon. Together with the lineages hROBIN1 of Haemoproteus attenuatus Valkiūnas, 1989 and hSFC1 of Haemoproteus balmorali Peirce, 1984, they formed a relatively well-supported clade A (Fig. 3, posterior probability of 0.77). However, all these lineages are divergent and readily distinguishable by morphology of their blood stages.
The cyt b lineages hMEAPI1 and hMEAPI2 were recorded in the same avian host species, but they differed significantly (genetic difference of 7.3%) and clustered separately in the phylogenetic tree (Fig. 3); these parasites show well distinguishable morphological characters (Fig. 1). Both parasites are even more divergent in the clpc gene (8.1%).
The lineage hWW1 clustered together with other so-called pale-stained haemoproteid species, with a high posterior probability of 0.96 (Fig. 3, clade B). It is worth noting that the mean genetic distance among cyt b lineages within this clade was low (1.3%), but all parasites have unique morphological characters, based on which they can be readily distinguished.
Discussion
In this study, we identified molecular markers (barcodes) based on mitochondrial and apicoplast genes for the identification of three common species of Haemoproteus, H. palloris n. sp. from the willow warbler and H. manwelli and H. gavrilovi from the European bee-eater. Morphological features of the blood stages and some life-history traits remain a basis for taxonomy and systematics of avian haemosporidian parasites (Valkiūnas, 2005; Atkinson, 2008). Contemporary taxonomic studies are usually accompanied by molecular data, mainly short sequences (of approximately 500 bp) obtained using PCR-based techniques (Merino et al., 2012; Mantilla et al., 2013; Dimitrov et al., 2014). This information markedly improves the identification of these parasites and is especially helpful in epidemiological, molecular ecology and phylogenetic research (Silveira et al., 2013; Clark et al., 2014; Outlaw & Ricklefs, 2014).
Haemoproteus manwelli and H. gavrilovi are common parasites in birds of the bee-eater family Meropidae (see Bennett, 1978; Valkiūnas, 2005). Our study generated the first DNA barcodes for Haemoproteus spp. parasitising birds of the Meropidae. Beadell et al. (2009) reported two cyt b lineages of unidentified Haemoproteus spp. (hMEBRE01, hMEBRE02,) and one of Plasmodium sp. (pACCTAC01) from the black-headed bee-eater Merops breweri and the rosy bee-eater Merops malimbicus Shaw sampled in Gabon. Ishtiaq et al. (2007) found one unidentified Haemoproteus sp. lineage (hMERORI01) and one Plasmodium sp. lineage (pSTASTR01) in the little green bee-eater Merops orientalis Latham sampled in Myanmar and India, respectively. In this study, we obtained valuable new cyt b and clpc lineages that will be useful for future researches through the GenBank and MalAvi online databases. In addition, we provide morphological description and image material for H. manwelli and H. gavrilovi and linked them with their cyt b and clpc sequences. Bee-eaters are relatively understudied host species of avian haemosporidians in Europe, which probably explains why sequences of these parasites have not been reported before.
In spite of sharing the same host species, H. manwelli and H. gavrilovi are clearly distinguishable both by their morphological features and by cyt b sequences (Fig. 3). It should be noted that the presence of volutin granules is a characteristic feature of H. gavrilovi gametocytes, but not of H. manwelli (compare Fig. 1A–H with Fig.1I–T). That is one of the most distinguishable features of these infections, which helps to separate them during co-infections. Volutin granules can be readily distinguished from pigment granules by their lesser light refraction and often bright-violet colour (Valkiūnas, 2005). Production of volutin has been reported in many avian Haemoproteus spp., and some haemoproteid species are characterised by a heavy volutin production; see for example the descriptions of Haemoproteus balmorali (parasite of Passseriformes, see Valkiūnas, 2005), Haemoproteus velans Coatney & Roudabush, 1937 (parasite of Piciformes, Dimitrov et al., 2014), Haemoproteus syrnii (Mayer, 1910) (parasite of Strigiformes, Karadjian et al., 2013; Bukauskaitė et al., 2015), Haemoproteus turtur Covaleda Ortega & Gállego Berenguer, 1950 and Haemoproteus columbae Kruse, 1890 (parasites of Columbiformes, Križanauskienė et al., 2013). The nature of the volutin granules observed in avian haemoproteids has not been determined. Volutin might have an important metabolic function, for instance by representing acidic cytoplasmic inclusions that have been observed in plants and some apicomplexan parasites (Karadjian et al., 2013). Despite the questionable value in the identification of many haemosporidian species (Greiner et al., 1977), here we found that heavy production of volutin granules has taxonomic value at the species level in some Haemoproteus spp. Numerous prominent volutin granules are permanently present in the gametocytes of H. gavrilovi, but not in H. manwelli. In other words, this study confirms that heavy production of volutin granules is a notable feature that is valuable in haemosporidian species delimitation when using microscopic methods. However, because the majority of avian haemoproteid species develop low amounts of volutin that can be difficult to visualise depending on the level of staining, this character can hardly be used in many taxonomic studies (Bishop & Bennett, 1989; Valkiūnas, 2005).
According to our phylogenetic analyses, parasites with pronounced volutin granules (H. attenuatus, H. balmorali, H. gavrilovi, H. turtur, H. columbae and H. syrnii) are not monophyletic; they appear in distant clades in the phylogenetic tree (Fig. 3). It seems that this feature evolves independently in these parasites. However, the clade A (Fig. 3) contains only parasite species that possess prominent volutin granules (i.e. H. attenuatus, H. balmorali and H. gavrilovi). It is likely that pronounced volutin production has evolved independently for Culicoides Latreille-transmitted (H. attenuatus, H. balmorali and H. gavrilovi) and for hippoboscid-transmitted (H. turtur, H. syrnii, H. columbae, and Haemoproteus iwa Work & Rameyer, 1996) parasites (Fig. 3). Nevertheless, it is worth nothing that some closely related species pairs, such as Haemoproteus multipigmentatus Valkiūnas, Santiago-Alarcon, Levin, Iezhova & Parker, 2010 and H. columbae or Haemoproteus sacharovi Novy & MacNeal, 1904 and H. turtur, did not share this morphological feature.
We were not able to detect haemosporidian parasites by PCR based methods in one sample in which gametocytes of H. manwelli were readily visible and the intensity of parasitaemia was relatively high (0.04%). It is usually believed that the PCR methods are more efficient than microscopic examination in the detection of chronic infections (Richard et al., 2002; Hellgren et al., 2004). However, we have shown that this is not always true. The unsuccessful PCR diagnostics in our sample might be due to poor quality or low concentration of the DNA template after extraction procedure (Richard et al., 2002). For instance, although we did not check the extraction quality by amplifying bird DNA, we did quantify DNA concentrations prior to PCR runs, and we confirmed that the DNA concentration was too low to be estimated by IMPLEN Nanophotometer P330 in that particular sample. However, there were two more samples with DNA concentration below the threshold of the machinery (IMPLEN Nanophotometer P330), but which successfully amplified and produced clean sequences.
The European bee-eaters are birds belonging to the order Coraciiformes which, according to the recent genomic data (Jarvis et al., 2014), are closely related to the woodpeckers (Piciformes). It is worth noting that the cyt b lineages hMEAPI1 and hMEAPI2 were grouped independently from each other, and they appeared in the clade of the haemoproteids parasitising other passerine hosts (Passeriformes) of the subgenus Parahaemoproteus. According to the mitochondrial cyt b gene phylogeny, it seems that H. manwelli and H. gavrilovi are more closely related to the parasites of passerine birds than to the parasites infecting non-passerine birds (Fig. 3 clade C). One possible explanation for their phylogenetic placement might be host shifting and later co-speciation of haemosporidians (Ricklefs et al., 2014). Alternatively, these two species might have evolved independently, and our phylogenetic estimation (Fig. 3) might suffer a bias due to limited sampling of haemoproteid parasites from non-passerine birds. Additional sampling of haemoproteids of birds of Coraciiformes is needed to address these questions.
The mitochondrial cyt b lineage hWW1 of the newly described H. palloris was reported for the first time in Swedish populations of willow warblers (Bensch & Åkesson, 2003). These authors analysed temporal and spatial variation in distribution of two cyt b lineages, hWW1 and hWW2, but their study applied only molecular methods without microscopic examination of blood samples. When morphological identification became crucial for defining species limits and host range of cyt b lineages, the lineage hWW2 was identified by Križanauskienė et al. (2006) as belonging to H. majoris. The lineage hWW1 did not previously have morphologically described characteristics, and this study provides valuable new description and measurement data for this parasite (Fig. 2; Table 1). Surprisingly, two molecular studies recorded this lineage in non-passerine birds, i.e. in birds of Anseriformes (Wood et al., 2013) and Upupiformes (Zehtindjiev et al., 2013). Based on available experimental data about vertebrate host specificity of Haemoproteus spp., such big jumps between divergent host taxa are unlikely (Valkiūnas, 2005; Atkinson, 2008). Although most of the host records of the lineage hWW1 are from old world warblers (former Sylviidae, now Sylvioidea superfamily), there are also several records of this parasite from African birds belonging to the Ploceidae and Estrildidae as well as from European birds of the Paridae and Muscicapidae (MalAvi database accessed on March 2015, see Bensch et al., 2009).
Based on the information available, it is difficult to rule out the possibility that this cyt b lineage has both a broad host range and geographical distribution, with transmission taking place both in Africa and Europe. However, shortcomings in PCR-based haemosporidian diagnostics make this conclusion difficult to draw. Molecular studies are insensitive in reading different stages of haemosporidian parasites and they are not useful for determining whether gametocytes of the lineage hWW1 occur in host blood that is PCR positive. In other words, molecular methods only require presence of parasite DNA, which can come from abortive parasite development, resulting in overestimation of host range when such DNA is detected from non-competent host species (Poulin & Keeney, 2008; Valkiūnas et al., 2014). It is therefore important to note that PCR based records of lineages in host species is not enough evidence of the bird species being a competent host. PCR can amplify DNA of sporozoites or tissue stages, which abort their development in avian hosts, but provide templates for DNA amplification. As such, there is increasing evidence that haemosporidian infections in non-competent vertebrate and invertebrate hosts result in abortive development of the parasites before they reach the stage of infectious gametocytes or sporozoites (Valkiūnas et al., 2013). It is possible that the records of parasite lineages across bird families and particularly orders for many haemosporidian species are remarkably broader than the ranges of their competent hosts. For example, gametocytes of Haemoproteus sp. were not found in the complementary blood slides from Eurasian hoopoe Upupa epops L. in the study of Zehtindjiev et al. (2013) while the same sample was PCR positive for hWW1 lineage. Because gametocytes of Leucocytozoon sp. were present in this bird (Karina Ivanova, personal communication), a non-specific amplification might explain the reason for false positive PCR-based diagnostic results (Cosgrove et al., 2006), but that could not explain successful sequencing of the hWW1 lineage afterwards.
According to available experimental data, avian Haemoproteus spp. do not complete life-cycles in birds belonging to different orders, suggesting they are specific on the level of host orders (Valkiūnas, 2005; Atkinson, 2008). It is interesting that PCR-based diagnostics show that there are more than 40 Haemoproteus cyt b lineages recorded from avian hosts belonging to different orders. For example, the lineage hQUERY01 was reported in birds of four orders, the lineage of hMW1 of Haemoproteus belopolskyi Valkiūnas, 1989 and hWW2 of H. majoris in three host orders, hSW1 of H. belopolskyi in two host orders (see MalAvi database). The vast majority of these reports came solely from molecular studies, and they need additional confirmation for the presence of mature gametocytes in the blood stream of the investigated hosts. The first possible explanation for unusual host records for lineage hWW1 might be the unknown fate sporozoites, which sometimes are present in the circulation, but abort their development (Valkiūnas et al., 2009). Secondly, several examples for abortive development of haemoproteid parasites in birds, indicate cases of blockage of infection development at tissue stages in non-adapted avian hosts (Olias et al., 2011; Cannell et al., 2013; Valkiūnas et al., 2014). Third, if the cyt b gene is relatively conservative and we can detect within-lineage morphological or genetic diversity using other more variable genes, then it is possible to have the same cyt b lineage reported in hosts of different orders. Such genetic variation was recorded in the merozoite surface protein 1 (MSP1) gene within globally distributed host generalist cyt b lineage pSGS1 of Plasmodium relictum (Grassi & Feletti, 1891) (see Hellgren et al., 2014). However, avian malaria parasites (Plasmodium spp.) are less specific and have broader host range and distribution than Haemoproteus spp. (Valkiūnas, 2005; Waldenström et al., 2002; Beadell et al., 2009). Hence, the chance to observe the same Haemoproteus sp. at the gametocyte stage in birds of different orders is hardly possible and needs confirmation. Our experience suggests that there is no evidence of morphological or molecular diversification among different isolates of the same haemoproteid cyt b lineage originating from different host species or regions. In contrast, there are many examples of intraspecific cyt b gene variation within the same morphospecies of avian haemoproteids (Valkiūnas et al., 2007; Iezhova et al., 2011; González et al., 2015). We encourage researchers in the field to use a combination of microscopic and molecular methods when claiming unusual host records that are supported solely by PCR methods.
The lineage hPFC1 of H. pallidus and the lineage hSYAT03 of H. pallidulus were the closest known lineages to lineage hWW1 (Fig. 3, clade B), with genetic divergences of 1.8% for both. This genetic similarity is in agreement with their similar morphological characters such as the pale-staining of the cytoplasm of macrogametocytes and the small size of pigment granules. Similar results were obtained in the study by Križanauskienė et al. (2006), in which H. pallidulus was not yet described, but the lineage hSYAT03 grouped with the lineage hWW1 in phylogenetic analyses. It seems that pale-stained parasites form a congruent monophyletic group with low cyt b genetic divergence, but with clearly distinguishable morphological features (Fig. 3, clade B).
It was difficult to develop molecular characterisation of H. palloris and H. majoris because their cyt b lineages (hWW1 and hWW2) exist in same avian host, the willow warbler (Bensch & Åkesson, 2003), and co-infections are possible. Haemoproteus majoris is especially difficult to identify (Valkiūnas, 2005), and it might be misidentified in case of low parasitaemia. There were no reported cases of co-infections with the cyt b lineages hWW1 or hWW2 in our samples, but it is known that PCR methods often underestimate co-infections in the wild bids (Valkiūnas et al., 2006; Martínez et al., 2009), and thus it is difficult to rule out that we could underestimate such co-infections by PCR. Infections with cyt b lineage hWW2 (five infected birds detected) were infrequent in our samples in comparison to the lineage hWW1 (52 infected birds) (data not shown). It is thus likely that we dealt with single infections of H. palloris in most of our complementary smears, and we can trust the morphological identification (Fig. 2).
Interestingly, the lineage hWW2 of H. majoris was more prevalent than the lineage hWW1 in the study by Bensch & Åkesson (2003) in Sweden, with significant differences reported in the geographical distribution of this parasite between years. The authors concluded that haemosporidian parasite prevalence in Swedish willow warblers varies both in time and space. Based on our results, we can assume that the willow warbler population passing through the eastern European flyway (including our study site in Bulgaria) mostly harbour the hWW1 lineage of H. palloris. We could not be sure that this is a stable pattern across years because our samples were mostly collected during autumn migration during one year. However, it seems that infections with the cyt b lineage hWW1 are rare in Western Europe among passerine birds (Cosgrove et al., 2008; Ventim et al., 2012), but quite prevalent in the mute swans Cygnus olor (Gmelin) breeding at the south coast of England (Wood et al., 2013). Unfortunately, none of the former studies have confirmation if gametocytes of the cyt b lineage hWW1 are present in the blood of the examined birds. The finding of the hWW1 lineage in mute swans is of theoretical interest because it was prevalent in this host, and parasites of passeriform birds have never been reported in anseriform birds before. Further confirmation by microscopic examination is required to prove if gametocytes of this parasite develop in mute swans and this bird is a competent host, which would demonstrate an unusual host shift for Haemoproteus spp.
Available data indicate that the cyt b lineage hWW1 is transmitted mainly in central and eastern Africa and, occasionally, in the temperate regions of Eurasia (Cosgrove et al., 2008; Palinauskas et al., 2013a). However, we recorded this lineage in birds that had hatched in the year of catching (data not shown), which means that the transmission also occurs in the breeding grounds or during migration of willow warbler in temperate regions of Europe. Our study shows that this infection is particularly common in the willow warblers migrating through east Bulgaria and confirms the transmission capabilities of the parasites due to observation of mature gametocytes in the blood stream.
The vectors of H. palloris remain unknown. Because this species belongs to subgenus Parahaemoproteus and its sequence clusters with Culicoides spp.-transmitted haemoproteids (Fig. 3), it is probable that this parasite is transmitted by blood-sucking insects of the same genus. A recent experimental studies showed that phylogenetic analyses based on parasite cyt b gene groups according to the different vectors used avian haemoproteids (Bukauskaitė et al., 2015). Based on the same arguments (Fig. 3), we predict that vectors transmitting H. manwelli and H. gavrilovi also are biting midges of Culicoides. Experimental studies are needed to determine which species of Culicoides biting midges are vectors for these parasites.
Haemoproteus palloris (hWW1) is especially similar to H. vacuolatus (hANLAT2) morphologically and phylogenetically (see the Remarks and Fig. 3, clade B), but the latter species has been recorded only in sedentary birds of tropical Africa (Valkiūnas et al., 2008b; Beadell et al., 2009; Loiseau et al., 2010). It is interesting that all lineages of clade B (Fig. 3) belong to this readily distinguishable morphospecies with low genetic divergence in the mitochondrial cyt b gene. It was shown that some cyt b lineages of Haemoproteus spp. with just two nucleotides difference (0.7%) could be readily identified based on morphological features of their gametocytes (Valkiūnas et al., 2008b). This is especially true for the so-called pale-stained parasites: H. pallidus, H. pallidulus, H. vacuolatus, H. concavocentralis and the newly-described H. palloris. There is no explanation why this pattern is so applicable for the group of pale-stained haemoproteid parasites and not so, for example, in species like Haemoproteus tartakovskyi Valkiūnas, 1986, H. belopolskyi, or H. majoris, which are densely stained with Giemsa. The majority of the pale-stained haemoproteid species have been reported in African migrants or resident bird species. It might be that they originally evolved in tropical climates and are adapted to tropical species of vectors, which remain unknown. Relatively recent host shifts and subsequent co-speciation might be a direct consequence of their diverse avian hosts belonging to the Sylviidae, Muscicapidae, Pycnonotidae, Fringillidae and Phylloscopidae.
In conclusion, this study describes three species of avian haemosporidians based on morphology of their blood stages and segments of their mitochondrial cyt b and apicoplast clpc genes, which can be used for molecular identification and diagnosis of these species. Illustrations of blood stages of a newly described species, H. palloris n. sp. (lineage hWW1) and two already known species, i.e. H. manwelli (lineage hMEAPI1) and H. gavrilovi (lineage hMEAPI2) are given, and phylogenetic analysis including closely related cyt b lineages suggest that traditional species taxonomy of avian haemoproteids is supported by molecular data (Fig. 3). Further work to increase the number of precise linkages between haemosporidian DNA sequences and their corresponding morphospecies is needed for better understanding host-parasite interactions, species delimitation and phylogeography. This study emphasises the value both of PCR-based diagnostics and microscopic examination of blood films in species identification and understanding host range of competent hosts for avian haemproteids.
References
Atkinson, C. T. (2008). Haemoproteus. In C. T. Atkinson, N. J. Thomas, & B. C. Hunter (Eds), Parasitic diseases of wild birds. Ames, IA: Wiley-Blackwell, pp. 13–35.
Bairlein, F. (1995). Manual of field methods. European-African songbird migration network. Institut für Vogelforschung, Wilhelmshaven.
Beadell, J. S., Covas, R., Gebhard, C., Ishtiaq, F., Melo, M., Schmidt, K., et al. (2009). Host associations and evolutionary relationships of avian blood parasites from West Africa. International Journal for Parasitology, 39, 257–266.
Beadell, J. S., Gering, E., Austin, J., Dumbacher, J. P., Peirce, M. A., Pratt, T. K., et al. (2004). Prevalence and differential host-specificity of two avian blood parasite genera in the Australo-Papuan region. Molecular Ecology, 13, 3829–3844.
Bennett, G. F. (1978). Avian Haemoproteidae. 8. The haemoproteids of the bee-eater family (Meropidae). Canadian Journal of Zoology, 56, 1721–1725.
Bennett, G. F., & Campbell, A. G. (1972). Avian Haemoproteidae. I. Description of Haemoproteus fallisi n. sp. and a review of the haemoproteids of the family Turdidae. Canadian Journal of Zoology, 50, 1269–1275.
Bensch, S., & Åkesson, S. (2003). Temporal and spatial variation of hematozoans in Scandinavian willow warblers. Journal of Parasitology, 89, 388–391.
Bensch, S., Hellgren, O., & Pérez-Tris, J. (2009). MalAvi: a public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Molecular Ecology Resources, 9, 1353–1358.
Bensch, S., Pérez-Tris, J., Waldenström, J., & Hellgren, O. (2004). Linkage between nuclear and mitochondrial DNA sequences in avian malaria parasites: multiple cases of cryptic speciation? Evolution, 58, 1617–1621.
Bensch, S., Stjernman, M., Hasselquist, D., Ostman, O., Hansson, B., Westerdahl, H., & Pinheiro, R. T. (2000). Host specificity in avian blood parasites: a study of Plasmodium and Haemoproteus mitochondrial DNA amplified from birds. Proceedings of the Royal Society B: Biological Sciences, 267(1452), 1583–1589.
Bishop, M. A., & Bennett, G. F. (1989). The haemoproteids of the avian order Strigiformes. Canadian Journal of Zoology, 67, 2676–2684.
Bukauskaitė, D., Žiegytė, R., Palinauskas, V., Iezhova, T. A., Dimitrov, D., Ilgūnas, M., et al. (2015). Biting midges (Culicoides, Diptera) transmit Haemoproteus parasites of owls: evidence from sporogony and molecular phylogeny. Parasites & Vectors, 8, 303.
Cannell, B. L., Krasnec, K. V., Campbell, K., Jones, H. I., Miller, R. D., & Stephens, N. (2013). The pathology and pathogenicity of a novel Haemoproteus spp. infection in wild Little Penguins (Eudyptula minor). Veterinary Parasitology, 197, 74–84.
Clark, N. J., Adlard, R. D., & Clegg, S. M. (2015). Molecular and morphological characterization of Haemoproteus (Parahaemoproteus) ptilotis, a parasite infecting Australian honeyeaters (Meliphagidae), with remarks on prevalence and potential cryptic speciation. Parasitology Research, 114, 1921–1928.
Clark, N. J., Clegg, S. M., & Lima, M. R. (2014). A review of global diversity in avian haemosporidians (Plasmodium and Haemoproteus: Haemosporida): new insights from molecular data. International Journal for Parasitology, 44, 329–338.
Coral, A. A., Valkiūnas, G., González, A. D., & Matta, N. E. (2015). In vitro development of Haemoproteus columbae (Haemosporida: Haemoproteidae), with perspectives for genomic studies of avian haemosporidian parasites. Experimental Parasitology, 157, 163–169.
Cosgrove, C. L., Day, K. P., & Sheldon, B. C. (2006). Coamplification of Leucocytozoon by PCR diagnostic tests for avian malaria: a cautionary note. Journal of Parasitology, 92, 1362–1365.
Cosgrove, C. L., Wood, M. J., Day, K. P., & Sheldon, B. C. (2008). Seasonal variation in Plasmodium prevalence in a population of blue tits Cyanistes caeruleus. Journal of Animal Ecology, 77, 540–548.
Darriba, D., Taboada, G. L., Doallo, R., & Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nature Methods, 9, 772.
Dimitrov, D., Zehtindjiev, P., Bensch, S., Ilieva, M., Iezhova, T. A., & Valkiūnas, G. (2014). Two new species of Haemoproteus Kruse, 1890 (Haemosporida, Haemoproteidae) from European birds, with emphasis on DNA barcoding for detection of haemosporidians in wildlife. Systematic Parasitology, 87, 135–151.
Drovetski, S. V., Aghayan, S. A., Mata, V. A., Lopes, R. J., Mode, N. A., Harvey, J. A., & Voelker, G. (2014). Does the niche breadth or trade-off hypothesis explain the abundance-occupancy relationship in avian Haemosporidia? Molecular Ecology, 23, 3322–3329.
Fallon, S. M., Ricklefs, R. E., Swanson, B. L., & Bermingham, E. (2003). Detecting avian malaria: an improved Polymerase Chain Reaction diagnostic. Journal of Parasitology, 89, 1044–1047.
Feldman, R. A., Freed, L. A., & Cann, R. L. (1995). A PCR test for avian malaria in Hawaiian birds. Molecular Ecology, 4, 663–673.
Ferraguti, M., Martínez-de la Puente, J., Muñoz, J., Roiz, D., Ruiz, S., Soriguer, R., & Figuerola, J. (2013). Avian Plasmodium in Culex and Ochlerotatus mosquitoes from southern Spain: effects of season and host-feeding source on parasite dynamics. PLoS One, 8, e66237.
Godfrey, R. D., Fedynich, A. M., & Pence, D. B. (1987). Quantification of haematozoa in blood smears. Journal of Wildlife Diseases, 23, 558–565.
González, A. D., Lotta, I. A., García, L. F., Moncada, L. I., & Matta, N. E. (2015). Avian haemosporidians from Neotropical highlands: Evidence from morphological and molecular data. Parasitology International, 64, 48–59.
Greiner, E. C., Mandal, A. K., & Nandi, N. C. (1977). Haemoproteus bennetti sp. n. and a review of the Haemoproteus from Picidae. Journal of Parasitology, 63, 651–656.
Hall, T. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95–98.
Hellgren, O., Atkinson, C. T., Bensch, S., Albayrak, T., Dimitrov, D., Ewen, J. G., et al. (2014). Global phylogeography of the avian malaria pathogen Plasmodium relictum based on MSP1 allelic diversity. Ecography, 38, 842–850.
Hellgren, O., Križanauskienė, A., Valkiūnas, G., & Bensch, S. (2007). Diversity and phylogeny of mitochondrial cytochrome B lineages from six morphospecies of avian Haemoproteus (Haemosporida: Haemoproteidae). Journal of Parasitology, 93, 889–896.
Hellgren, O., Kutzer, M., Bensch, S., Valkiūnas, G., & Palinauskas, V. (2013). Identification and characterization of the merozoite surface protein 1 (msp1) gene in a host-generalist avian malaria parasite, Plasmodium relictum (lineages SGS1 and GRW4) with the use of blood transcriptome. Malaria Journal, 12, 381.
Hellgren, O., Waldenström, J., & Bensch, S. (2004). A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium and Haemoproteus from avian blood. Journal of Parasitology, 90, 797–802.
Iezhova, T. A., Dodge, M., Sehgal, R. N. M., Smith, T. B., & Valkiūnas, G. (2011). New avian Haemoproteus species (Haemosporida: Haemoproteidae) from African birds, with a critique of the use of host taxonomic information in hemoproteid classification. Journal of Parasitology, 97, 682–694.
Ishtiaq, F., Gering, E., Rappole, J. H., Rahmani, A. R., Jhala, Y. V., Dove, C. J., et al. (2007). Prevalence and diversity of avian hematozoan parasites in Asia: a regional survey. Journal of Wildlife Diseases, 43, 382–398.
Jarvis, E. D., Mirarab, S., Aberer, A. J., Li, B., Houde, P., Li, C., et al. (2014). Whole-genome analyses resolve early branches in the tree of life of modern birds. Science, 346, 1320–1331.
Karadjian, G., Puech, M.-P., Duval, L., Chavatte, J.-M., Snounou, G., & Landau, I. (2013). Haemoproteus syrnii in Strix aluco from France: morphology, stages of sporogony in a hippoboscid fly, molecular characterization and discussion on the identification of Haemoproteus species. Parasite (Paris, France), 20, 32.
Križanauskienė, A., Hellgren, O., Kosarev, V., Sokolov, L., Bensch, S., & Valkiunas, G. (2006). Variation in host specificity between species of avian hemosporidian parasites: evidence from parasite morphology and cytochrome B gene sequences. Journal of Parasitology, 92, 1319–1324.
Križanauskienė, A., Iezhova, T. A., Sehgal, R. N. M., Carlson, J. S., Palinauskas, V., Bensch, S., & Valkiūnas, G. (2013). Molecular characterization of Haemoproteus sacharovi (Haemosporida, Haemoproteidae), a common parasite of columbiform birds, with remarks on classification of haemoproteids of doves and pigeons. Zootaxa, 3613, 85–94.
Križanauskienė, A., Pérez-Tris, J., Palinauskas, V., Hellgren, O., Bensch, S., & Valkiūnas, G. (2010). Molecular phylogenetic and morphological analysis of haemosporidian parasites (Haemosporida) in a naturally infected European songbird, the blackcap Sylvia atricapilla, with description of Haemoproteus pallidulus sp. nov. Parasitology, 137, 217–227.
Lauron, E. J., Oakgrove, K. S., Tell, L. A., Biskar, K., Roy, S. W., & Sehgal, R. N. M. (2014). Transcriptome sequencing and analysis of Plasmodium gallinaceum reveals polymorphisms and selection on the apical membrane antigen-1. Malaria Journal, 13, 382.
Li, J., Wirtz, R. A., McConkey, G. A., Sattabongkot, J., Waters, A. P., Rogers, M. J., & McCutchan, T. F. (1995). Plasmodium genus-conserved primers for species identification and quantitation. Experimental Parasitology, 81, 182–190.
Loiseau, C., Iezhova, T., Valkiūnas, G., Chasar, A., Hutchinson, A., Buermann, W., et al. (2010). Spatial variation of haemosporidian parasite infection in African rainforest bird species. Journal of Parasitology, 96, 21–29.
Mantilla, J. S., González, A. D., Valkiūnas, G., Moncada, L. I., & Matta, N. E. (2013). Description and molecular characterization of Plasmodium (Novyella) unalis sp. nov. from the Great Thrush (Turdus fuscater) in highland of Colombia. Parasitology Research, 112, 4193–4204.
Martínez, J., Martínez-de la Puente, J., Herrero, J., Del Cerro, S., Lobato, E., Rivero-de Aguilar, J., et al. (2009). A restriction site to differentiate Plasmodium and Haemoproteus infections in birds: on the inefficiency of general primers for detection of mixed infections. Parasitology, 136, 713–722.
Martinsen, E. S., Perkins, S. L., & Schall, J. J. (2008). A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): evolution of life-history traits and host switches. Molecular Phylogenetics and Evolution, 47, 261–273.
Matta, N. E., Pacheco, M. A., Escalante, A. A., Valkiūnas, G., Ayerbe-Quiñones, F., & Acevedo-Cendales, L. D. (2014). Description and molecular characterization of Haemoproteus macrovacuolatus n. sp. (Haemosporida, Haemoproteidae), a morphologically unique blood parasite of black-bellied whistling duck (Dendrocygna autumnalis) from South America. Parasitology Research, 113, 2991–3000.
Merino, S., Hennicke, J., Martínez, J., Ludynia, K., Torres, R., Work, T. M., et al. (2012). Infection by Haemoproteus parasites in four species of frigatebirds and the description of a new species of Haemoproteus (Haemosporida: Haemoproteidae). Journal of Parasitology, 98, 388–397.
Olias, P., Wegelin, M., Zenker, W., Freter, S., Gruber, A. D., & Klopfleisch, R. (2011). Avian malaria deaths in parrots, Europe. Journal of Infectious Diseases, 171, 950–952.
Ondrejicka, D. A., Locke, S. A., Morey, K., Borisenko, A. V., & Hanner, R. H. (2014). Status and prospects of DNA barcoding in medically important parasites and vectors. Trends in Parasitology, 30, 582–591.
Outlaw, D. C., & Ricklefs, R. E. (2014). Species limits in avian malaria parasites (Haemosporida): how to move forward in the molecular era. Parasitology, 2, 1–10.
Palinauskas, V., Iezhova, T. A., Križanauskienė, A., Markovets, M. Y., Bensch, S., & Valkiūnas, G. (2013a). Molecular characterization and distribution of Haemoproteus minutus (Haemosporida, Haemoproteidae): a pathogenic avian parasite. Parasitology International, 62, 358–363.
Palinauskas, V., Križanauskienė, A., Iezhova, T. A., Bolshakov, C. V., Jönsson, J., Bensch, S., & Valkiūnas, G. (2013b). A new method for isolation of purified genomic DNA from haemosporidian parasites inhabiting nucleated red blood cells. Experimental Parasitology, 133, 275–280.
Perkins, S. L., & Schall, J. J. (2002). A molecular phylogeny of malarial parasites recovered from cytochrome b gene sequences. Journal of Parasitology, 88, 972–978.
Poulin, R., & Keeney, D. B. (2008). Host specificity under molecular and experimental scrutiny. Trends in Parasitology, 24, 24–28.
Richard, F. A., Sehgal, R. N. M., Jones, H. I., & Smith, T. B. (2002). A Comparative Analysis of PCR-Based Detection Methods for Avian Malaria. Journal of Parasitology, 88, 819–822.
Richardson, D. S., Jury, F. L., Blaakmeer, K., Komdeur, J., & Burke, T. (2001). Parentage assignment and extra-group paternity in a cooperative breeder: the Seychelles warbler (Acrocephalus sechellensis). Molecular Ecology, 10, 2263–2273.
Ricklefs, R. E., & Fallon, S. M. (2002). Diversification and host switching in avian malaria parasites. Proceedings of the Royal Society B: Biological Sciences, 269, 885–892.
Ricklefs, R. E., Outlaw, D. C., Svensson-Coelho, M., Medeiros, M. C. I., Ellis, V. A., & Latta, S. (2014). Species formation by host shifting in avian malaria parasites. Proceedings of the National Academy of Sciences of the United States of America, 111, 14816–14821.
Ronquist, F., & Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19, 1572–1574.
Silveira, P., Belo, N. O., Lacorte, G. A., Kolesnikovas, C. K. M., Vanstreels, R. E. T., Steindel, M., et al. (2013). Parasitological and new molecular-phylogenetic characterization of the malaria parasite Plasmodium tejerai in South American penguins. Parasitology International, 62, 165–171.
Svensson, L. (1992). Identification Guide to European Passerines (4th ed., p. 368). Stockholm: British Trust for Ornithology.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., & Kumar, S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular biology and evolution, 30, 2725–2729.
Valkiūnas, G. (2005). Avian malaria parasites and other haemosporidia (p. 946). New: CRC PRESS Boca Raton.
Valkiūnas, G., Bensch, S., Iezhova, T. A., Križanauskienė, A., Hellgren, O., & Bolshakov, C. V. (2006). Nested cytochrome B polymerase chain reaction diagnostics underestimate mixed infections of avian blood haemosporidian parasites: microscopy is still essential. Journal of Parasitology, 92, 418–422.
Valkiūnas, G., Iezhova, T. A., Križanauskienė, A., Palinauskas, V., Sehgal, R. N. M., & Bensch, S. (2008a). A comparative analysis of microscopy and PCR-based detection methods for blood parasites. Journal of Parasitology, 94, 1395–1401.
Valkiūnas, G., Iezhova, T. A., Loiseau, C., Chasar, A., Smith, T. B., & Sehgal, R. N. M. (2008b). New species of haemosporidian parasites (Haemosporida) from African rainforest birds, with remarks on their classification. Parasitology Research, 103, 1213–1228.
Valkiūnas, G., Iezhova, T. A., Loiseau, C., & Sehgal, R. N. M. (2009). Nested cytochrome B polymerase chain reaction diagnostics detect sporozoites of hemosporidian parasites in peripheral blood of naturally infected birds. Journal of Parasitology, 95, 1512–1515.
Valkiūnas, G., Kazlauskienė, R., Bernotienė, R., Palinauskas, V., & Iezhova, T. A. (2013). Abortive long-lasting sporogony of two Haemoproteus species (Haemosporida, Haemoproteidae) in the mosquito Ochlerotatus cantans, with perspectives on haemosporidian vector research. Parasitology Research, 112, 2159–2169.
Valkiūnas, G., Križanauskienė, A., Iezhova, T. A., Hellgren, O., & Bensch, S. (2007). Molecular phylogenetic analysis of circumnuclear hemoproteids (Haemosporida: Haemoproteidae) of sylvid birds, with a description of Haemoproteus parabelopolskyi sp. nov. Journal of Parasitology, 93, 680–687.
Valkiūnas, G., Palinauskas, V., Ilgūnas, M., Bukauskaitė, D., Dimitrov, D., Bernotienė, R., et al. (2014). Molecular characterization of five widespread avian haemosporidian parasites (Haemosporida), with perspectives on the PCR-based detection of haemosporidians in wildlife. Parasitology Research, 113, 2251–2263.
Ventim, R., Morais, J., Pardal, S., Pérez-Tris, J., Ramos, J. A., & Mendes, L. (2012). Host-parasite associations and host-specificity in haemoparasites of reed bed passerines. Parasitology, 139, 310–316.
Waldenström, J., Bensch, S., Kiboi, S., Hasselquist, D., & Ottusson, U. (2002). Cross-species infection of blood parasites between resident and migratory songbirds in Africa. Molecular Biology and Evolution, 11, 1545–1554.
Waldenström, J., Bensch, S., & Ostman, O. (2004). A new nested polymerase chain reaction method very efficient in detecting Plasmodium and Haemoproteus infections from avian blood. Journal of Parasitology, 90, 191–194.
WHO. (2014). World Malaria Report 2014. Geneva. http://apps.who.int/iris/bitstream/10665/144852/2/9789241564830_eng.pdf?ua=1. Accessed 14 January 2015.
Wood, M. J., Childs, D. Z., Davies, A. S., Hellgren, O., Cornwallis, C. K., Perrins, C. M., & Sheldon, B. C. (2013). The epidemiology underlying age-related avian malaria infection in a long-lived host: the mute swan Cygnus olor. Journal of Avian Biology, 44, 12.
Zehtindjiev, P., Ivanova, K., Mariaux, J., & Georgiev, B. B. (2013). First data on the genetic diversity of avian haemosporidians in China: cytochrome b lineages of the genera Plasmodium and Haemoproteus (Haemosporida) from Gansu Province. Parasitology Research, 112, 3509–3515.
Acknowledgements
We are grateful to Karina Ivanova for her help during microscopic examination of material and to Strahil Peev, Konstantin Valchev, Stefan Braianavsci and Iovelina Bedeva for their help in the field work. This is report number 59 for Kalimok field station.
Funding
This study was supported by the European Union Structural Funds project “Postdoctoral Fellowship Implementation in Lithuania” (VP-3.1-ŠMM-01-V-02-004) and the postdoctoral project grant DO02-277 to MI from the Bulgarian Science Fund.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All applicable institutional, national and international guidelines for the care and use of animals were followed. The research was conducted under permissions 213/26.06.2009 and 433/07.03.2012 issued from the Ministry of Evironment and Waters of Bulgaria.
Rights and permissions
About this article
Cite this article
Dimitrov, D., Iezhova, T.A., Zehtindjiev, P. et al. Molecular characterisation of three avian haemoproteids (Haemosporida, Haemoproteidae), with the description of Haemoproteus (Parahaemoproteus) palloris n. sp.. Syst Parasitol 93, 431–449 (2016). https://doi.org/10.1007/s11230-016-9638-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11230-016-9638-8