Abstract
Two new species of Haemoproteus Kruse, 1890 (Haemosporida, Haemoproteidae) are described: Haemoproteus (Parahaemoproteus) homovelans n. sp. from Grey-faced Woodpecker, Picus canus Gmelin, and Haemoproteus (Parahaemoproteus) concavocentralis n. sp. recorded in Hawfinch, Coccothraustes coccothraustes (Linnaeus), both sampled in Bulgaria. The morphology of the gametocytes and their host-cells are described and mitochondrial cytochrome b (cyt b) gene sequences are generated. Haemoproteus homovelans possesses circumnuclear gametocytes lacking volutin granules. This parasite is particularly similar to Haemoproteus velans Coatney & Roudabush, 1937 also possessing circumnuclear gametocytes that are, however, overfilled with volutin. Haemoproteus concavocentralis can be readily distinguished from all described avian haemoproteids due to the presence of an unfilled concave space between the central part of advanced gametocytes and erythrocyte nucleus. Bayesian phylogenetic analyses of 40 haemosporidian cyt b lineages showed close relationships of H. concavocentralis (hHAWF2) with a group of Haemoproteus spp. possessing gametocytes that are pale-stained with Giemsa. The lineage hPICAN02 of H. homovelans clustered with parasites infecting non-passerine birds. Phylogenetic analyses support the current subgeneric classification of the avian haemoproteids and suggest that cyt b lineage hPIPUB01 (GenBank EU254552) has been incorrectly assigned to Haemoproteus picae Coatney & Roudabush, 1937, a common parasite of corvid birds (Passeriformes). This study emphasises the importance of combining molecular techniques and light microscopy in the identification and field studies of avian haemosporidian parasites. Future development of barcodes for molecular identification of haemoproteids will allow better diagnostics of these infections, particularly in veterinary studies addressing insufficiently investigated tissue pathology caused by these parasites.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Haemoproteids (Haemosporida, Haemoproteidae) represent the most diverse group among avian blood parasites, with over 140 species described to date. In the last decade, knowledge about the diversity of this group of parasites has increased rapidly due to the implementation of molecular methods for identification and more intensive research being carried out in tropical areas (Valkiūnas et al., 2008c; Bensch et al., 2009; Iezhova et al., 2010, 2011; Levin et al., 2011, 2012; Merino et al., 2012). The great genetic diversity of haemosporidian parasites has encouraged taxonomists to look more closely at parasites with similar morphology and, as a result, several new species have recently been described from temperate regions of Europe (Valkiūnas et al., 2007; Križanauskienė et al., 2010, 2012).
Species of the genus Haemoproteus Kruse, 1890 are closely related to the agents of malaria belonging to the genus Plasmodium Marchiafava & Celli, 1885 (Haemosporida, Plasmodiidae), which cause diseases in animals and humans. Sexual process and sporogony of Haemoproteus spp. take place in biting midges (Ceratopogonidae) and louse flies (Hippoboscidae) which transmit these parasites (Valkiūnas, 2005). Haemoproteid infections can cause direct and indirect pathologies affecting avian host fitness, breeding success, moulting, predation risks, and sometimes can be even lethal for the birds (Atkinson et al., 1988; Marzal et al., 2005, 2013; Ferrell et al., 2007; Møller & Nielsen, 2007). Several outbreaks associated with mortalities due to haemoproteid infections were recently described as an abortive development in non-adapted avian host (Olias et al., 2011; Cannell et al., 2013). Such abortive infections are difficult to diagnose because gametocytes are not present in the blood stream, but tissue stages can be determined using histological sections and PCR-based methods. However, identification of Haemoproteus spp. is hardly possible based only on the morphology of tissue stages (exoerythrocytic meronts) (Valkiūnas, 2005; Atkinson, 2008). Molecular approaches can help distinguish different Haemoproteus lineages (sequences), but PCR-based identification of the majority of described haemoproteid species still needs development e.g. it would be helpful for studies addressing distribution, vectors, areas of transmission and many other issues. Morphological characterisation of Haemoproteus spp. lineages is essential for better understanding of their molecular epidemiology, particularly during abortive development in avian hosts (Olias et al., 2011; Cannell et al., 2013; Levin et al., 2013). Studies addressing the development of Haemoproteus spp. barcodes are still insufficient (Valkiūnas et al., 2008a).
Haemoproteids are characterised by a long-lasting chronic stage of infection, with relatively high intensity of parasitemia that usually facilitates morphological identification of the species of Haemoproteus in wild-caught birds, which is rare in natural Plasmodium spp. infections (Valkiūnas, 2005). On the other hand, the lack of erythrocytic merogony makes the experimental transmission of haemoproteids by subinoculation of infected blood in susceptible hosts impossible. This is an obstacle for experimental research with Haemoproteus spp. Additionally, difficulties in infecting and maintaining vectors of Haemoproteus spp. in the laboratory also complicate experimental studies of haemoproteids (Valkiūnas et al., 2002; Garvin et al., 2003). Nevertheless, the enhanced possibility for molecular characterisation of Haemoproteus spp. from natural infections is essential for taxonomic research of these parasites.
Current systematics and taxonomy of haemoproteid parasites is based mainly on morphological characters of their blood stages (gametocytes) and the information available about vertebrate-host specificity of the parasites (Valkiūnas, 2005). Molecular methods have revealed great genetic diversity, with over 470 mitochondrial cytochrome b (cyt b) lineages of Haemoproteus spp. currently deposited in MalAvi database (http://mbio-serv2.mbioekol.lu.se/Malavi/index.html, see Bensch et al., 2009). Many of these lineages might represent different parasite species (Bensch et al., 2004, 2009). However, it should be noted that the rapidly increasing number of molecular studies result not only in marked accumulation of genetic information, but also in accumulation of incorrect parasite identifications in GenBank (Valkiūnas et al., 2008a). Furthermore, the majority of DNA sequences in GenBank, especially those obtained from wildlife, have been identified only to the generic level, and sometimes at even higher levels of classification. Currently, it is still unclear if closely related but distinct cyt b lineages of haemosporidians represent biological species or different levels of intraspecific variation (Iezhova et al., 2010).
Hellgren et al. (2007) suggested that cyt b lineages of Haemoproteus spp. with a genetic divergence higher than 5% are expected to be morphologically differentiated in most cases. However, this does not mean that lineages with divergence lower than 5% in this gene do not differ morphologically. There are examples of morphologically readily distinguishable species with cyt b divergences of 1% or even less (Valkiūnas et al., 2008a; Križanauskienė et al., 2010; Levin et al., 2012). Additional studies on the morphology of haemoproteids in complementary blood slides should be conducted. Development of DNA barcodes of haemosporidian species would bring new opportunities for better understanding parasite taxonomy, life history, distribution and phylogeny.
This study aims to provide morphological description and molecular characterisation of two new haemoproteid species found in wild birds from Southeast Europe. We obtained data on the morphology of the parasites and submitted their cyt b sequences to GenBank and MalAvi databases. We also (i) compare the novel morphological and sequence information with the data available for other avian haemosporidian parasites, and (ii) discuss obstacles and advantages of using PCR-based information in molecular detection and identification of haemosporidian parasites in wildlife.
Materials and methods
Study site and collection of the blood samples
Among the complementary blood slides from 12 new Haemoproteus spp. cyt b lineages found by Dimitrov et al. (2010), we identified two undescribed parasites and their cyt b lineages (hPICAN02 and hHAWF2), which exhibited readily distinguishable novel morphology. Cyt b lineage hPICAN02 was found in the blood of a single sample from the Grey-faced Woodpecker, Picus canus Gmelin, captured in Brodilovo, Southeast Bulgaria (n = 1; 42°04′N, 27°51′E). The lineage hHAWF2 was identified in six of nine examined Hawfinches, Coccothraustes coccothraustes (Linnaeus), at Kalimok Biological Station, Northeast Bulgaria (n = 7; 44°00′N, 26°26′E); River Kamchia, East Bulgaria (n = 1; 43°07′N, 26°47′E); and Brodilovo, Southeast Bulgaria (n = 1; 42°04′N, 27°51′E).
Birds were captured by mist nets in deciduous forest habitats during spring–summer in 2007–2009. From each bird, several blood drops were taken and immediately used for preparation of two or three blood smears. Approximately 25 μl of blood was collected for DNA analysis in 500 μl SET buffer (0.15 M NaCl, 0.05 M Tris, 0.001 M EDTA, pH 8.0) and stored at ambient temperature in the field and later at −20°C in the laboratory. The blood slides were air-dried, fixed in absolute methanol for one minute in the field and stained with Giemsa solution in the laboratory, as described by Valkiūnas et al. (2008b).
Examination of the blood films and parasite morphology
Blood films were examined for 10–15 min at low magnification (×400), and then at least 100 fields were studied at high magnification (×1000), as described by Valkiūnas et al. (2008b). Olympus BX51 and Zeiss Axio Imager M2 light microscopes equipped with Olympus DP12 and ProgRes c10 plus digital cameras and imaging software DPSOFT and ProgRes CapturePro v2.8.0 were used to examine the blood slides and prepare illustrations. We used calibrated Image-Pro Plus software to take measurements. To identify the parasites, we used morphological and morphometric features according to Valkiūnas (2005). Additionally, to confirm the morphological identification of the new species, we compared our material with type-specimens of morphologically or genetically similar parasites, i.e. Haemoproteus velans Coatney & Rudabush, 1937 deposited at Queensland Museum, Queensland, Australia (accession number 45241) (Fig. 1Q–T) and of Haemoproteus vacuolatus Valkiūnas, Iezhova, Loiseau, Chasar, Smith & Sehgal, 2008 deposited at the Nature Research Centre, Vilnius, Lithuania (accession number 42415 NS) (Fig. 2Q–T).
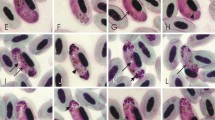
Gametocytes of Haemoproteus homovelans n. sp. from the blood of Grey-faced Woodpecker Picus canus (A–P) and Haemoproteus velans from the blood of Northern Flicker Colaptes auratus (Q–T): A–D, young gametocytes; E–L, Q, S, macrogametocytes; M–P, R, T, microgametocytes. Short simple arrows indicate unfilled spaces between gametocyte and erythrocyte nucleus; long simple arrows indicate volutin granules; short triangle arrows indicate parasite nuclei; long triangle arrows indicate unfiled space resembling vacuoles at erythrocyte poles; triangle arrowheads indicate pigment granules. Giemsa-stained thin blood films. Scale-bar: 10 μm
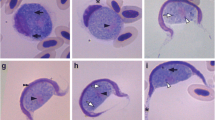
Gametocytes of Haemoproteus concavocentralis n. sp. from the blood of Hawfinch Coccothraustes coccothraustes (A–P) and of Haemoproteus vacuolatus from the blood of Yellow-whiskered Greenbul Andropadus latirostris (Q–T): A, young gametocyte; B–L, Q, S, mature macrogametocytes; M–P, R, T, microgametocytes. Short simple arrows indicate unfilled spaces between gametocyte and erythrocyte nucleus; short triangle arrows indicate parasite nuclei; long triangle arrows indicate prominent single vacuoles; triangle arrowheads indicate pigment granules. Giemsa-stained thin blood films. Scale-bar: 10 μm
Intensity of infection was estimated as a percentage by actual counting the number of parasites per 1,000 red blood cells or per 10,000 red blood cells in light infections (i.e. <0.1%), as recommended by Godfrey et al. (1987).
Extraction of DNA, PCR and sequencing
DNA was extracted by a standard phenol-chloroform protocol (Sambrook et al., 2002) and quantified by spectrophotometer (Spectrome Genesys 10 Bio). Diluted to 25 ng/μl, the total DNA was used as a template in PCR assays for detection of the parasites. We used primers and temperature profiles as described in Hellgren et al. (2004). The method consists of a nested PCR assay that amplifies a part of the parasite mitochondrial cyt b gene in two steps, first an initial PCR (primers HaemNFI/HaemNR3; 570 bp excluding primers) that amplifies cyt b lineages of Haemoproteus, Plasmodium and Leucocytozoon Berestneff, 1904, followed by a second step that amplifies lineages of the genera Plasmodium and Haemoproteus (primers HaemF/HaemR2; 479 bp excluding primers). By amplifying the parasite DNA in two PCRs the sensitivity of the screening is increased (Hellgren et al., 2004; Waldenström et al., 2004). Samples that showed positive amplification on a 2% agarose gel were sequenced using the procedures described by Bensch et al. (2000). Amplified fragments were sequenced in both directions with primers HaemF and HaemR2. We used a dye terminator cycle sequencing kit (Big Dye Terminator Ready Reaction Mix, Applied Biosystems Inc., Foster City, CA, USA) and the samples were loaded on an ABI PRISM ™ 3100 sequencing robot (Applied Biosystems, Florida, USA).
Sequences were edited and aligned using BioEdit (Hall, 1999). The appearance of double peaks in the sequence was considered a co-infection. Co-infection was only detected in one Hawfinch; parasites from this bird were excluded from the morphological description.
Phylogenetic analysis
In total, 40 cyt b lineages were used in the final alignment for Bayesian analysis. We used 36 sequences of haemosporidian parasites (30 for Haemoproteus spp., five for Plasmodium spp. and one for Leucocytozoon spp.), where species were microscopically identified and the sequences deposited in GenBank and MalAvi. Additionally, we used four cyt b lineages of Haemoproteus spp. which were reported in birds from order Piciformes in MalAvi database. The cyt b lineage of Leucocytozoon schoutedeni Rodhain, Pons, Vandenbranden & Bequaert, 1913, was used as an outgroup.
Bayesian phylogeny was constructed using MrBayes version 3.1.2 (Ronquist & Huelsenbeck, 2003). We used the General Time Reversible model including invariable sites and variation among sites (GTR+I+G) selected by the software MrModeltest 2.3 (Nylander, 2004; software available from http://www.abc.se/~nylander/). Gaps and missing data in the alignment were discarded prior to analyses. Two simultaneous runs were conducted with a sample frequency of every 100th generation over 10 million generations; 25% of the trees were discarded as ‘burn-in’ period. The remaining trees were used to construct a majority rule consensus tree. The phylogenies were visualised using Tree View 1.6.6. (software available from http://evolution.genetics.washington.edu/phylip/software.html).
The sequence divergence between different lineages was calculated with Jukes-Cantor model of substitution, with all substitutions weighted equally, implemented in the programme MEGA version 5 (Tamura et al., 2011) in which pairwise deletion was selected. The pairwise deletion option removed from the analysis sites containing missing data or alignment gaps.
MalAvi reference codes (see Bensch et al., 2009) and GenBank accession numbers of all cyt b lineages are given in Fig. 3.
Bayesian phylogeny of 40 cytochrome b (cyt b) gene lineages of positively identified species of avian pigment-forming haemosporidian parasites. Cyt b lineage of Leucocytozoon schoutedeni was used as an outgroup. New species are given in bold. Codes of the lineages (according MalAvi database, http://mbio-serv2.mbioekol.lu.se/Malavi/index.html) are given before GenBank accession numbers and species names of parasites. Nodal support values near branches indicate posterior clade probabilities. Light grey boxes indicate groups of closely related species of haemoproteids: A, Parahaemoproteus, B, Haemoproteus. Dark grey boxes indicate groups of closely related species within subgenus Parahaemoproteus: C, pale-stained parasites, D, non-passeriform parasites
Haemoproteus ( Parahaemoproteus ) homovelans n. sp.
Type-host: Grey-faced Woodpecker Picus canus Gmelin (Piciformes, Picidae). Female, adult bird caught on 13 July 2008.
Type-locality: Mixed deciduous forests of beech and oak, shrubs and grasslands near River Veleka, Strandja Nature Park, in the region of village of Brodilovo, Burgas District, Bulgaria (42°04′40″N, 27°51′28″E; 21 m above sea level).
Site of infection: Mature erythrocytes; no other data.
Prevalence: One of one bird.
Type-specimens: Hapantotype (accession number 48757NS, ex Picus canus; parasitemia intensity approximately 0.1 %, 13 July 2008, Strandja Nature Park, collected by M. Ilieva) is deposited in the Institute of Ecology of Nature Research Centre, Vilnius, Lithuania. Parahapantotypes are deposited in the U. S. National Parasite Collection, Beltsville, Maryland, USA (accession number USNPC 107271.00) and the Queensland Museum, Queensland, Australia (accession number G465688). In the hapantotype one trypomastigote of Trypanosoma sp. (Kinetoplastida: Trypanosomatidae Doflein, 1901) was recorded.
Distribution: Based on MalAvi database this parasite and its lineage hPICAN02 have been recorded only in the type-host in Bulgaria.
Representative DNA sequence: Mitochondrial cyt b gene, lineage hPICAN02 (479 bp, GenBank accession no. GU085195).
Etymology: The species name refers to the morphological and morphometric similarity of the new species with Haemoproteus velans Coatney & Roudabush, 1937, a parasite infecting birds of the order Piciformes.
Description (Fig. 1A–P)
Young gametocytes (Fig. 1A–D). Develop in mature erythrocytes. Earliest forms seen anywhere in infected erythrocytes, predominantly occupy sub-central or polar position in infected erythrocytes; often rod-like or comma-like shaped (Fig. 1A, D). Volutin granules absent. Pigment granules small (<0.5 μm), poorly visible. Outline even or amoeboid (Fig. 1A–D). Young gametocytes do not displace erythrocyte nuclei laterally (Fig. 1A–D).
Macrogametocytes (Fig. 1E–L). Grow along the erythrocyte nucleus, enclosing the nucleus with ends; gametocyte extends longitudinally along the nucleus, with one end often surrounding nucleus asymmetrically in relation to erythrocyte poles (Fig. 1E, F). Growing forms often (in approximately 80% of gametocytes) do not touch erythrocyte nucleus, resulting in presence of more or less evident unfilled space (a cleft), which is usually seen between parasite and erythrocyte nucleus (Fig. 1F–I); this cleft disappears in fully-grown gametocytes. Outline becomes more amoeboid as gametocyte grows (Fig. 1G, H). Some mature gametocytes (approximately 5%) do not encircle erythrocyte nucleus, but markedly displace it laterally (Fig. 1J). Fully-grown gametocytes encircle nucleus completely and occupy all cytoplasmic space in erythrocyte. Unfiled space resembling vacuole sometimes seen between circumnuclear gametocytes and poles of infected erythrocyte (Fig. 1K). Cytoplasm stains dark blue, heterogeneous in appearance. Parasite nucleus compact, oval or sometimes of irregular shape, central or sub-central in position. Nucleolus not seen. Pigment granules small, <0.5 μm, usually roundish in growing gametocytes (Fig. 1E–H); as gametocyte develops, they assume a rod-like shape and reach medium size, 0.5–1.0 μm. Pigment granules randomly scattered throughout cytoplasm. Volutin granules absent. Infected erythrocytes hypertrophied in length, width and area (Table 1).
Microgametocytes (Fig. 1M–P). General configuration as for macrogametocytes with the usual haemosporidian sexual dimorphic characters. Parasite nucleus stains pink, diffuse, of variable form and often with unclear margins. Volutin granules absent. Number of pigment granules lower than in macrogametocytes (Table 1).
Remarks
Among parasites of birds belonging to the Piciformes, gametocytes of H. homovelans n. sp. are particularly similar to those of H. velans and H. cornuata Bennett & Nandi, 1981 (Valkiūnas 2005). The new species is most similar to H. velans due to patterns of growth and predominant circumnuclear fully-grown gametocytes; this is reflected in the species name. Haemoproteus velans can be readily distinguished from H. homovelans due presence of numerous prominent volutin granules and their clearly distinct clumps both in macro- and microgametocytes (compare Fig. 1E–P with Fig. 1Q–T). Additionally, pigment granules are markedly masked and poorly visible in gametocytes of H. velans, but they are readily visible in H. homovelans (see Fig. 1E–P). Patterns of growth of gametocytes of H. cornuata are also similar to H. homovelans, but circumnuclear gametocytes do not develop in the former species.
Haemoproteus ( Parahaemoproteus ) concavocentralis n. sp.
Type-host: Hawfinch Coccothraustes coccothraustes (Linnaeus) (Passeriformes, Fringillidae). The type-specimen is from a male juvenile bird caught on 20 June 2007 at Kalimok Biological Station. Parahapantotype material came from a male adult bird sampled on 18 May 2009 at Strandja Nature Park, near the village of Brodilovo, Burgas District, Bulgaria (42°04′40″N, 27°51′28″E; 21 m above sea level), collected by D. Dimitrov.
Type-locality: Planted forest of black locust (Robinia pseudoacacia) and ash trees (Fraxinus spp.) at the territory of the Kalimok Biological Station, region of the village of Nova Cherna, Silistra District, Bulgaria (44°00′40″N, 26°26′14″E, 18 m above sea level).
Site of infection: Mature erythrocytes; no other data.
Prevalence: In the type-locality, the prevalence was 5 out of 7 birds (71.4%); overall prevalence was 6 out of 9 birds (66.7%) in all samples collected in Bulgaria.
Type-specimens: Hapantotype (accession number 48756NS, ex Coccothraustes coccothraustes, 20 June 2007, Kalimok Biological Station, collected by D. Dimitrov, intensity of parasitemia approximately 0.3 %) is deposited in the Institute of Ecology of Nature Research Centre, Vilnius, Lithuania. Parahapantotypes are deposited in the U. S. National Parasite Collection, Beltsville, Maryland, USA (accession number USNPC 107272.00), Queensland Museum, Queensland, Australia (accession number G465689) and Institute of Biodiversity and Ecosystem Research, Sofia (accession number Protozoa-2013-0003). Co-infection with Leucocytozoon spp. (similar to L. fringillinarum Woodcock, 1910 and L. dubreuili Mathis & Léger, 1911) is present in the hapantotype material. Co-infection with Haemoproteus tartakovskyi Valkiūnas, 1986 is present in parahapantotypes.
Distribution: Based on MalAvi database this parasite and its lineage hHAWF2 have been recorded only in Bulgaria. It is probably restricted to the Hawfinch populations in their range in Southeast Europe.
Representative DNA sequence: Mitochondrial cyt b gene lineage hHAWF2 (479 bp, GenBank accession no. GQ396708).
Etymology: The species name reflects the concave shape of the central part of advanced gametocytes (Fig. 2B–G, M–N), the most distinctive feature of H. concavocentralis.
Description (Fig. 2A–P)
Young gametocytes (Fig. 2A). Only one young gametocyte was found in a mature erythrocyte in the type-material (Fig. 2A).
Macrogametocytes (Fig. 2B–L). Develop in mature erythrocytes. Cytoplasm pale-blue, heterogeneous in appearance, volutin granules absent. Outline even or slightly irregular at the terminal gametocyte edges. Several vacuoles or vacuole-like spaces of different size and position present in the majority of gametocytes. Gametocyte grows along the nucleus of infected erythrocyte; encloses the nucleus with its ends, but does not encircle it completely. Growing gametocyte usually adheres to erythrocyte envelope but does not touch erythrocyte nucleus resulting in presence of a broad cleft between gametocyte and erythrocyte nucleus. Cleft broader in gametocyte centre and narrower at gametocyte edges (Fig. 2B–G) resulting in a concave shape of the growing parasite (Fig. 2D–G). As parasite develops, gametocyte fills up space on the poles of erythrocyte and then gradually adheres to erythrocyte nucleus close to its poles (Fig. 2D–G). A clear unfilled space between gametocyte and erythrocyte nucleus is maintained at this development stage (Fig. 2F, G). Thin portion of cytoplasm appears close to erythrocyte nucleus; it moves along the nucleus (Fig. 2C, G), gradually increases in size and finally the cleft between the parasite and the nucleus disappears. Fully-grown gametocyte completely fills up the space between erythrocyte nucleus and envelope closely appressing erythrocyte nucleus and markedly displacing it laterally (Fig. 2L). Parasite nucleus relatively small (Table 1), of variable form and position, usually sub-central and often located close to erythrocyte envelope. Nucleolus not seen. Pigment granules roundish or oval, small (<0.5 μm) and medium (0.5–1.0 μm) in size, usually randomly scattered throughout cytoplasm. Influence of growing gametocyte on infected erythrocyte not pronounced. Fully-grown gametocyte markedly displaces erythrocyte nucleus laterally and hypertrophies infected cell in length and area (Table 1).
Microgametocytes (Fig. 2M–P): General configuration and pattern of growth as macrogametocyte with usual haemosporidian sexually dimorphic characters. Parasite nucleus stains pale-pink, diffuse, of variable form and often with unclear margins.
Remarks
Data on the pattern of development and the position of gametocytes in infected erythrocytes are essential for identification of this species. A readily distinguishable space between the growing gametocyte and the erythrocyte nucleus is a unique character for H. concavocentralis n. sp. This space gives gametocytes a concave shape (Fig. 2D–G) and this is reflected in the species name. Due to this character H. concavocentralis can be readily distinguished from all described species of haemoproteids of passeriform birds.
Based on the phylogenetic analysis (Fig. 3, clade C) H. concavocentralis is closely related to H. vacuolatus. The existence of one clear circular vacuole, which is present in the cytoplasm of each advanced macrogametocyte is a characteristic feature of H. vacuolatus (Fig. 2Q–T). Such vacuoles were not seen in H. concavocentralis. Additionally, H. concavocentralis can be readily distinguished from H. vacuolatus due to (i) unique patterns of growth (see above), and (ii) marked vacuolisation of the cytoplasm.
During identification of H. concavocentralis, attention also should be paid to the pale-blue staining of the cytoplasm in macrogametocytes. This character helps to distinguish this parasite from H. tartakovskyi and Haemoproteus fringillae (Labbé, 1894), common parasites of the Hawfinch.
Morphological and molecular examination of the blood samples and phylogenetic analysis
Morphological and molecular examination of the samples used for designation of the type-specimens of H. homovelans (hPICAN02) and H. concavocentralis (hHAWF2) showed the presence of single infections. This was confirmed by screening of entire blood smears and sequencing of the PCR products from both directions. The hapantotype material used for the morphological description of the lineage hHAWF2 was from a bird with co-infection of H. concavocentralis and Leucocytozoon sp. There are different primers used for molecular identification of parasites belonging to Leucocytozoon spp. that were not applied in this study and in the present case only the lineage hHAWF2 was detected.
A second bird was co-infected with H. concavocentralis and H. tartakovskyi; blood films from this bird were used for taking measurements and were deposited as parahapantotype material. The haemoproteid infection in this bird comprised mainly fully grown mature gametocytes of H. concavocentralis but also light parasitemia of H. tartakovskyi (0.02%). PCR amplification did not detect the infection with H. tartakovskyi; only the lineage hHAWF2 of H. concavocentralis was present on the electrophoregrams. However, both parasite species were readily distinguishable under light microscope and we easily selected mature gametocytes of H. concavocentralis (hHAWF2) to take measurements.
We confirmed Haemoproteus spp. infections by light microscopy examination in four of six Hawfinches infected with lineage hHAWF2. In two of these birds, we recorded only Leucocytozoon sp. and Plasmodium sp. on the blood slides. Of the remaining four birds, three were co-infected with H. concavocentralis and H. tartakovskyi and one carried an additional unidentified Plasmodium infection. However, no co-infections in these birds were detected by PCR.
The genetic divergence between cyt b lineages of H. concavocentralis (hHAWF2) and H. tartakovskyi (lineages hHAWF1 and hSISKIN1) was relatively high, i.e. 4.5 and 4.8% respectively; both parasites were located rather far from each other in the phylogenetic tree (Fig. 3). Although the lineage hHAWF2 clustered together with the lineage hANLA1 of H. vacuolatus (Fig. 3, clade C), the genetic divergence between them (1.8%) was the same as the divergence between the lineages hPFC1 of H. pallidus Valkiūnas & Iezhova, 1991 and hSYAT03 of H. pallidulus Križanauskienė, Pérez-Tris, Palinauskas, Hellgren, Bensch & Valkiūnas, 2010. However, the morphological features of H. vacuolatus were more similar to H. concavocentralis (hHAWF2) than to both H. pallidus (hPFC1) and H. pallidulus (hSYAT03).
The intensity of haemoproteid infection was approximately 0.1% in the type-preparations of H. homovelans; all stages of gametocyte development were present. Cyt b lineage hPICAN02 formed a well-supported clade (with nodal support value of 0.98) with other lineages parasitising birds of the Columbiformes, Strigiformes and Piciformes (Fig. 3, clade D). The most closely related cyt b lineage was hMODO1 of H. sacharovi Novy & MacNeal, 1904 with genetic divergence of 2.3% between these lineages. However, both parasites exhibit significant morphological differences.
Discussion
Species identification of avian haemosporidian parasites is a difficult task in wildlife due to the following main reasons: (i) common light natural infections that might be a consequence of the methods of bird sampling (mist-netting and trapping) that predominantly select actively moving, healthy individuals with light parasitemia; (ii) common co-infections of parasites belonging to the same or different genera; (iii) insufficient training in taxonomy of researchers to identify species; and (iv) application of different protocols for molecular identification and labelling of the parasite lineages by different researches. These difficulties should be addressed because studies on parasite diversity and species richness are of great importance for further analyses on host–parasite interactions such as host specificity, pathogenicity, distribution and co-evolution.
Traditional taxonomy and systematics of haemosporidian parasites are predominantly based on morphological features of the blood stages in the vertebrate hosts observed under light microscopy (Garnham, 1966; Valkiūnas, 2005). The implementation of molecular methods for detection and identification of haemosporidian parasites (Bensch et al., 2000, 2004; Perkins & Schall, 2002; Ricklefs & Fallon, 2002; Beadell et al., 2004; Hellgren et al., 2004; Waldenström et al., 2004; Martinsen et al., 2008) has revealed remarkable genetic diversity among these hematozoans and has markedly supplemented traditional taxonomy. Further taxonomic research, focused on the linkages between different parasite species and their genetic lineages is essential for parasite barcoding (Martinsen et al., 2006, 2008; Hellgren et al., 2007; Valkiūnas et al., 2007; Križanauskienė et al., 2006, 2010, 2012).
Recent morphological descriptions of new haemosporidian species are usually accompanied by DNA sequence information and phylogenetic hypotheses about the relationships of the novel species with already described parasites. In this study, H. homovelans n. sp. and H. concavocentralis n. sp. were attributed to the subgenus Parahaemoproteus because cyt b lineages of these parasites cluster (Fig. 3, clade A) with the lineages transmitted by biting midges (Ceratopogonidae) (Križanauskienė et al., 2013). The lineages of the hippoboscid-transmitted species Haemoproteus columbae Kruse, 1890, Haemoproteus multipigmentatus Valkiūnas, Santiago-Alarcon, Levin, Iezhova, & Parker, 2010, Haemoproteus iwa Work & Rameyer, 1996 and Haemoproteus jenniae Levin, Valkiūnas, Iezhova, O’Brien & Parker, 2012 which belong to the subgenus Haemoproteus, are positioned in a separate clade with high nodal support (clade B in Fig. 3). This group appears to be a sister clade to the species of subgenus Parahaemoproteus, as also revealed by Iezhova et al. (2011) and Križanauskienė et al. (2013). According to recent phylogenetic studies (Santiago-Alarcon et al., 2010; Iezhova et al., 2011; Levin et al., 2012; Križanauskienė et al., 2013), Parahaemoproteus and Haemoproteus are closely related sister groups of avian haemoproteids, so the traditional subgeneric classification of avian haemoproteids (Levine & Campbell, 1971; Valkiūnas, 2005) remains valid.
In our samples, we reported three individual birds with co-infection with H. concavocentralis and H. tartakovskyi, but none with infection of H. fringillae. It is important to note that co-infection was not detected in the same birds by PCR. Hence, we confirm the results from previous studies that have applied general PCR protocols and often underestimated co-infections in the wild birds (Valkiūnas et al., 2006; Martínez et al., 2009). This is an obstacle for studies that rely solely on PCR-based methods for assessment of haemosporidian diversity. PCR screening alone is not advised in studies of haemosporidian parasites, particularly in such species as the Hawfinch because co-infections were reported in over 80% of infected individuals of this species in Europe (Valkiūnas et al., 2003).
Important advantages of the PCR-based diagnostics in comparison to light microscopy diagnostics of haemosporidian parasites are: (i) high sensitivity to detect light infections; (ii) accurate identification of light chronic infections; (iii) opportunities to identify species lineages based on tissue stages; (iv) opportunities to construct phylogenetic hypotheses; and (v) possibilities to apply molecular identification which can be used easily by non-specialists in taxonomy of haemosporidians (Bensch et al., 2000, 2004; Hellgren et al., 2004; Waldenström et al., 2004). However, all currently used general PCR protocols (Perkins & Schall, 2002; Hellgern et al., 2004; Waldenström et al., 2004; Martinsen et al., 2008) usually markedly underestimate co-infections which are common in wildlife and even predominate in some bird populations (Valkiūnas et al., 2003, 2006; Martínez et al., 2009). It is worth mentioning that PCR-based methods also detect circulating haemosporidian sporozoites, whose fate is unclear in the birds (Valkiūnas et al., 2009). Because PCR may detect sporozoites and also abortive haemosporidian infections (Cannell et al., 2013; Levin et al., 2013), conclusions regarding the specificity and distribution of haemosporidian lineages in wildlife should be made with caution. In order to be accepted as a successfully developing lineage of haemosporidians, such PCR-based information should be supported with the detection of blood stages (gametocytes) of the parasites. There is a crucial need of a synthesis of the information provided by the tools of traditional parasitology and molecular biology, particularly in the field research of blood parasites.
Vector species of H. homovelans and H. concavocentralis need to be identified, but the close phylogenetic relationships of their cyt b lineages with the lineages of ceratopogonid-transmitted parasites (Fig. 3, clade A) show that the biting midges (Culicoides spp.) should be suspected as possible vectors. It is worth mentioning that the recent molecular survey of whole bodies of biting midges recorded the lineage hHAWF2 in Culicoides circumscriptus (Kieffer) indicating that this midge could be a vector of H. concavocentralis (see Bobeva et al., 2013). The genetic distances among the cyt b lineages of the two new species described here and the lineages of hippoboscid-transmitted parasites are more than 10%; it is unlikely that these parasites could be transmitted by hippoboscid flies.
Based on gametocyte morphology, H. homovelans n. sp. is especially similar to H. velans which has been recorded in the same avian host, the Grey-faced Woodpecker, but at a distant geographical location, Southeast Asia (Thailand) (Greiner et al., 1977). The new species can be readily distinguished from H. velans due to several morphological features, which are summarised in the Remarks. It is worth mentioning that H. velans has also been reported from the Syrian Woodpecker, Dendrocopus syriacus (Hemprich & Ehrenberg), in South Bulgaria (Shurulinkov & Golemansky, 2002). However, the latter study was not accompanied by illustrations or sequence information, so the report of H. velans in Syrian Woodpecker requires confirmation. In the notes to the description of H. velans, Shurulinkov & Golemansky (2002) mentioned that gametocytes observed in their material lack volutin granules and differ from the original description of H. velans. From this point of view, the haemoproteid species from the Syrian Woodpecker is more similar to H. homovelans. Collection of additional material for molecular characterisation of H. velans from its type-host, the Northern Flicker, Colaptes auratus (Linnaeus), and from the type-locality (North America, Nebraska, USA) is required for further phylogenetic comparison between H. homovelans and H. velans.
This study provides the first information for molecular characterisation of haemoproteid parasites of birds belonging to the Piciformes; cyt b lineage hPICAN02 can be used for molecular diagnostics of H. homovelans. Martinsen et al. (2008) identified one Haemoproteus cyt b lineage (hPIPUB01, GenBank accession no. EU254552) isolated from the Downy Woodpecker, Picoides pubescens (Linnaeus), as Haemoproteus picae Coatney & Roudabush, 1937, a widespread parasite of birds from the family Corvidae of the Passeriformes (Valkiūnas, 2005). According to our phylogeny, cyt b lineage hPIPUB01 appeared in clade D (Fig. 3), with genetic divergence of 3.2% between the lineage hPICAN02 of H. homovelans, a parasite of non-passeriform birds. It is probable that this is an incorrectly developed DNA barcode because the authors did not provide morphological evidence for the identification of this lineage. Further studies are needed for identification of haemoproteids of the Downy Woodpecker.
The clearly visible unfilled concave space between the growing gametocytes and the erythrocyte nuclei (Fig. 2B–G) is a unique morphological feature of H. concavocentralis; it can be used for distinguishing this parasite from all other described species of haemoproteids parasitising birds (Valkiūnas, 2005). It was unexpected to find a new species of Haemoproteus in a well-studied avian host such as the Hawfinch (Peirce, 1981; Valkiūnas et al., 2003; Palinauskas et al., 2007), particularly at as high prevalence and with so readily distinguishable morphology. This is probably due to the restricted area of distribution of this parasite in Southeast Europe, a suggestion that needs confirmation from larger spatial study of different European populations of Hawfinches. Haemoproteus tartakovskyi and H. fringillae are the most common haemoproteid parasites in C. coccothraustes (see Valkiūnas et al., 2003) but gametocytes of both species are markedly different from those of H. concavocentralis (see Valkiūnas, 2005) (see Remarks). Furthermore, the type-host of H. concavocentralis, the Hawfinch is mainly sedentary widespread bird on Balkan Peninsula (BWPi 2.0, 2006) suggesting that this infection is endemic in the region.
Lineage hHAWF2 clustered together with four so-called pale-stained parasites: Haemoproteus pallidus, H. pallidulus, H. minutus Valkiūnas & Iezhova, 1992 and H. vacuolatus (Fig. 3, clade C). In comparison with other Haemoproteus spp., the cytoplasm of macrogametocytes of these species stains pale with Giemsa, probably due to the relatively low density of some cytoplasmic structures in the macrogametocytes (endoplasmic reticulum, ribosomes, osmiophilic bodies) (Valkiūnas, 2005). Interestingly, all these parasites are members of the same strongly-supported clade (Fig. 3, clade C) with a small mean genetic divergence (1.3%) among their cyt b lineages. However, blood stages of these parasites are readily distinguishable from each other (Valkiūnas, 2005; Valkiūnas et al., 2008a; Križanauskienė et al., 2010; Palinauskas et al., 2013) and possess one common feature, i.e. the cytoplasm of both macro- and microgametocytes assume similar pale-blue staining. We observed this character clearly on the slides with co-infection of H. concavocentralis and H. tartakovskyi. This is a characteristic feature of all parasites from clade C (Fig. 3) but is a rare feature in avian haemoproteids.
Parasites of the genus Haemoproteus have been often considered as relatively benign, with minimal or no effect on avian hosts (Bennett et al., 1993; Gilman et al., 2007). This is probably true in evolutionary adapted host–parasite systems (Valkiūnas, 2005). However, mortality and severe pathologies were recorded during abortive Haemoproteus spp. development at tissue stage in non-adapted hosts, for instance, captive parrots in Europe and Little Penguins, Eudyptula minor (J. R. Forster), in Australia (Olias et al., 2011; Cannell et al., 2013). It remains unclear how often avian haemosporidians cause lethal disease in wildlife (Levin et al., 2013). Unfortunately, it is difficult or even impossible to use the morphology of the exoerythrocytic tissue stages alone for accurate diagnosis of Haemoproteus spp. infections (Valkiūnas, 2005; Atkinson, 2008). Due to difficulties in identification of haemosporidian species from histological preparations of tissues, recent studies (Olias et al., 2011; Cannell et al., 2013) applied PCR-based techniques to tissue samples and reported cyt b lineages of Haemoproteus spp. in dead birds.
In conclusion, the present study describes two new species, develops DNA barcodes for their molecular diagnostics, and identifies their phylogenetic relationships within subgenus Parahaemoproteus. This study also emphasises the value of molecular and microscopy methods in the identification of avian haemosporidians and descriptions of new species. The development of barcodes for molecular diagnostics of avian haemosporidian parasites is an urgent task, particularly because of the huge biodiversity of these avian pathogens, the common co-infections in the wildlife, and the severe pathology caused by some species. Importantly, the molecular diagnostics of haemoproteid parasites is becoming an essential tool in veterinary studies, particularly in determining parasites in tissue samples from necropsies of birds (Olias et al., 2011; Cannell et al., 2013). Identification of haemosporidian parasites, which have abortive development in avian hosts, but cause lethal diseases, is a new field of interest that reveals the pathogenic potential of some haemoproteid species that have been traditionally considered to be relatively benign.
References
Atkinson, C. T. (2008). Haemoproteus. In: Atkinson, C. T., Thomas, N. J., & Hunter B. C. (Eds) Parasitic Diseases of Wild Birds. Ames, Iowa, USA: Wiley-Blackwell, pp. 13–35.
Atkinson, C. T., Forrester, D. J., & Greiner, E. C. (1988). Pathogenicity of Haemoproteus meleagridis (Haemosporina: Haemoproteidae) in experimentally infected domestic turkeys. Journal of Parasitology, 74, 228–239.
Beadell, J. S., Gering, E., Austin, J., Dumbacher, J. P., Peirce, M. A., Pratt, T. K., Atkinson, C., & Fleischer, R. C. (2004). Prevalence and differential host specificity of two avian blood parasite genera in the Australo-Papuan region. Molecular Ecology, 13, 3829–3844.
Bennett, G. F., Peirce, M. A., & Ashford, R. W. (1993). Avian haematozoa: Mortality and pathogenicity. Journal of Natural History, 27, 993–1001.
Bennett, G. F., & Campbell, A. G. (1972). Avian Haemoproteidae. III. Description of Haemoproteus fallisi n. sp. and a review of the haemoproteids of the family Turdidae. Canadian Journal of Zoology, 50, 1269–1275.
Bensch, S., Stjernman, M., Hasselquist, D., Ostman, O., Hansson, B., Westerdahl, H., & Pinheiro, R. T. (2000). Host specificity in avian blood parasites: a study of Plasmodium and Haemoproteus mitochondrial DNA amplified from birds. Proceedings of the Royal Society B, 267, 1583–1589.
Bensch, S., Pérez-Tris, J., Waldenström, J., & Hellgren, O. (2004). Linkage between nuclear and mitochondrial DNA sequences in avian malaria parasites: multiple cases of cryptic speciation? Evolution, 58, 1617–1621.
Bensch, S., Hellgren, O., & Pérez-Tris, J. (2009). MalAvi: a public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Molecular Ecology Resources, 9, 1353–1358.
Bobeva, A. B., Zehtindjiev, P., Bensch, S., & Radrova, J. (2013). A survey of biting midges of the genus Culicoides Latreille, 1809 (Diptera: Ceratopogonidae) in NE Bulgaria, with respect to transmission of avian haemosporidians. Acta Parasitolgica 58, 585–591.
BWPi 2.0. (2006). The Birds of the Western Palaearctic Interactive, 2006 Upgrade. DVD Birdguides, Shrewsbury.
Cannell, B. L., Krasnec, K. V., Campbell, K., Jones, H. I., Miller, R. D., & Stephens, N. (2013). The pathology and pathogenicity of a novel Haemoproteus spp. infection in wild Little Penguins (Eudyptula minor). Veterinary Parasitology, 197, 74–84.
Dimitrov, D., Zehtindjiev, P., & Bensch, S. (2010). Genetic diversity of avian blood parasites in SE Europe: Cytochrome b lineages of the genera Plasmodium and Haemoproteus (Haemosporida) from Bulgaria. Acta Parasitologica, 55, 201–209.
Ferrell, S. T., Snowden, K., Marlar, A. B., Garner, M., & Lung, N. P. (2007). Fatal hemoprotozoal infections in multiple avian species in a zoological park. Journal of Zoo and Wildlife Medicine, 38, 309–316.
Garnham, P. C. C. (1966). Malaria parasites and other Haemosporidia. Oxford: Blackwell Scientific Publications.
Garvin, M. C., Homer, B. L., & Greiner, E. C. (2003). Pathogenicity of Haemoproteus danilewskyi, Kruse, 1890, in blue jays (Cyanocitta cristata). Journal of Wildlife Diseases, 39, 161–169.
Gilman, S., Blumstein, D. T., & Foufopoulos, J. (2007). The effect of hemosporidian infections on white-crowned sparrow singing behaviour. Ethology, 113, 437–445.
Godfrey, R. D., Fedynich, A. M., & Pence, D. B. (1987). Quantification of hematozoa in blood smears. Journal of Wildlife Diseases, 23, 558–565.
Greiner, E. C., Mandal, A. K., & Nandi, N. C. (1977). Haemoproteus bennetti sp. n. and a review of the haemoproteids from the Picidae (Woodpeckers). Journal of Parasitology, 63, 651–656.
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor v. 5.0.9. Nucleic Acids Symposium, 41, 95–98.
Hellgren, O., Waldenström, J., & Bensch, S. (2004). A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium and Haemoproteus from avian blood. Journal of Parasitology, 90, 797–802.
Hellgren, O., Križanauskienė, A., Valkiūnas, G., & Bensch, S. (2007). Diversity and phylogeny of mitochondrial cytochrome b lineages from six morphospecies of avian Haemoproteus (Haemosporida: Haemoproteidae). Journal of Parasitology, 93, 889–896.
Iezhova, T. A., Valkiūnas, G., Loiseau, C., Smith, T. B., & Sehgal, R. N. M. (2010). Haemoproteus cyanomitrae sp. nov. (Haemosporida, Haemoproteidae) from a widespread African songbird, the olive sunbird Cyanomitra olivacea. Journal of Parasitology, 96, 137–143.
Iezhova, T. A., Dodge, M., Sehgal, R. N. M., Smith, T. B., & Valkiūnas, G. (2011). New avian Haemoproteus species (Haemosporida: Haemoproteidae) from African birds, with a critique of the use of host taxonomic information in Hemoproteid classification. Journal of Parasitology, 97, 682–694.
Križanauskienė, A., Hellgren, O., Kosarev, V., Sokolov, L., Bensch, S., & Valkiūnas, G. (2006). Variation in host specificity between species of avian haemosporidian parasites: evidence from parasite morphology and cytochrome b gene sequences. Journal of Parasitology, 92, 1319–1324.
Križanauskienė, A., Pérez-Tris, J., Palinauskas, V., Bensch, S., & Valkiūnas, G. (2010). Molecular phylogenetic and morphological analysis of haemosporidian parasites (Haemosporida) in a naturally infected European songbird, the blackcap Sylvia atricapilla, with description of Haemoproteus pallidulus sp. nov. Parasitology, 137, 217–227.
Križanauskienė, A., Iezhova, T. A., Chernetsov, N., Palinauskas, V., & Valkiūnas, G. (2012). Haemoproteus nucleocondensus n. sp. (Haemosporida, Haemoproteidae) from a Eurasian songbird, the Great Reed Warbler Acrocephalus arundinaceus. Zootaxa, 3441, 36–46.
Križanauskienė, A., Iezhova, T. A., Sehgal, R. N. M., Carlson, J. S., Palinauskas, V., Bensch, S., & Valkiūnas, G. (2013). Molecular characterization of Haemoproteus sacharovi (Haemosporida, Haemoproteidae), a common parasite of columbiform birds, with remarks on classification of haemoproteids of doves and pigeons. Zootaxa, 3613, 085–094.
Levin, I. I., Valkiūnas, G., Santiago-Alarcon, D., Cruz, L. L., Iezhova, T. A., O’Brien, S. L., Hailer, F., Dearborn, D., Scheiber, E. A., Fleischer, R. C., Ricklefs, R. E., & Parker, P. G. (2011). Hippoboscid-transmitted Haemoproteus parasites (Haemosporida) infect Galapagos Pelecaniform birds: Evidence from molecular and morphological studies, with a description of Haemoproteus iwa. International Journal for Parasitology, 41, 1019–1027.
Levin, I. I., Valkiūnas, G., Iezhova, T. A., O’Brien, S. L., & Parker, P. G. (2012). Novel Haemoproteus species (Haemosporida: Haemoproteidae) from the swallow-tailed gull (Laridae), with remarks on the host range of hippoboscid-transmitted avian hemoproteids. Journal of Parasitology, 98, 847–854.
Levin, I. I., Zwiers, P., Deem, S. L., Geest, E. A., Higashiguchi, J. M., Iezhova, T. A., Jiménez-Uzcátegui, G., Kim, D. H., Morton, J. P., Perlut, N. G., Renfrew, R. B., Sari, E. H. R., Valkiūnas, G., & Parker, P. G. (2013). Multiple lineages of avian malaria parasites (Plasmodium) in the Galapagos Islands and evidence for arrival via migratory birds. Conservation Biology, 27, 1366–1377.
Levine, N. D., & Campbell, G. R. (1971). A check-list of the species of the genus Haemoproteus (Apicomplexa, Plasmodiidae). Journal of Parasitology, 18, 475–484.
Martínez, J., Martínez-De La Puente, J., Herrero, J., Del Cerro, S., Lobato, E., Rivero-De Aguilar, J., Vásquez, R. A., & Merino, S. (2009). A restriction site to differentiate Plasmodium and Haemoproteus infections in birds: on the inefficiency of general primers for detection of mixed infections. Parasitology, 136, 713–722.
Martinsen, E. S., Paperna, I., & Schall, J. J. (2006). Morphological versus molecular identification of avian Haemosporidia: an exploration of three species concepts. Parasitology, 133, 279–288.
Martinsen, E. S., Perkins, S., & Schall, J. J. (2008). A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): evolution of life-history traits and host switches. Molecular Phylogenetics and Evolution, 47, 261–273.
Marzal, A., De Lopes, F., Navarro, C., & Møller, A. P. (2005). Malarial parasites decrease reproductive success: an experimental study in a passerine bird. Oecologia, 142, 541–545.
Marzal, A., Reviriego, M., Hermosell, I. G., Balbontín, J., Bensch, S., Relinque, C., Rodríguez, L., Garcia-Longoria, L., & de Lope, F. (2013). Malaria infection and feather growth rate predict reproductive success in house martins. Oecologia, 171, 853–861.
Merino, S., Hennicke, J., Martínez, J., Ludynia, K., Torres, R., Work, T. M., Stroud, S., Masello, J. F., & Quillfeldt, P. (2012). Infection by Haemoproteus parasites in four species of frigatebirds and the description of a new species of Haemoproteus (Haemosporida, Haemoproteidae). Journal of Parasitology, 98, 388–397.
Møller, A. P., & Nielsen, J. T. (2007). Malaria and risk of predation: a comparative study of birds. Ecology, 88, 871–881.
Nylander, J. A. A. (2004). MrModeltest v2. Evolutionary Biology Centre, Uppsala, Program distributed by the author. Available from: http://www.abc.se/~nylander/. Accessed 18 February 2012.
Olias, P., Wegelin, M., Zenker, W., Freter, S., Gruber, A. D., & Klopfleisch, R. (2011). Avian Malaria Deaths in Parrots, Europe. Emergent Infection Diseases, 17, 950–952.
Palinauskas, V., Kosarev, V., Shapoval, A., Bensch, S., & Valkiūnas, G. (2007). Comparison of mitochondrial cytochrome b lineages and morphospecies of two avian malaria parasites of the subgenera Haemamoeba and Giovannolaia (Haemosporida: Plasmodiidae). Zootaxa, 1626, 39–50.
Palinauskas, V., Iezhova, T. A., Križanauskienė, A., Markovets, M. Y., Bensch, S., & Valkiūnas, G. (2013). Molecular characterization and distribution of Haemoproteus minutus (Haemosporida, Haemoproteidae): a pathogenic avian parasite. Parasitology International, 62, 358–363.
Peirce, M. A. (1981). Distribution and host–parasite check-list of the haematozoa of birds in Western Europe. Journal of Natural History, 15, 419–458.
Perkins, S. L., & Schall, J. J. (2002). A molecular phylogeny of malarial parasites recovered from cytochrome b gene sequences. Journal of Parasitology, 88, 972–978.
Ricklefs, R. E., & Fallon, S. M. (2002). Diversification and host switching in avian malaria parasites. Proceeding of the Royal Society B, 269, 885–892.
Ronquist, F., & Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19, 1572–1574.
Sambrook, J., Fritch, F. J., & Maniatis, T. (2002). Molecular cloning: a laboratory manual. Cold Spring Harbor, New York, USA: Cold Spring Harbor Laboratory Press.
Santiago-Alarcon, D., Outlaw, D. C., Ricklefs, R. E., & Parker, P. G. (2010). Phylogenetic relationships of haemosporidian parasites in New World Columbiformes, with emphasis on the endemic Galapagos dove. International Journal for Parasitology, 40, 463–470.
Shurulinkov, P., & Golemansky, V. (2002). Haemoproteids (Haemosporida: Haemoproteidae) of wild birds in Bulgaria. Acta Protozoologica, 41, 359–374.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., & Kumar, S. (2011). MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution, 28, 2731–2739.
Valkiūnas, G. (2005). Avian malaria parasites and other haemosporidia. Boca Raton, Florida: CRC Press.
Valkiūnas, G., Liutkevičius, G., & Iezhova, T. A. (2002). Complete development of three species of Haemoproteus (Haemosporida, Haemoproteidae) in the biting midge Culicoides impunctatus (Diptera: Ceratopogonidae). Journal of Parasitology, 88, 864–868.
Valkiūnas, G., Iezhova, T. A., & Shapoval, A. P. (2003). High prevalence of blood parasites in Hawfinch Coccothraustes coccothraustes. Journal of Natural History, 37, 2647–2652.
Valkiūnas, G., Bensch, S., Iezhova, T. A., Križanauskienė, A., Hellgren, O., & Bolshakov, C. V. (2006). Nested cytochrome b PCR diagnostics underestimate mixed infections of avian blood haemosporidian parasites: microscopy is still essential. Journal of Parasitology, 92, 418–422.
Valkiūnas, G., Križanauskienė, A., Iezhova, T. A., Hellgren, O., & Bensch, S. (2007). Molecular phylogenetic analysis of circumnuclear hemoproteids (Haemosporida: Haemoproteidae) of Sylviid birds, with a description of Haemoproteus parabelopolskyi sp. nov. Journal of Parasitology, 93, 680–687.
Valkiūnas, G., Atkinson, C. T., Bensch, S., Sehgal, R. N. M., & Ricklefs, R. E. (2008a). Parasite misidentifications in GenBank: how to minimise their number? Trends in Parasitology, 24, 247–248.
Valkiūnas, G., Iezhova, T. A., Križanauskienė, A., Palinauskas, V., Sehgal, R. N. M., & Bensch, S. (2008b). A comparative analysis of microscopy and PCR-based detection methods for blood parasites. Journal of Parasitology, 94, 1395–1401.
Valkiūnas, G., Iezhova, T. A., Loiseau, C., Chasar, A., Smith, T. B., & Sehgal, R. N. M. (2008c). New species of haemosporidian parasites (Haemosporida) from African rainforest birds, with remarks on their classification. Parasitology Research, 103, 1213–1228.
Valkiūnas, G., Iezhova, T. A., Loiseau, C., & Sehgal, R. N. M. (2009). Nested cytochrome b polymerase chain reaction diagnostics detect sporozoites of hemosporidian parasites in peripheral blood of naturally infected birds. Journal of Parasitology, 95, 1512–1515.
Waldenström, J., Bensch, S., Hasselquist, D., & Östman, Ö. (2004). A new nested polymerase chain reaction method very efficient in detecting Plasmodium and Haemoproteus infections from avian blood. Journal of Parasitology, 90, 191–194.
Acknowledgements
The authors are grateful to Dr. Robert Adlard and Dr. Mal Bryant, Queensland Museum and Science Centre, Australia for providing the type-material of H. velans. We thank Dr. Vaidas Palinauskas for assistance in the laboratory. We are grateful to both anonymous reviewers for the valuable comments and suggestions. Part of the laboratory work was possible because of the facilities created in the Institute of Biodiversity and Ecosystem Research during the project CEBDER funded by the Bulgarian National Science Fund. This study was supported by the European Union Structural Funds project “Postdoctoral Fellowship Implementation in Lithuania” (VP-3.1-ŠMM-01-V-02-004).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dimitrov, D., Zehtindjiev, P., Bensch, S. et al. Two new species of Haemoproteus Kruse, 1890 (Haemosporida, Haemoproteidae) from European birds, with emphasis on DNA barcoding for detection of haemosporidians in wildlife. Syst Parasitol 87, 135–151 (2014). https://doi.org/10.1007/s11230-013-9464-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11230-013-9464-1