Abstract
Haemoproteus spp. and Plasmodium spp. are blood parasites that occur in birds worldwide. Identifying the species within this group is complex, especially in wild birds that present low parasitemia when captured, making morphological identification very difficult. Thus, the use of alternative tools to identify species may be useful in the elucidation of the distribution of parasites that circulate in bird populations. The objectives of this study were to determine the prevalence and parasitemia of the genera Plasmodium and Haemoproteus in Tachyphonus coronatus in the Atlantic Forest, Brazil, and to evaluate the molecular diversity, geographic distribution, and specificity of these parasites based on coalescent species delimitation methods. Microscopic analysis, PCR, cyt b gene sequencing, phylogenetic analysis and coalescent species delimitation using single-locus algorithms were performed (Poisson tree process (PTP) and multi-rate Poisson tree process (MPTP) methods). The analyses were performed in 117 avian host individuals. The prevalence was 55.5% for Plasmodium and 1.7% for Haemoproteus, with a mean parasitemia of 0.06%. Twenty-five Plasmodium and two Haemoproteus lineages were recovered. The MPTP method recovered seven different evolutionarily significant units (ESUs) of Plasmodium and one of Haemoproteus, whereas PTP presented fourteen ESUs of Plasmodium and one of Haemoproteus. The MPTP was more consistent with current taxonomy, while PTP overestimated the number of lineages. These ESUs are widely distributed and have already been found in 22 orders of birds that, all together, inhabit every continent, except Antarctica. The computational methods of species delimitation proved to be effective in cases where the classification of Haemosporida based just on morphology is insufficient.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The protozoans in the Haemosporida order (Alveolata, Apicomplexa) are blood parasites that occur in birds and other vertebrates and are distributed worldwide. The species of the genera Plasmodium and Haemoproteus are the most well studied (Valkiūnas 2005). These parasites have a complex life cycle with an asexual stage, which occurs in the vertebrate host, and a sexual stage, which occurs in haematophagous dipterans (Insecta: Diptera) (Garnham 1966). Morphological characteristics, such as the presence of pigments, gametocyte shape, host cell nucleus displacement, parasite position in cells, type of exoerythrocytic schizogony, and vertebrate host, have traditionally been used to describe species in this group. To date, the use of these morphological characters has led to the description of 55 Plasmodium and more than 140 Haemoproteus species (Dimitrov et al. 2014; Valkiūnas and Iezhova 2018). In recent years, the accumulation of mitochondrial cytochrome b gene (cyt b) sequences from avian haemosporidian available in databases has created a discussion regarding the current number of existing species. In the MalAvi database, created for avian haemosporidian parasites, approximately 2800 lineages of Plasmodium and Haemoproteus based on the cyt b gene have been deposited, in which the majority are not attributed to any morphologically known species (http://mbio-serv2.mbioekol.lu.se/Malavi/, Bensch et al. 2009). Because of an exponential increase in the number of nucleotide sequences in databases, many studies have used differences in at least one base pair (single-nucleotide polymorphism; SNP) in the cyt b gene to delimit new lineages (Bensch et al. 2004; Clark and Clegg 2017). However, these parasites exhibit slow rates of mitochondrial evolution (Ricklefs and Outlaw 2010); therefore, the use of the cyt b gene as DNA barcode for the estimation of species limits in haemosporidians should be performed with caution.
A consensus on how to delimit the haemosporidian species based on genetic sequences does not exist yet (Perkins 2014); however, researchers suggest that an interaction of species concepts should be used. Perkins (2000) was one of the first to apply the three main classes of concepts of species: morphological species concept (MSC), biological species concept (BSC), and phylogenetic species concept (PSC). This author stated that, ideally, these three classes should be integrated at the specific level. Although the concept of morphological similarity has been used for many years (Garnham 1966; Valkiūnas 2005), species identification in the Haemosporida order has been a challenge for researchers. Complications arise from the distortion of morphological characteristics in blood smears, such as the occurrence of cryptic species, and by the low parasitemia of many haemosporidian lineages in wild birds. Additionally, the advancement of molecular approaches has helped to reduce specialist taxonomists, which compromises morphological studies, such as the production of low-quality slides and few professionals trained to perform this analysis. The biological species concept is difficult to use because of the complex life cycle of these parasites and the lack of information regarding reproductive isolation between defined species (Perkins 2000; Outlaw and Ricklefs 2014).
Regarding the phylogenetics species concept (PSC), recently, new methods based on the coalescent model have been shown to be effective in discovering cryptic biodiversity and in establishing patterns for species delimitation for various taxa (Pons et al. 2006; Fujita et al. 2012; Musher and Cracraft 2018; Galen et al. 2018). The PSC defines a species as a grouping of organisms with patterns of ancestry and descent, which differ from other clusters (Eldredge and Cracraft 1980). Computational species delimitation algorithms can delimit or validate evolutionary significant units (ESUs) (Ryder 1986) from genetic sequences, and can be powerful in the identification and assignment of probabilities to reproductively isolated lineages (Fujita et al. 2012; Galen et al. 2018; Mello et al. 2018). To date, the use of these methods to inform species limits in microbial symbionts has been rare. They have only been used to delimit ESUs for Leucocytozoon (Haemosporida) using nuclear genes, which presented high species richness in Alaska birds (Galen et al. 2018). Thus, a coalescent species delimitation analysis using only the cyt b gene has not been conducted for haemosporidians.
The Brazilian Atlantic Forest has one of the richest avifauna in the world (Mittermeier et al. 2005), with approximately 891 bird species. Among these, 213 are endemic and 233 are on the endangered species list (MMA 2020). The status of these avifauna has resulted from a set of environmental degradation processes resulting from activities such as extraction, mining, agriculture and urban occupation (Galindo-Leal and Câmara 2003; Dean 2004; Lima 2014). Currently, this ecosystem is reduced to isolated forest remnants that are 8.5 to 12% of its original extent (Ribeiro et al. 2005).
The information generated from bird studies can be used as an indicator of environmental health and as an indicator of ecosystems biodiversity because of their high species richness, their taxonomic and systematic knowledge, and their sensitivity to environmental changes (Furness and Greenwood 1993; Turner 1996; Bierregaard Jr and Stouffer 1997; Piratelli et al. 2008). Thus, studies on avian haemosporidians are extremely important because of the increasing in knowledge regarding species diversity and distribution, the possibility of determining the pathogenicity in the hosts, and the contribution to conservation programs (Garnham 1966; Valkiūnas 2005; Olias et al. 2011; Vanstreels et al. 2015; Ilgūnas et al. 2016; Valkiūnas et al. 2017a).
The ruby-crowned tanager (Tachyphonus coronatus) is a bird species in the Thraupidae family that has several natural habitats, including the subtropical or tropical moist lowland rainforests, subtropical or tropical mountain rainforests, and highly degraded ancient forests (BirdLife International 2018). This species has a generalist habit and an extremely wide range, covering areas of southern Brazil and the Atlantic Forest, reaching Argentina and Paraguay (Sigrist 2014). Thus, T. coronatus is a valuable representative of birds in ecological studies, because this species has not reached the limits of vulnerability, under the criterion of range size and population tendency, presenting a state of “conservation of little concern or stable” (BirdLife International 2018).
We did not find another study in the literature that applied the coalescent species delimitation analysis, using two different algorithms based on a single-locus (Poisson tree processes [PTP] and multi-rate Poisson tree processes [MPTP]) for Plasmodium and Haemoproteus species. The limits of species recovered by the coalescent methods also were used to evaluate the richness, distribution and specificity of the avian haemosporidians. Prevalence and parasitemia were evaluated to understand the haemosporidian dynamics in a population of ruby-crowned tanager, Tachyphonus coronatus Vieillot, 1822 (Passeriformes: Thraupidae), in the Atlantic Forest of Minas Gerais, Brazil.
Material and methods
Area of study and blood collection
Birds were captured from five different fragments of the Atlantic Forest in the municipality of Juiz de Fora, Minas Gerais, southeast of Brazil (21° 45′ 51″ S, 43 21′ 01″ W) from 2013 to 2015 (36 months). The climate of the region is classified as a humid subtropical climate (CWA) according to Köppen (1970), and experiences a cold and dry season between May and September, and a hot and humid season between October and April. Mist nets were used to capture the birds, which were weighed and tagged with marking rings numbered to avoid resampling, and then were weighed. The identification of bird species was performed according to Ridgely and Tudor (2009) and Sigrist (2014) based on morphometric analyses and phenotypic characteristics, such as size, plumage colour, beak and legs.
Blood samples from 117 individuals of T. coronatus were collected by brachial vein puncture and one drop (approximately 5 μL) was used for the preparation of blood smears for microscopic analysis. The volume of blood collected did not exceed 1% of the total weight of the animal. Approximately 30 μL of blood was stored at − 20 °C for molecular analyses. After the sampling procedures, birds were immediately released. All sample collection procedures were evaluated and approved by the Animal Experiment Ethics Committee of the Federal University of Juiz de Fora (042/2012) and by the System of Authorization and Information on Biodiversity (SISBIO) (29268-3).
Microscopic analysis
Blood smears were prepared for each collected bird. The blood smears were dried and fixed in absolute methanol for 3 min at the collection site and stained with Giemsa (Merck, Darmstadt, Germany) in the laboratory, according to Valkiūnas et al. (2008a). An Olympus BX-51 light microscope (Olympus, Tokyo, Japan), equipped with an Olympus Evolt E-330 digital camera, was used to examine the slides at a magnitude of × 600 for 20 min. One hundred microscopic fields were observed at a magnitude of × 1000 to estimate the prevalence of infection (Bush et al. 1997). Parasitemia was determined by counting infected erythrocytes in 10,000 erythrocytes (Godfrey et al. 1987).
Molecular methods
Total DNA was extracted from 20 μL of the blood sample that was positive by microscopic analysis using the Wizard® Genomic DNA Purification Kit (Promega®, São Paulo, Brazil) according to the manufacturer’s recommendations. After DNA extraction, the DNA was quantified in a spectrophotometer (Nanodrop ND-2000 ®, Thermo Scientific, Wilmington, DE, USA) and the DNA samples were standardized to a concentration of 25 ng/μL. The samples were stored in triplicate at − 20 °C until molecular analyses were performed.
A nested PCR was used to amplify a 479-bp fragment from the haemosporidian cytb gene. In the first reaction, the primers HaemNFI (5′-CATATATTAAGAGAAITATGGAG-3′) and HaemNR3 (5′-ATAGAAAGATAAGAAATACCATTC-3′) were used to amplify the cytb gene of Plasmodium spp., Haemoproteus spp. and Leucocytozoon spp. (Hellgren et al. 2004). The PCR was performed in a final volume of 25 μL, using 1X GoTaq® Green Master Mix (Promega, MA, USA), 1 μM of each primer and 100 ng of genomic DNA. In the second reaction, the primers HaemF (5′-ATGGTGCTTTCGATATATGCATG-3′) and HaemR2 (5′-GCATTATCTGGATGTG ATAATGGT-3′) were used to amplify the cytb gene of Plasmodium/Haemoproteus (Bensch et al. 2000). The second reaction was conducted in a final volume of 25 μL, containing 1X GoTaq ® Green Master Mix, 0.4 μM of each primer and 2 μL of the first reaction. The thermocycling conditions of both reactions were 94 °C for 3 min, 35 cycles of 94 °C for 30 s, 52 °C for 30 s, 72 °C for 45 s and a final extension at 72 °C for 10 min.
If co-infections of Plasmodium/Haemoproteus were observed upon microscopic examination, an additional nested PCR (used to amplify parasites of the Haemoproteus) was performed using the primer pair 3760F (5′-ATGGAGTGGGTGTTTTAGAT-3′) and 4292Rw (5′-TGGAATAACATGTARAGGAGT-3′) in the first reaction, and the primers HML (5′-GCTACTGGTGCTACATTTGT-3′) and HMR (5′-CCT AAA GGA TTA GAG CTA CC-3′) in the second reaction (Merino et al. 2008). The first reaction was conducted in a final volume of 25 μL, containing 1X GoTaq ® Green Master Mix (Promega, MA, USA), 1.0 μM of primers, and 100 ng of total DNA. The second reaction was performed under the same conditions, modifying only the primers and the template. In this second reaction, 0.4 μM of the primers and 1 μL of the first reaction were used. The thermocycling conditions were the same as those used by Merino et al. (2008). Ultra-pure water was used a negative control for both protocols. Genomic DNA from Plasmodium gallinaceum was extracted from a cell culture and used as a positive control for both protocols. PCR amplicons were separated by 2% agarose gel electrophoresis, stained with Blue Green Loading Dye I (LGC Biotechnology, Cotia, São Paulo, Brazil), and visualized under ultraviolet light.
The amplified products were purified using the QIAquick® Purification Kit (Qiagen®, São Paulo, Brazil), and subjected to bidirectional sequencing using the HaemF/HaemR2 or HML/HMR primers (Bensch et al. 2000; Merino et al. 2008) on a 3130xL Genetic Analyzer Sequencer (Applied Biosystems®, Carlsbad, CA, USA), following the manufacturer’s instructions.
Phylogenetic analysis
Phylogenetic reconstructions were performed using a dataset containing 27 sequences of the cytb gene of haemosporidian blood parasites obtained in the present work, and a set of Plasmodium and Haemoproteus morphospecies sequences available from the MalAvi database (n = 116) in November 2019 (Bensch et al. 2009)(Supplementary Table S3). The species Leucocytozoon buteonis, Leucocytozoon fringillinarum and Leucocytozoon quynzae were chosen as the outgroup. Sequences were aligned in MAFFT (Katoh et al. 2017) with default options and then visually inspected. After removing poorly aligned positions in the flanking regions with Gblocks (Talavera and Castresana 2007), a matrix with 479 bp was obtained. Each sequence that contained at least one different nucleotide was considered a new lineage, named according to Bensch et al. (2009) and deposited in the MalAvi and Genbank databases. Haemosporida phylogeny was conducted under a maximum likelihood (ML) and Bayesian inference (BI) frameworks. ML topology was inferred via RAxML (Stamatakis 2014), using the GTR + GAMMA + I model with four gamma categories (Tavaré 1986; Yang 1994); this was chosen as the best substitution model in JmodelTest, implemented in MEGA7 (Kumar et al. 2016). Clade support was assessed with the bootstrap convergence criterion of RAxML (Stamatakis 2014) with 500 pseudo-replicates. Bayesian inference analyses were performed using MrBayes v. 3.2 (Ronquist et al. 2012) and the evolutionary model was the GTR + GAMMA + I. Two simultaneous and independent Markov chain Monte Carlo simulations were performed using three heated chains and one cold chain (N chains = 4) with a sample and print frequency of 500 and a diagnostic frequency of 5000 for 1,000,000 generations or until the average standard deviation of the split frequencies was below 0.01, indicating the convergence of the two independent runs. All trees remaining after discarding the burn-in (25%) were used in the calculation of posterior probabilities using a majority rule consensus.
Computational delimitation of species
The species delimitation Poisson tree process (PTP) and multi-rate Poisson tree process (MPTP) approaches were used to delimit evolutionarily significant units (ESUs). These tools are based on the coalescence theory and differ mainly in speciation modeling. The PTP models speciation in units of substitutions per sites and assumes that the number of substitutions between species is greater than the number of substitutions within species (Zhang et al. 2013). The MPTP is a version of the PTP that improves estimation in phylogenies that have different inter- and intraspecific rates. The speciation rate may be constant between sister species, but the intraspecific coalescence rate and genetic diversity can vary significantly even among sister species; therefore, MPTP accounts for the different rates of branching events within each delimited species (Kapli et al. 2017). The dataset used to delimit haemosporidian ESUs was assembled in a particular way to avoid sampling bias; therefore, the analyses for Plasmodium spp. and Haemoproteus spp. were performed separately. All cytb gene sequences greater than 430 bp in length (“long sequences”) that were deposited in the MalAvi database were used, along with the novel sequences produced in the present study. The Plasmodium spp. dataset comprised of 906 sequences (26 from the present study and 880 available in the MalAvi database), whereas the Haemoproteus spp. dataset comprised 984 sequences (two from the present study and 982 available in the MalAvi database). The species L. buteonis, L. fringillinarum and L. quynzae were chosen as the outgroup. Phylogenetic tree inferences were conducted using the ML method, as described in the previous subsection. The PTP and MPTP methods to delimit ESUs were employed on both rooted phylogenies obtained in RAxML (Plasmodium and Haemoproteus datasets), and both methods were implemented in the MPTP Webservice (http://mptp.h-its.org/#/tree), using the default parameters. The outgroup was removed before all analyses.
Geographic distribution and host specificity analyses
From the results obtained by the species delimitation method with the best performance in this study, the “hosts and sites” in the MalAvi database was queried to record the geographical coordinates of the locations where the lineages grouped in each ESU which was sampled. We used the geographic coordinates of the sampling country for lineages that had no coordinates recorded in the database. With these data, we built geographic maps in the ArcMap 10.3 (Santos 2009) using the global distribution of the ESUs and a network graph representing the distribution of these significant units by order of avian hosts.
Results
Prevalence and intensity of infection
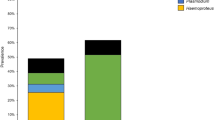
Samples were collected from 117 specimens of T. coronatus in the Atlantic Forest of Minas Gerais, Brazil. Overall, the prevalence of Plasmodium spp. and Haemoproteus spp. was 56.4% (66 individuals). Plasmodium genus was recorded in 65 individuals (55.5%) and Haemoproteus in two individuals (1.7%). Only one individual exhibited co-infection. Mean parasitemia was 0.06%. Most of the infected birds (60%) had low parasitemia (0.01–0.05%) and only 13% had a parasite intensity greater than 0.1%.
Phylogenetic reconstruction
From 66 positive samples from the microscopic examination, 27 lineages of the cytb gene of parasites were recovered. Phylogenetic reconstruction from the dataset with morphospecies sequences available in the MalAvi database showed that 25 lineages from this study clustered with the Plasmodium spp. lineages and 2 with the Haemoproteus spp. lineages (Fig. 1). Among the lineages of Plasmodium spp., nine are new lineages (two lineages were obtained from a co-infected bird) and the other remaining samples were identified in previous studies (MalAvi database). For Haemoproteus, both are new lineages. The Haemoproteus spp. were recovered as a sister group of representatives of the Plasmodium spp. The lineages obtained in this study formed six different clusters (Fig. 1). The TCCA94 lineage grouped as a sister group with the lineage of the P. elongatum (clade 1) (genetic distance: 0.3%) and the lineage TCSB1033 with a sister group of the lineages of P. matutinum (genetic distance: 2.4%) and P. lutzi (clade 2) (genetic distance: 2.4%). Clade 3 recovered in a single cluster with nine lineages obtained in this study (TCJB29, TCJB161, TCJB851, TCJB472, TCJB1855, TCJB1569, TCJB1952, TCJB1021 and TCJB53). In the fourth clade (clade 4), the TCJB79-2 lineage had P. unalis (genetic distance: 1.4%) as a sister group. No known morphospecies lineage grouped within the TCJB730 and TCJB1821 lineages, which formed clade 5, while the sixth clade (clade 6) grouped nine other lineages obtained in the present study (TCJB904, TCCH216, TCJB57, TCJB617, TCJB642, TCJB1596, TCJB1789, TCJB1070 and TCJB1011) with the P. nucleophilum lineage (Mean distance genetic: 0%). The seventh cluster (clade 7) grouped the TCJB1650 and TCJB79-1 lineages with P. cathemerium emerging as the sister group, and had a genetic distance of 1.4% and 2.4%, respectively. The two lineages (TCJB20 and TCJB75) that clustered with Haemoproteus (clade 8) represented H. ilanpapernai as a sister group, with a genetic distance 2.5% and 2.1%, respectively (Fig. 1). Here we use the alternative names of the lineages, and their corresponding names in MalAvi database are supplied in Supplementary Table S1.
Phylogenetic tree inferred by maximum likelihood and Bayesian analyses based on 479 bp of the mitochondrial gene cytochrome b (cytb) of avian haemosporidian morphospecies. Only branch values (ML/BI) above 70% are presented and indicated near the nodes. The values close to the nodes represent the bootstrap values followed by the Bayesian posterior probability. The sequences obtained in the present study are highlighted in bold. (-) represents < 70% bootstrap or posterior probability values. Leucocytozoon lineages were used as outgroups to root the tree. Bar represents 6 substitutions per 100 nucleotide positions
Species delimitation
The species delimitation analyses produced estimates that ranged from 118 species (MPTP) to 450 species (PTP) for Plasmodium lineages. For Haemoproteus lineages, the methods produced between 65 species (MPTP) and 436 species (PTP) (Table 1). In relation to Plasmodium spp., both of the species delimitation algorithms presented a high frequency of match profiles (when the ESUs agreed with the taxonomy classification), and a low number of lumper profiles (groups having more than one taxonomically defined species under a single name). However, the algorithm PTP exhibited more splitter cases (when taxonomically defined species were recovered in different ESUs) than did MPTP for the Plasmodium lineages. Furthermore, an elevated number of lineages that did not have morphological characterization associated was identified after the application of the species delimitation methods because they were grouped into clusters with a morphospecies (Table 1). In this case, the frequency of identification lineages was higher for the MPTP analysis (462 lineages identified) than the PTP analysis (210 lineages identified). The analyses of Haemoproteus spp. showed that, although the PTP method presented more match profiles than did MPTP, the number of splitter profiles recovered by PTP was more frequent than that of MPTP (Table 1). However, the PTP presented fewer cases of lumper profiles than did MPTP for the Haemoproteus lineages. Regarding the lineages obtained in this study, MPTP grouped 25 Plasmodium lineages in seven ESUs (Fig. 2, Supplementary Table S2). Of the seven ESUs, four of them presented lineages associated with P. cathemerium, P. nucleophilum, P. unalis and P. elongatum (Supplementary Table S2). For Haemoproteus lineages, the MPTP method produced more lumper cases than did PTP, which formed one single ESU, a correspondent with nine morphospecies (Fig. 3, Supplementary Table S2). The PTP was a splitter in relation to the MPTP method for Plasmodium lineages, inferring 14 ESUs. The main difference was the division of one ESU, which united nine lineages in the present study using the MPTP method, into seven ESUs in the PTP (Fig. 2, Supplementary Table S2). Both methods agreed with the association of ESU 39 (MPTP) and ESU 134 (PTP) with the morphospecies P. nucleophilum and ESU 103 (MPTP) and ESU 392 (PTP) with P. elongatum. For Haemoproteus lineages, the PTP method inferred one single ESU with our two lineages, without association with morphospecies (Fig. 3, Supplementary Table S2).
Species distribution and host specificity
The eight ESUs delimited by MPTP, obtained from T. coronatus, were recorded in 22 orders of birds (Fig. 4) that are collectively are distributed among all continents, except Antarctica (Fig. 5). The order Passeriformes not only had the highest number of ESUs, but was the only order that was represented all ESUs delimited in this work (Fig. 4). Among continents, most of the seven ESUs of Plasmodium were recorded mostly in South America, mainly in the northern and south-eastern regions of Brazil (Fig. 5). The highest occurrence of the ESU containing Haemoproteus lineages occurred in North America, although lineages of this ESU have already been recorded in the northern region of Brazil. The eight delimited species were recorded in both captive and wild birds.
Network graph representing the orders of birds parasitized by the haemosporidian species delimited in the present study by MPTP. Each colour represents a species; “n” represents the number of times that species was recorded in general. The numbers in parentheses represent the number of times each species of haemosporidian was recorded in each the orders of birds
Discussion
Information on haemosporidians of birds in Brazil has advanced substantially in recent years. However, studies have concentrated on a few regions and have been generally related to the diagnosis and ecology of these parasites (Ferreira et al. 2016; Fecchio et al. 2017; Tostes et al. 2017; Tostes et al. 2018; Fecchio et al. 2018; De Oliveira et al. 2019; Fecchio et al. 2019b, 2019c). Records of haemosporidian parasites in T. coronatus were assessed only in São Paulo and in the Atlantic Forest of Minas Gerais, both with a diagnostic purpose (Bennett and Lopes 1980; Woodworth-Lynas et al. 1989; Ribeiro et al. 2005; Fecchio et al. 2019a). Thus, the present study is the pioneer in the characterization of molecular diversity, phylogenetic relationships, and species delimitation of T. coronatus parasites.
The prevalence of haemosporidian recorded in this study was quite high compared with the prevalence found in other bird species in the Neotropical region, which may be related to several factors such as habitat, host genetics, the presence of vectors, their distribution, and vector transmission capacity (Bensch et al. 2007; Glaizot et al. 2012). T. coronatus is a widely distributed bird in Brazil and commonly found in the Atlantic Forest, where the diversity of birds and dipterans is very high (Guedes 2012; Lima 2014). This factor contributes to the wide and diverse contact of these birds with other hosts and vectors and consequently a larger number of individuals potentially infected by haemosporidian parasites.
The different Plasmodium and Haemoproteus lineages (n = 27) identified in T. coronatus and the phylogenetic reconstruction suggest that a high diversity of haemosporidian parasites exists in this bird population. Most lineages clustered in clade 3, which had a sister cluster with P. matutinum and P. lutzi species and in clade 6, included P. nucleophilum in the cluster, indicating that these lineages are widespread in the region. In addition, P. lutzi and P. nucleophilum have been reported to parasitize other bird species in the Atlantic Forest, including some that reside within the same region where the specimens of this study were captured (Chagas et al. 2013, 2017; Lacorte et al. 2013; Tostes et al. 2017; Oliveira et al. 2019), which suggests low host specificity, although it is necessary to investigate whether these records constitute abortive infections or not. Other lineages clustered with P. elongatum and P. unalis and two others emerged as a sister group to P. cathemerium, suggesting that these species were also present in the studied region. Recent studies have recorded P. unalis in Turdus sp. at the same site where the ruby-crowned tanager was captured (Tostes et al. 2018). Additionally, P. elongatum was recorded in species in the Anatidae and Phasianidae families from the São Paulo Zoo, which is located within the Atlantic Forest (Chagas et al. 2017). Furthermore, Lacorte et al. (2013) presented evidence for the presence of P. elongatum lineages in Turdus sp. from the Atlantic Forest. P. cathemerium was recorded in penguins from Bahia, Espirito Santo and Santa Catarina rehabilitation centers (Vanstreels et al. 2015); however, there are no records of P. cathemerium in endemic wild bird of Brazil.
The two Haemoproteus lineages formed clade 8, which had the H. ilanpapernai as a sister group. This parasite was described in a bird species found in Southeast Asia (Karadjian et al. 2014). However, it was previously identified as H. syrnii (Karadjian et al. 2013). Although the description of H. ilanpapernai has been described using molecular and morphological approaches, the morphometric data have not been not fully detailed, which suggests the need for improvements in parasite morphology to clarify the correct taxonomic position of this haemosporidian.
The inconsistencies in the traditional taxonomy when phylogenetic reconstruction was used highlight the difficulty in identifying or delimiting avian haemosporidian species. For example, H. sacharovi and H. turtur, traditionally classified in the Haemoproteus subgenus, branched out into a clade with species from the Parahaemoproteus subgenus. These species have appeared in clades with Parahaemoproteus in several phylogenies of avian haemosporidians (Martinsen et al. 2008; Santiago-Alarcon et al. 2010; Valkiūnas et al. 2010a; Križanauskienė et al. 2013). Plasmodium emerged as a monophyletic group, but the phylogenetic position of the subgenus disagreed with traditional classification. Plasmodium tejerai, P. matutinum and P. lutzi species, of the subgenus Haemamoeba, were grouped far from the other representative species of this group. A similar result was detected in the molecular description of P. matutinum, which showed a close phylogenetic relationship with P. tejerai, but a distant relationship with other Haemamoeba species (Valkiūnas et al. 2017). Incongruities were also observed in P. megaglobularis, which is classified morphologically as a representative of the subgenus Novyella; however, its sequence clustered with representatives of the subgenus Haemamoeba. This disagreement was also found in the original description of the species (Valkiūnas et al. 2008b), contradicting the division of the Plasmodium subgenus, which was based on traditional taxonomy from morphological characters. Thus, phylogenetic analyses based on the cyt b gene for molecular identification at genus and subgenus level of haemosporidian blood parasites should be applied with caution.
Previous studies recommend that simultaneous analysis of DNA sequences and morphological characteristics should be performed to identify and classify avian haemosporidians (Clark et al. 2014; Braga et al. 2011; Outlaw and Ricklefs 2014; Valkiūnas and Iezhova 2018; Valkiūnas et al. 2010b). However, in wildlife, the identification of these parasites at the species level is a difficult task. This occurs mainly because of the presence of mild infections that are very common in wild animals and may be a consequence of bird sampling methods (e.g. mist netting), wherein the healthy individuals who move actively even with low parasitemia are most often captured (Valkiūnas 2005; Dimitrov et al. 2014). In addition to morphological and molecular approaches, important issues related to “limits of species,” such as the need for trained parasitologists, improvements in the method of specifying lineages in molecular databases, and development of criteria for defining species, have been discussed by experts (Perkins et al. 2011; Littlewood 2012; Outlaw and Ricklefs 2014).
Criteria for delimiting species of avian haemosporidian have been suggested, and include an agreement between multi-locus sequence data (Falk et al. 2011) and absolute levels of sequence divergence (Bensch et al. 2004; Valkiūnas et al. 2009a). However, the major challenge for researchers interested in haemosporidian systematics is the development of genetic markers (Perkins 2014). The lack of new genetic markers makes it difficult to work with multigene phylogenies. Absolute values of genetic divergence were proposed as a method for species delimitation (Hellgren et al. 2007; Valkiūnas et al. 2009a, 2009b; Outlaw and Ricklefs 2014); however, because of the limited understanding of mtDNA gene evolution rates, little is known about how to interpret the divergence between lineages using this approach (Pacheco et al. 2017). Studies have shown there are changes in species limits when comparing the nuclear and mitochondrial genomes. Small nucleotide substitutions in mitochondrial and nuclear genes have recovered different patterns of diversity (Nilsson et al. 2016; Galen et al. 2018).
The use of integrated analyses with the application of multi-locus delimitation methods has been considered a promising tool in the search for diversity and parasitic evolution (Galen et al. 2018). While new specific genetic markers are not yet developed, haemosporidian lineages based on the analysis of the cyt b gene are being added in the GenBank and MalAvi databases at a high rate (Bensch et al. 2009; Perkins 2014), making this gene a useful tool to define species, operational taxonomic units, or evolutionary significant units (ESUs) for this group of parasites (Outlaw and Ricklefs 2014). Barcode methods are of particular importance for large-scale research and promotes rapid species discovery and biodiversity estimates (Hofmann et al. 2019). Additionally, they are computationally less demanding and present better performance with a large amount of data, as is the case in the present work. Thus, the use of computational methods for species delimitation may be an adequate alternative to handle the large volume of deposited sequences within databases, where most of the sequences are identified only at the genus level.
Herein, two coalescent species delimitation methods were applied and their performance was analyzed to infer limits of species in haemosporidian lineages from a species of bird in Brazil’s Atlantic Forest. The MPTP method was congruent with the traditional taxonomy when delimiting ESUs containing only one Plasmodium morphospecies: P. cathemerium, P. nucleophilum, P. unalis and P. elongatum. The other three ESUs containing Plasmodium lineages in T. coronatus which did not cluster with lineages of morphologically known species suggest that new haemosporidian species are circulating in the bird population. Thus, conducting a thorough research study is important to uncover the diversity of haemoparasites that may be widespread in the region. When clustering different morphospecies into a single ESU, the MPTP method acted as a “lumper delimiter” (which defines the species broadly, grouping more than one taxonomically defined species under a single name). The clustering of Haemoproteus lineages in the same ESU with nine different morphospecies lineages shows the great difficulty of delimiting species in this group, where genetic divergence between species is minimal. In cases like this, computational methods for delimiting species may be insufficient, requiring greater gene sampling and the use of multigene analysis to infer the species limits for this group. Additionally, the MPTP identified a high frequency of lineages that were not previously identified because it grouped these lineages into clades containing morphospecies already deposited in the database.
The PTP method, in turn, proved to be a splitter for Plasmodium and Haemoproteus lineages, overestimating the diversity in both datasets. In the latter, the two lineages obtained from T. coronatus that formed a single ESU with nine other morphospecies using the MPTP methods, formed a single ESU without the grouping of any lineage. These results suggest that the computational species delimitation method used for the genus Plasmodium will not always be the best method to use for Haemoproteus genus. We observed that in this study MPTP presented better results for Plasmodium lineages than did PTP, especially if we considered the diversity previously estimated for the group. Regarding Haemoproteus, for example, 66 morphospecies are currently deposited in the MalAvi database, and fusing the PTP method, the diversity of this group consisted of 456 lineages, which suggests an estimate that is far from reality.
The MPTP method presented a more accurate performance in delimiting species limits for a dataset with the chosen molecular marker (cytb), presenting a low frequency of splitter and lumper profiles. Although the MPTP method was a lumper in relation to the current taxonomy, it allowed the identification of at least eight haemosporidian species in T. coronatus. Importantly, these methods support species delimitation in the current taxonomy but do not replace classical taxonomy based on morphological characteristics. In cases that the morphological classification is inadequate, the use of these methods is an alternative. According to Carstens et al. (2013), a conservative position must be taken when inferring species in delimitation studies, because in most scenarios, it is better not to delimit species than to falsely create entities, placing excessive limits on biodiversity. Furthermore, especially for Plasmodium lineages, the MPTP method was shown to have reliable results, with the number of delimited species close to the total number of taxonomic species in studies of diversity and conservation (Blair and Bryson Jr 2017; Correa et al. 2017; Kapli et al. 2017).
In this context, the distribution of these possible species in avian hosts worldwide was estimated in this study using the MPTP results, because it was the method that produced more realistic results regarding the number of morphospecies described in the literature and did not group many morphospecies in the same clade (lumper profile). Because the PTP proved to be more of a splitter method, it could overestimate the diversity and make the specificity and geographic distribution of the parasites less reliable, which negatively affects the importance of the conservation of these hosts. The wide distribution of Plasmodium spp. and Haemoproteus spp. in a varied bird species shows a low specificity in this group of parasites, which were traditionally considered to be highly specific (Valkiūnas 2005). These haemosporidian blood parasites can cause serious damage to avian hosts (Atkinson and LaPointe 2009). In addition to blood pathologies, various organs may be damaged by exoerythrocytic meronts, which grow on reticular endothelial cells and may be present throughout the organs of susceptible hosts (Garnham 1966; Valkiūnas 2005). Studies provide evidence that damage to organs by common lineages of haemosporidians causes disease and even mortality (Dinhopl et al. 2015; Ilgūnas et al. 2016; Palinauskas et al. 2016). Thus, the great allocation of haemosporidian species in T. coronatus deserves attention, because this bird can act as a reservoir and facilitate the transmission to other bird species, which may be more sensitive to the parasite.
Delimitation of species using computational algorithms can be an excellent tool for the integrative taxonomy of avian haemosporidian. It may also be valuable for the studies of diversity and distribution, in addition to aiding in the identification of cryptic species. A large number of cytb gene sequences from these parasites deposited in the databases forms a true DNA barcode for this group of parasites. Due to the controversies in Haemosporida classification and because most of the sequences are not attributed to morphological species, these methods may aid in the inference of significant taxonomic units because they are easily applied to a wide variety of organisms (Kapli et al. 2017). Additionally, it may represent the only viable approach to estimating biodiversity, which seems to be impossible to infer in macroecological studies of wildlife. Studies suggest that methods using single-locus barcodes provide meaningful clusters that are closest to taxonomically recognized species. These methods are useful for studies that approximate species estimation or when more thorough systematic research is practically impossible (Monaghan et al. 2009; Esselstyn et al. 2012; Ratnasingham and Hebert 2013; Kapli et al. 2017). It is worth noting that single-locus methods have been used for other groups as a primary method for inferring species limits and require refinement of the methods using an multi-locus approach (Leria et al. 2020). Thus, it is extremely important to sequence more markers for a large number of lineages in Haemosporida.
In this study, the haemosporidian diversity in an endemic bird of South America and the distribution of the supposed species found in this bird were estimated, as well as in other birds worldwide. Although the method used was a lumper, a high level of diversity was detected. Although the T. coronatus was the specific case, these results should not be neglected because this parasitological scenario may be occurring throughout the Atlantic Forest. Thus, studies that compare methods of computational delimitation could help to improve the results and bring to new perspectives to the integrative taxonomy of haemosporidian parasites, which is of great importance for the implementation of conservation programs. Furthermore, delimiting species of avian malaria parasites is important to associate with ecological and macroevolutionary processes (Outlaw and Ricklefs 2014). More studies that explore the diversification dynamics of avian haemosporidians would be useful to better understand the biological process of this group.
References
Atkinson CT, LaPointe DA (2009) Introduced avian diseases, climate change, and the future of Hawaiian honeycreepers. J Avian Med Surg 23:53–63. https://doi.org/10.1647/2008-059.1
Bennett G, Lopes OS (1980) Blood parasites of some birds from São Paulo state, Brazil. Mem Inst Oswaldo Cruz 75:117–134. https://doi.org/10.1590/S0074-02761980000100012
Bensch S, Stjernman M, Hasselquist D, Ostman O, Hansson B, Westerdahl H, Pinheiro RT (2000) Host specificity in avian blood parasites: a study of Plasmodium and Haemoproteus mitochondrial DNA amplified from birds. Proc R Soc B Biol Sci 267:1583–1589. https://doi.org/10.1098/rspb.2000.1181
Bensch S, Pérez-Tris J, Waldenström J, Hellgren O (2004) Linkage between nuclear and mitochondrial DNA sequences in avian malaria parasites: multiple cases of cryptic speciation? Evolution 58:1617–1621. https://doi.org/10.1111/j.0014-3820.2004.tb01742.x
Bensch S, Waldenström J, Jonzén N, Westerdahl H, Hansson B, Sejberg D, Hasselquist D (2007) Temporal dynamics and diversity of avian malaria parasites in a single host species. J Anim 76:112–122. https://doi.org/10.1111/j.1365-2656.2006.01176.x
Bensch S, Hellgren O, Peréz-Tris J (2009) MalAvi: a public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol Ecol Resour 9:1353–1358. https://doi.org/10.1111/j.1755-0998.2009.02692.x
Bierregaard Jr RO, Stouffer PC (1997) Understory birds and dynamic habitat mosaics in Amazonian rainforests. In Laurance WF, Bierregaard, RO., Jr., Tropical forest remnants, ecology, management, and conservation of fragmented communities. Chicago: The University of Chicago Press, pp 138–155
BirdLife International (2018) IUCN Red List for birds. Downloaded from http://www.birdlife.org. Acess in November 28, 2019
Blair C, Bryson RW Jr (2017) Cryptic diversity and discordance in single-locus species delimitation methods within horned lizards (Phrynosomatidae: Phrynosoma). Mol Ecol Resour 17:1168–1182. https://doi.org/10.1111/1755-0998.12658
Braga EM, Silveira P, Belo NO, Valkiūnas G (2011) Recent advances in the study of avian malaria: an overview with an emphasis on the distribution of Plasmodium spp. in Brazil. Mem Inst Oswaldo Cruz 106:3–11. https://doi.org/10.1590/S0074-02762011000900002
Bush AO, Lafferty KD, Lotz JM, Shostak AW (1997) Parasitology meets ecology on its own terms: Margolis et al. revisited. J Parasitol 83:575–583. https://doi.org/10.2307/3284227
Carstens BC, Pelletier TA, Reid NM, Satler JD (2013) How to fail at species delimitation. Mol Ecol 22:4369–4383. https://doi.org/10.1111/mec.12413
Chagas CRF, Valkiūnas G, Nery CVC, Henrique PC, Gonzalez IHL, Monteiro EF, Guimarães LO, Romano CM, Kirchgatter K (2013) Plasmodium (Novyella) nucleophilum from Egyptian goose in São Paulo Zoo, Brazil: microscopic confirmation and molecular characterization. Int J Parasitol Wildl 2:286–291. https://doi.org/10.1016/j.ijppaw.2013.09.008
Chagas CR, Valkiūnas G, de Oliveira GL, Monteiro EF, Guida FJ, Simões RF, Rodrigues PT, de Albuquerque Luna EJ, Kirchgatter K (2017) Diversity and distribution of avian malaria and related haemosporidian parasites in captive birds from a Brazilian megalopolis. Malar J 16:83. https://doi.org/10.1186/s12936-017-1729-8
Clark NJ, Clegg SM (2017) Integrating phylogenetic and ecological distances reveals new insights into parasite host specificity. Mol Ecol 26(11):3074–3086. https://doi.org/10.1111/mec.14101
Clark NJ, Clegg SM, Lima MR (2014) A review of global diversity in avian haemosporidians (Plasmodium and Haemoproteus: Haemosporida): new insights from molecular data. Int J Parasitol 44:329–338. https://doi.org/10.1016/j.ijpara.2014.01.004
Correa C, Vásquez D, Castro-Carrasco C, Zúñiga-Reinoso Á, Ortiz JC, Palma RE (2017) Species delimitation in frogs from South American temperate forests: the case of Eupsophus, a taxonomically complex genus with high phenotypic variation. PLoS One 12:e0181026. https://doi.org/10.1371/journal.pone.0181026
De Oliveira L, Cedrola F, Senra MVX, Scopel KKG, Martinele I, Tostes R, Dias RJP, D’Agosto M (2019) Polymorphism evidence in Plasmodium (Haemamoeba) lutzi Lucena, 1939 (Apicomplexa, Haemosporida) isolated from Brazilian wild birds. Parasitol Int 70:70–76
Dean W. (2004) A ferro e fogo: a história e a devastação da Mata Atlântica brasileira. São Paulo
Dimitrov D, Zehtindjiev P, Bensch S, Ilieva M, Iezhova T, Valkiūnas G (2014) Two new species Haemoproteus Kruse, 1890 (Haemosporida, Haemoproteidae) from European birds, with emphasison DNA barcoding for detection of haemosporidians in wildlife. Syst Parasitol 87:135–151. https://doi.org/10.1007/s11230-013-9464-1
Dinhopl N, Nedorost N, Mostegl MM, Weissenbacher-Lang C, Weissenböck H (2015) In situ hybridization and sequence analysis reveal an association of Plasmodium spp. with mortalities in wild passerine birds in Austria. Parasitol Res 114:1455–1462. https://doi.org/10.1007/s00436-015-4328-z
Eldredge N, Cracraft J (1980) Phylogenetic patterns and the evolutionary process. Method and theory in comparative biology. Columbia University Press, New York, p 349
Esselstyn JA, Evans BJ, Sedlock JL, Anwarali Khan FA, Heaney LR (2012) Single-locus species delimitation: a test of the mixed Yule–coalescent model, with an empirical application to Philippine round-leaf bats. Proc R Soc B Biol Sci 279:3678–3686. https://doi.org/10.1098/rspb.2012.0705
Falk BG, Mahler DL, Perkins SL (2011) Tree-based delimitation of morphologically ambiguous taxa: a study of the lizard malaria parasites on the Caribbean island of Hispaniola. Int J Parasitol 41:967–980. https://doi.org/10.1016/j.ijpara.2011.05.004
Fecchio A, Pinheiro R, Felix G, Faria IP, Pinho JB, Lacorte GA, Braga EM, Farias IP, Aleixo A, Tkach VV, Collins MD, Bell JA, Weckstein JD (2017) Host community similarity and geography shape the diversity and distribution of haemosporidian parasites in Amazonian birds. Ecog 00:01–10. https://doi.org/10.1111/ecog.03058
Fecchio A, Bell JA, Collins MD, Farias IP, Trisos CH, Tobias JA, Tkach VV, Weckstein JD, Ricklefs RE, Batalha-Filho H (2018) Diversification by host-switching and dispersal shaped the diversity and distribution of avian malaria parasites in Amazonia. Oikos 127:1233–1242
Fecchio A, Wells K, Bell JA, Tkach VV, Lutz HL, Weckstein JD, Clegg SM, Clark NJ (2019a) Climate variation influences host specificity in avian malaria parasites. Ecol Lett 22:547–557. https://doi.org/10.1111/ele.13215
Fecchio A, Bell JA, Bosholn M, Vaughan JA, Tkach VV, Lutz HL, Cueto VR, Gorosito CA, González-Acuña D, Stromlund C, Kvasager D, Comiche KJM, Kirchgatter K, Pinho JB, Berv J, Anciães M, Fontana CS, Zyskowski K, Sampaio S, Dispoto JH, Galen SC, Weckstein JD, Clark NJ (2019b) An inverse latitudinal gradient in infection probability and phylogenetic diversity for Leucocytozoon blood parasites in New World birds. J Anim Ecol 89:423–435. https://doi.org/10.1111/1365-2656.13117
Fecchio A, Bell JA, Pinheiro RBP, Cueto VR, Gorositio CA, Lutz HL, Gaiotti MG, Paiva LV, França LF, Toledo-Lima G, Toletino M, Pinho JB, Fontana CS, Grande JM, Santillán MA, Caparroz R, Roos AL, Kohler G, Bessa R, Nogueira W, Moura T, Nolasco EC, Comiche KJM, Kirchgatter K, Guimarães LO, Dispoto JH, Marinia MA, Tkach VV, Weckstein JD, Batalha-Filho H, Collins MD (2019c) Avian host composition, local speciation, and dispersal drive the regional assembly of avian malaria parasites in South American birds. Mol Ecol 28:2681–2693
Ferreira FCJ, Rodrigues RA, Sato Y, Borges MAZ, Braga EM (2016) Searching for putative avian malaria vectors in a seasonally dry tropical forest in Brazil. Parasit Vectors 9:587. https://doi.org/10.1186/s13071-016-1865-y
Fujita MK, Leaché AD, Burbrink FT, McGuire JA, Moritz C (2012) Coalescent-based species delimitation in an integrative taxonomy. Trends Ecol Evol 27:480–488
Furness RJ, Greenwood J (1993) Birds as monitors of environmental change. Chapman and Hall, London
Galen SC, Nunes R, Sweet PR, Perkins SL (2018) Integrating coalescent species delimitation with analysis of host specificity reveals extensive cryptic diversity despite minimal mitochondrial divergence in the malaria parasite genus Leucocytozoon. BMC Evol Biol 18(1):128
Galindo-Leal C, Câmara IG (2003) Atlantic Forest hotspot status: na overview. In: Galindo-Leal C, Câmara IG (eds) The Atlantic Forest of South America: biodversity status, threats, and outlook. Island Press, Washington
Garnham PC (ed) (1966) Malaria parasites and other Haemosporidia. Blackwell Scientific Publications, Oxford
Glaizot O, Fumagalli L, Iritano K, Lalubin F, Van Rooyen J, Christe P (2012) High prevalence and lineage diversity of avian malaria in wild populations of great tits (Parus major) and mosquitoes (Culex pipiens). PLoS One 7:e34964. https://doi.org/10.1371/journal.pone.0034964
Godfrey RD, Fedynich AM, Pence DB (1987) Quantification of hematozoa in blood smears. J Wildl Dis 23:558–565. https://doi.org/10.7589/0090-3558-23.4.558
Guedes ML (2012) Culicidae (Diptera) no Brasil: relações entre diversidade, distribuição e enfermidades. Oecol Aust 16:283–296. https://doi.org/10.4257/oeco.2012.1602.07
Hellgren O, Waldenström J, Bensch S (2004) A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium, and Haemoproteus from avian blood. J Parasitol 90:797–802. https://doi.org/10.1645/GE-184R1
Hellgren O, Križanauskienė A, Valkiūnas G, Bensch S (2007) Diversity and phylogeny of mitochondrial cytochrome b lineages from six morphospecies of avian Haemoproteus (Haemosporida: Haemoproteidae). J Parasitol 93:889–896. https://doi.org/10.1645/GE-1051R1.1
Hofmann EP, Nicholson KE, Luque-Montes IR, Köhler G, Cerrato-Mendoza CA, Medina-Flores M, Wilson LD, Townsend JH (2019) Cryptic diversity, but to what extent? Discordance between single-locus species delimitation methods within mainland anoles (Squamata: Dactyloidae) of northern Central America. Front Genet 10:1–13
Ilgūnas M, Bukauskaitė D, Palinauskas V, Iezhova TA, Dinhopl N, Nedorost N, Weissenbacher-Lang C, Weissenböck H, Valkiūnas G (2016) Mortality and pathology in birds due to Plasmodium (Giovannolaia) homocircumflexum infection, with emphasis on the exoerythrocytic development of avian malaria parasites. Malar J 15:256. https://doi.org/10.1186/s12936-016-1310-x
Kapli P, Lutteropp S, Zhang J, Kobert K, Pavlidis P, Stamatakis A, Flouri T (2017) Multi-rate Poisson tree processes for single-locus species delimitation under maximum likelihood and Markov chain Monte Carlo. Bioinformatics 33:1630–1638. https://doi.org/10.1093/bioinformatics/btx025
Karadjian G, Puech MP, Duval L, Chavatte JM, Snounou G, Landau I (2013) Haemoproteus syrnii in Strix aluco from France: morphology, stages of sporogony in a hippoboscid fly, molecular characterization and discussion on the identification of Haemoproteus species. Parasite 20:32. https://doi.org/10.1051/parasite/2013031
Karadjian G, Martinsen E, Duval L, Chavatte JM, Landau I (2014) Haemoproteus ilanpapernai n. sp. (Apicomplexa, Haemoproteidae) in Strix seloputo from Singapore: morphological description and reassignment of molecular data. Parasite 21:17. https://doi.org/10.1051/parasite/2014018
Katoh K, Rozewicki J, Yamada KD (2017) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform bbx108:1–7. https://doi.org/10.1093/bib/bbx108
Köppen W (1970) Roteiro para classificação climática. Mimeo
Križanauskienė A, Iezhova TA, Sehgal RNM, Carlson JS, Palinauskas V, Bensch S, Valkiūnas G (2013) Molecular characterization of Haemoproteus sacharovi (Haemosporida, Haemoproteidae), a common parasite of columbiform birds, with remarks on classification of haemoproteids of doves and pigeons. Zootaxa 3613:85–94. https://doi.org/10.11646/zootaxa.3616.1.7
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Lacorte GA, Felix GM, Pinheiro RR, Chaves AV, Almeida-Neto G, Neves FS, Leite LO, Santos FRM, Braga EM (2013) Exploring the diversity and distribution of neotropical avian malária parasites: a molecular survey from Southeast Brazil. PLoS One 8:e57770. https://doi.org/10.1371/journal.pone.0057770
Leria L, Vila-Farré M, Álvarez-Presas M, Sánchez-Gracia A, Rozas J, Sluys R, Riutort M (2020) Cryptic species delineation in freshwater planarians of the genus Dugesia (Platyhelminthes, Tricladida): extreme intraindividual genetic diversity, morphological stasis, and karyological variability. Mol Phylogenet Evol 143:1–24
Lima LM (2014) Birds of the Atlantic Forest: richness, status, composition, endemism, and conservation. Dissertation, Universidade de São Paulo
Littlewood DTJ (2012) Systematics as a cornerstone of parasitology: overview and preface. Parasitology 138:1633–1637. https://doi.org/10.1017/S0031182011001533
Martinsen ES, Perkins S, Schall JJ (2008) A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): evolution of life-history traits and host switches. Mol Phylogenet Evol 47:261–273. https://doi.org/10.1016/j.ympev.2007.11.012
Mello B, Vilela JF, Schrago CG (2018) Conservation phylogenetics and computational species delimitation of Neotropical primates. Biol Conserv 217:397–406
Merino S, Moreno J, Vásquez RA, Martínez J, Sánchez-monsálvez I, Estades CF, Ippi S, Sabat P, Rozzi R, Mcgehee S (2008) Haematozoa in forest birds from southern Chile: latitudinal gradients in prevalence and parasite lineage richness. Austral Ecol 33:329–340. https://doi.org/10.1111/j.1442-9993.2008.01820.x
Mittermeier RA, Robles Gil P, Hoffmann M, Pilgrim J, Brooks T, Mittermeier CG, Lamoreux J, Fonseca GAB (2005) Hotspots revisited: Earth’s biologically richest and most endangered terrestrial ecoregions. CEMEX, Washington DC
MMA, Ministry of the Environment, Brazil. https://www.mma.gov.br; Accessed on May, 2020
Monaghan MT, Wild R, Elliot M, Fujisawa T, Balke M, Inward DJ, Lees DC, Ranaivosolo R, Eggleton P, Barraclough TG, Vogler AP (2009) Accelerated species inventory on Madagascar using coalescent-based models of species delineation. Syst Biol 58:298–311. https://doi.org/10.1093/sysbio/syp027
Musher LJ, Cracraft J (2018) Phylogenomics and species delimitation of a complex radiation of Neotropical suboscine birds (Pachyramphus). Mol Phylogenet Evol 118:204–221
Nilsson E, Taubert H, Hellgren O, Huan X (2016) Multiple cryptic species of sympatric generalists within the avian blood parasite Haemoproteus majoris. J Evol Biol 29:1812–1826
Olias P, Wegelin M, Zenker W, Freter S, Gruber AD, Klopfleisch R (2011) Avian malaria deaths in parrots. Europe Emerg Infect Dis 17:950–952
Oliveira L, Cedrola F, Senra MVX, Scopel K, Martinela I, Tostes RC, Dias RJP, D’Agosto M (2019) Polymorphism evidence in Plasmodium (Haemamoeba) lutzi Lucena, 1939 (Apicomplexa, Haemosporida) isolated from Brazilian wild birds. Parasitol Int 70:70–76
Outlaw DC, Ricklefs RE (2014) Species limits in avian malaria parasites (Haemosporida): how to move forward in the molecular era. Parasitol 141:1223–1232. https://doi.org/10.1017/S0031182014000560
Pacheco MA, Matta NE, Valkiūnas G, Parker PG, Mello B, Stanley CE Jr, Lentino M, Garcia-Amado MA, Cranfield M, Kosakovsky Pond SL, Escalante AA (2017) Mode and rate of evolution of haemosporidian mitochondrial genomes: timing the radiation of avian parasites. Mol Biol Evol 35:383–403. https://doi.org/10.1093/molbev/msx285
Palinauskas V, Žiegytė R, Iezhova TA, Ilgūnas M, Bernotienė R, Valkiūnas G (2016) Description, molecular characterisation, diagnostics and life cycle of Plasmodium elongatum (lineage pERIRUB01), the virulent avian malaria parasite. Int J Parasitol 46:697–707. https://doi.org/10.1016/j.ijpara.2016.05.005
Perkins SL (2000) Species concepts and malaria parasites: detecting a cryptic species of Plasmodium. Proc R Soc B Biol Sci 267:2345–2350. https://doi.org/10.1098/2Frspb.2000.1290
Perkins SL (2014) Malaria’s many mates: past, present, and future of the systematics of the order Haemosporida. J Parasitol 100:11–25. https://doi.org/10.1645/13-362.1
Perkins SL, Martinsen ES, Falk BG (2011) Do molecules matter more than morphology? Promises and pitfalls in parasites. Parasitology 138:1664–1674. https://doi.org/10.1017/S0031182011000679
Piratelli AJ, Sousa SD, Corrêa JS, Andrade VT, Ribeiro RY, Avelar L, Oliveira EF (2008) Searching for bioindicators of forest fragmentation: passerine birds in the Atlantic forest of southeastern Brazil. Braz J Biol 68:259–268
Pons J, Barraclough TG, Gomez-Zurita J, Cardoso A, Duran DP, Hazell S, Kamoun S, Sumlin WD, Vogler AP (2006) Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst Biol 55:595–609
Ratnasingham S, Hebert PD (2013) A DNA-based registry for all animal species: the Barcode Index Number (BIN) system. PLoS One 8:e66213. https://doi.org/10.1371/journal.pone.0066213
Ribeiro SF, Sebaio F, Branquinho FC, Marini MA, Vago AR, Braga EM (2005) Avian malaria in Brazilian passerine birds: parasitism detected by nested PCR using DNA from stained blood smears. Parasitol 130:261–267. https://doi.org/10.1017/S0031182004006596
Ricklefs RE, Outlaw DC (2010) A molecular clock for malaria parasites. Science 329:226–229
Ridgely RS, Tudor G (2009) Field guide to the songbirds of South America: the passerines. University of Texas Press, Austin
Ronquist F, Teslenko M, Van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542
Ryder OA (1986) Species conservation and systematics: the dilemma of subspecies. Trends Ecol Evol 1:9–10. https://doi.org/10.1016/0169-5347(86)90059-5
Santiago-Alarcon D, Outlaw DC, Ricklefs RE, Parker PG (2010) Phylogenetic relationships of haemosporidian parasites in New World Columbiformes, with emphasis on the endemic Galapagos dove. Int J Parasitol 40:463–470. https://doi.org/10.1016/j.ijpara.2009
Santos RP (2009) Introdução ao ArcGis: Conceitos e Comandos. Brasília, DF
Sigrist T (2014) Avifauna Brasileira - Guia de Campo Avis Brasilis, 4th edn. Avis Brasilis, São Paulo
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1213. https://doi.org/10.1093/bioinformatics/btu033
Talavera G, Castresana J (2007) Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56:564–577. https://doi.org/10.1080/10635150701472164
Tavaré S (1986) Some probabilistic and statistical problems in the analysis of DNA sequences. Lectures Math Life Sci (Amer Math Soc) 17:57–86
Tostes R, Dias RJP, Martinele I, Senra MVX, D’Agosto M, Massard CL (2017) Multidisciplinary re-description of Plasmodium (Novyella) paranucleophilum in Brazilian wild birds of the Atlantic Forest kept in captivity. Parasitol Res 116:1887–1897. https://doi.org/10.1007/s00436-017-5465-3
Tostes D, Dias RJP, Oliveira L, Senra MV, Massard CL, D’Agosto M (2018) Molecular and morphological characterization of a Brazilian lineage of Plasmodium (Novyella) unalis in Turdus spp. (Passeriformes) of the Atlantic Forest, with remarks on new hosts and high genetic variation. J Parasitol 104:70–78. https://doi.org/10.1645/16-189
Turner IM (1996) Species loss in fragments of tropical rain forest: a review of the evidence. J Appl Ecol 33:200–209
Valkiūnas G (2005) Avian malaria parasites and other Haemosporidia. CRC Press, Boca Raton
Valkiūnas G, Iezhova TA (2018) Keys to the avian malaria parasites. Malar J 17:212. https://doi.org/10.1186/s12936-018-2359-5
Valkiūnas G, Iezhova TA, Križanauskienė A, Palinauskas V, Sehgal RNM, Bensch S (2008a) A comparative analysis of microscopy and PCR-based detection methods for blood parasites. J Parasitol 94:1395–1401. https://doi.org/10.1645/GE-1570.1
Valkiūnas G, Iezhova TA, Loiseau C, Chasar A, Thomas BS, Sehgal RNM (2008b) New species of haemosporidian parasites (Haemosporida) from African rainforest birds, with remarks on their classification. Parasitol Res 103:1213–1228. https://doi.org/10.1007/s00436-008-1118-x
Valkiūnas G, Iezhova TA, Loiseau C, Sehgal R (2009a) Nested cytochrome b polymerase chain reaction diagnostics detect sporozoites of hemosporidian parasites in peripheral blood of naturally infected birds. J Parasitol 95:1512–1515. https://doi.org/10.1645/GE-2105.1
Valkiūnas G, Iezhova TA, Loiseau C, Smith TB, Sehgal RNM (2009b) New malaria parasites of the subgenus Novyella in African rainforest birds, with remarks on their high prevalence, classification and diagnostics. Parasitol Res 104:1061–1077. https://doi.org/10.1007/s00436-0081289-5
Valkiūnas G, Santiago-Alarcon D, Levin II, Iezhova TA, Parker PG (2010a) A new Haemoproteus species (Haemosporida: Haemoproteidae) from the endemic Galapagos dove Zenaida galapagoensis, with remarks on the parasite distribution, vectors, and molecular diagnostics. J Parasitol 96:783–792. https://doi.org/10.1645/GE-2442.1
Valkiūnas G, Sehgal RN, Iezhova TA, Hull AC (2010b) Identification of Leucocytozoon toddi group (Haemosporida: Leucocytozoidae), with remarks on the species taxonomy of leucocytozoids. J Parasitol 96:170–177. https://doi.org/10.1645/GE-2109.1
Valkiūnas G, Ilgūnas M, Bukauskaitė D, Palinauskas V, Bernotienė R, Iezhova TA (2017) Molecular characterization and distribution of Plasmodium matutinum, a common avian malaria parasite. Parasitology 144:1726–1735. https://doi.org/10.1017/S0031182017000737
Valkiūnas G, Pendl H, Olias P (2017a) New Haemoproteus parasite of parrots, with remarks on the virulence of haemoproteids in naive avian hosts. Acta Trop 176:256–262
Vanstreels RE, da Silva-Filho RP, Kolesnikovas CK, Bhering RC, Ruoppolo V, Epiphanio S, Amaku M, Ferreira Junior FC, Braga EM, Catão-Dias JL (2015) Epidemiology and pathology of avian malaria in penguins undergoing rehabilitation in Brazil. Vet Res 46:1–12. https://doi.org/10.1186/s13567-015-0160-9
Woodworth-Lynas CB, Caines JR, Bennett GF (1989) Prevalence of avian Haematozoa in São Paulo state, Brazil. Mem Inst Oswaldo Cruz 84:515–526. https://doi.org/10.1590/S007402761989000400009
Yang Z (1994) Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J Mol Evol 39:306–314. https://doi.org/10.1007/BF00160154
Zhang J, Kapli P, Pavlidis P, Stamatakis A (2013) A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29:2869–2876. https://doi.org/10.1093/bioinformatics/btt499
Acknowledgements
We thank all students of the Laboratório de Protozoologia (LabProto) and Laboratório de Artrópodes Parasitos (LAP)—who helped in laboratory work and fieldwork—and Dr. Ralph Maturano for identification of bird’s species. We thank all avian haemosporidian researchers that provided their sequences to MalAvi.
Funding
This study was financially supported by the Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG/PPM/2016; PPM-00734-16) and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, grant numbers 1525/2011), and a fellowship was granted to LO (PhD) and to HAS and RJPD (Bolsa de Produtividade de Pesquisa PQ) from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures and sampling methods of this study involving animals were in accordance with the ethical standards of the institution or practice at which the studies were conducted and were evaluated and approved by the Comitê de Ética e Experimentação Animal da Universidade Federal de Juiz de Fora (protocolo no. 42/2012) and by the Sistema de Autorização e Informação em Biodiversidade (SISBIO) (no. 29268-3).
Additional information
Section Editor: Larissa Howe
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 466 kb)
Rights and permissions
About this article
Cite this article
Oliveira, L., Dias, R.J.P., Rossi, M.F. et al. Molecular diversity and coalescent species delimitation of avian haemosporidian parasites in an endemic bird species of South America. Parasitol Res 119, 4033–4047 (2020). https://doi.org/10.1007/s00436-020-06908-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-020-06908-9