Abstract
Drought is a major abiotic stress in crop yield and its inevitable consequence is the increased production of reactive oxygen species (ROS) and cell damage. To reduce excessive ROS accumulation in soybean, AtYUCCA6 gene was transformed via Agrobacterium-mediated transformation. About 3% of transformation efficiency was generated from five batches of the transformation experiment. Eighteen transgenic plants were produced with PPT resistance and analyzed for introgression of AtYUCCA6. T-DNA insertion and expression were confirmed by PCR, Southern blot and reverse transcriptase-PCR. In the drought tolerance tests with transgenic lines #2, #3, and #5, all three lines were less affected by drought treatment and survived in the water-deficit conditions while non-transgenic plants did not survive under the same drought condition. The physiological aspects of transgenic lines were also much stronger than NT plants by showing higher chlorophyll content and lower ion leakage during water-deficit conditions (p < 0.01), indicating the prevention of cell-membrane damage. Measurement of transpiration rate on detached leaves from transgenic plants showed nearly 10% less water loss. Finally, 3 transgenic lines (#2, #3, and #5) were investigated for ROS accumulation by DAB staining of detached leaves under water-deficit conditions. Unlikely NT plants with severe dark browning after 14 days of drought treatment, transgenic lines #2, #3, and #5 did not show significant browning.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As a sessile organism, plants are constantly exposed to external adverse environments, such as drought, salinity, heat, cold, and heavy metals. Drought is a major abiotic stress that causes severe problems in crop growth and yield (Santner and Estelle 2009; Manavalan et al. 2009; Zhang et al. 2010). One inevitable consequence of drought stress is the increase of toxic molecules, reactive oxygen species (ROS), in various cell compartments, such as chloroplasts, peroxisomes, and mitochondria (Cruz de Carvalho 2008; Blomster et al. 2011; Chan et al. 2016).
Plants produce various phytohormones, which play important roles in regulating growth and development as well as the response to diverse environmental stresses. Auxin, a plant hormone, regulates many aspects of physiological and developmental process in plants, including cell division, expansion, and differentiation, seed dormancy, lateral root formation, floral organ formation, and tropic responses (Kim et al. 2007; Iglesias et al. 2010; Park et al. 2013; Shi et al. 2014; Cha et al. 2015; Cheng et al. 2015). Indole-3-acetic acid (IAA), the major auxin, is synthesized via tryptophan (Trp)-dependent and Trp-independent pathways. In Arabidopsis, the Trp-dependent IAA biosynthetic pathway can proceed via four well-defined routes that produce indole-3-pyruvic acid (IPA), indole-3-acetaldoxime (IAOx), indole-3-acetamide (IAM), and tryptamine (TAM) as intermediates (Zhao 2012). YUCCA, belonging to plant flavin monooxygenase (FMO) family, directly catalyzes the conversion of IPA to IAA, which is considered as a predominant pathway (Zhao 2014). Arabidopsis possesses 11 YUCCA gene families, and the proteins play important roles in auxin (IAA) biosynthesis and plant development (Kim et al. 2011, 2013a, b; Dai et al. 2013; Park et al. 2013; Cha et al. 2015; Ke et al. 2015). Overexpression of AtYUCCA6 in Arabidopsis and potato was found to improve IAA-related phenotypes and increase drought tolerance by controlling toxic ROS accumulation under drought stress (Kim et al. 2013a, b; Cha et al. 2015). Despite to these dominant agronomical traits of YUCCA family genes, genetic transformation has not been applied to crop soybeans so far. Therefore, the development of drought-tolerant soybean varieties will contribute not only to coping with environmental changes, but also to practical use in agriculture.
Soybean (Glycine max L.) is one of the most significant sources of vegetable oil and plant-derived protein for food. Transformation is an optimal technique for developing soybean varieties with desired traits that are difficult to obtain from traditional breeding. Agrobacterium-mediated transformation has been efficiently used to introduce foreign genes into soybeans (Verma et al. 2014; Du et al. 2016; Li et al. 2017). Many researchers have used cotyledonary nodes by germinating seeds as the explants for soybean transformation (Hinchee et al. 1988; Di et al. 1996; Zhang et al. 1999). Instead of germinating soybean seeds, soybean transformation using “half-seed” explants has been improved (Paz et al. 2006). Using half-seed explants is a preferred method for soybeans, because it is less time-consuming, is simple to use, and allows easy explant preparation. There are important factors to increase the efficiency of the transformation process, such as simple wounding, treatment by sonication, vacuuming, and additional use of thiol compounds in the co-cultivation medium. Thiol compounds, including l-cysteine, sodium thiosulfate, and dithiothreitol (DTT), can inhibit the extensive tissue browning or cell death in the wounded area of a shoot pad and significantly improve T-DNA transfer and the frequency of transformed cells. (Olhoft et al. 2003; Dan 2008; Verma et al. 2014; Kim et al. 2013a, b, 2016, 2017a, b, 2018).
In this study, soybean plants overexpressing AtYUCCA6 were produced by Agrobacterium-mediated transformation with the expectation they would be stress-tolerant because of the reduction of ROS. AtYUCCA6 overexpression exhibited low amounts of ROS accumulation and better drought tolerance in transgenic soybean plants than in wild-type plants.
Materials and methods
Vector construction and Agrobacterium preparation
The AtYUCCA6 cDNA open reading frame was amplified from its original vector (provided by Dr. D J Yun at Konkuk University, Republic of Korea) using AtYUCCA6-F primer (5′-ATGGAAGGTAAACTAGCACATGAC-3′) and AtYUCCA6-R primer (5′-TCAATTCCCACCACAATCACTCTC-3′). The desired destination vector, pPZP-3′PinII-Bar (provided by Dr. J K Kim at Myongji University, Republic of Korea) was used for the vector construction. The resultant plasmid, pPZP-3′PinII-Bar-AtYUCCA6 (Fig. S1), was transformed into Agrobacterium tumefaciens strain EHA105 for soybean transformation and subsequently cultured on solid YEP media [10 g l−1 yeast extract, 5 g l−1 NaCl, 10 g l−1 peptic peptone, and 1.0% (w/v) plant agar, pH 7.0] containing 50 mg l−1 spectinomycin and 25 mg l−1 rifampicin at 28 °C for 2 days. A single colony was chosen and grown in 20 ml of liquid YEP medium containing 50 mg l−1 spectinomycin and 25 mg l−1 rifampicin at 28 °C for a day until OD600 reached between 0.6 and 0.8. Competent cells were then prepared by mixing equal volumes of 30% (v/v) glycerol. Aliquots of competent cells were frozen and kept at − 70 °C until used.
Soybean transformation
Mature soybean seeds of Korean cultivar Kwangankong were used in Agrobacterium-mediated soybean transformation by following the method described by Kim et al. (2012, 2013a, b, 2016, 2017a, b, 2018). Five batches of the transformation experiment were carried out with 120–130 soybean seeds each time. To identify putative transformants expressing the Bar gene, two trifoliate leaves from T0 plants were screened using an herbicide paint assay. The upper surface of a leaf was painted with the mixture of 100 mg l−1 PPT and Tween 20 using a brush. The response to this herbicide assay was screened after 3–5 days of PPT leaf painting. Plants with PPT resistance were grown in a greenhouse until maturity, and T1 seeds were harvested (Fig. S2).
Confirmation of transgene in transgenic plants
Genomic DNA was extracted from leaf tissues of non-transgenic (NT) and transgenic plants using cetyltrimethylammonium bromide. The polymerase chain reaction (PCR) analysis was performed using KOD FX (TOYOBO, Osaka, Japan) according to the manufacturer’s instructions with a thermal cycler (Takara, Japan). The primer sets were designed in the regions of AtYUCCA6 (5′-AGGTAAACTAGCACATGACCACCG-3′/5′-TCAATTCCCACCACAATCACTCTC-3′) and Bar (5′-AGACAAGCACGGTCAACTTCCGTA-3′/5′-CCGGCAGGCTGAAGTCCAGC-3′) genes. To evaluate the T-DNA insertion into the plant genome, additional primers of DNA from Bar gene to left border (LB) (5′-AGACAAGCACGGTCAACTTCCGTA-3′/5′-TGGCAGGATATATTGTGGTGTAAA-3′) and from right border (RB) to AtYUCCA6 gene (5′-GTTTACCCGCCAATATATCCTGTCA-3′/5′-TCAATTCCCACCACAATCACTCTC-3′) were used to amplify both end regions of the vector.
For Southern blot analysis, 20 µg of genomic DNA from NT and transgenic plants were digested overnight using HindIII, fractionated on 0.8% (w/v) agarose gel by electrophoresis, then transferred onto Hybond N+ nylon membrane (Amersham Pharmacia, USA). Hybridization, washing, and detection were performed using a digoxigenin (DIG)-labeled DNA probe and a chemiluminescent system (Roche, Germany) according to the manufacturer’s instructions. The DIG-labeled probe was prepared by PCR amplification with the Bar primers (5′- AACTTCCGTACCGAGCCGCA-3′/5′-TCGTAGGCGTTGCGTGCCTT-3′).
RNA analysis of transgenic plants
Total RNAs were isolated from both NT and transgenic T0 plants using plant RNA purification reagent (Invitrogen, USA) according to the manufacturer’s instructions. Reverse transcriptase-PCR (RT-PCR) was conducted using the RT-PCR Remix Kit (Genetbio, Korea) according to the manufacturer’s instructions. The primer sets used in the RT-PCR were as follows: AtYUCCA6, 5′-AGGTAAACTAGCACATGACCACCG-3′/5′-TCAATTCCCACCACAATCACTCTC-3′; Bar, 5′-AGACAAGCACGGTCAACTTCCGTA-3′/5′-CCGGCAGGCTGAAGTCCAGC-3′. The constitutive TUB (5′- TGAGCAGTTCACGGCCATGCT/5′-CTCGGCAGTGGCATCCTGGT-3′) was used as an internal control to normalize the amount of leaf RNA in the soybeans.
Drought-stress treatment in transgenic plants
To analyze the drought tolerance in AtYUCCA6 transgenic plants, NT and transgenic plants (T2) were grown in the same volume of soil and identical containers in a growth chamber with the conditions of 25 °C, 18 h light/6 h dark, and 60% humidity for 3 weeks, until the leaves on two nodes were fully expanded. Under the same conditions of plant growth, the phenotype of drought-stressed plants was monitored by the exposure of no-irrigation for 14 days and resuming irrigation for 3 days after the end of the drought treatment.
Measurement of total chlorophyll and relative ion leakage
Total chlorophyll from leaves of NT and transgenic plants after drought-stress treatment was isolated in 80% (v/v) acetone. The chlorophyll content was calculated using a spectrophotometer, as described by Wu et al. (2008). Statistical analysis was also performed using the Excel t test program to confirm significant differences.
The extent of ion leakage from NT and transgenic plants after the drought-stress treatment was measured by means of its conductivity. One gram of leaf samples was soaked in 10 ml of distilled water for 24 h at room temperature. The conductivity of the solution (Lt) was measured using an EC-400L conductivity meter (Istek, Korea). The leaf samples were then returned to the solution in the tubes, which were sealed and incubated at 95 °C for 20 min. The solution (L0) was then cooled to room temperature, and conductivity was re-measured. The Lt/L0 × 100 values were calculated and used to evaluate the relative electrolyte leakage (Fan et al. 1997). Statistical analysis was also performed using the Excel t test program to confirm significant differences.
Examination of transpiration rate
NT and transgenic plants were grown under the same conditions, including identical containers, the same volume of soil, a long day photoperiod (18 h light/6 h dark), and 60% humidity in the growth chamber. The fully expanded leaves from two nodes were detached and weighed for 200 min at 40-min intervals and compared to the initial weight on a sterile bench in an extractor hood. Statistical analysis was performed using the Excel t test program to confirm significant differences.
Detection of ROS content in transgenic plants
Drought treatment was applied to NT and transgenic plants for 14 days under the same conditions as mentioned above. The leaves of similar developmental stages were then detached from randomly chosen sites, immersed in 3,3′-diaminobenzidine (DAB) staining solution (1 mg ml−1, pH 3.8; Sigma, USA) for 4 h, incubated in 100% ethanol at 95 °C for 15 min until chlorophyll was cleared, and then observed with a stereomicroscope. Quantitative analysis of DAB staining was performed using image analysis software (ImageJ 1.52a; Java 1.8.0_112) (Sekulska-Nalewajko et al. 2016).
Results
Production of transgenic soybean plants using Agrobacterium-mediated transformation
To produce transgenic soybean plants, pPZP-3′PinII-Bar-AtYUCCA6 plasmid (Fig. S1) was used for the soybean transformation with half-seed explants of the Korean soybean cultivar Kwangankong (Fig. S2), following the modified protocol described by Kim et al. (2012, 2013a, b, 2016, 2017a, b). About 3% of transformation efficiency was generated from five batches in the transformation experiment, in which 100–120 soybean seeds were used each time. Eighteen transgenic plants were produced with PPT resistance. Among them, eight well-grown and early harvested transgenic plants (lines #2, #3, #4, #5, #9, #10, #12, and #14) were selected to confirm the integration of the transgene, and these lines were examined using PCR with AtYUCCA6 and Bar primers to amplify the DNA fragments of 1281 bp and 552 bp in size, respectively. In addition, T-DNA insertion was also confirmed by amplifying both end regions of the vector construct (Fig. 1). All eight transgenic lines showed the expected amplification of transgene sequences. To analyze the transcription level of AtYUCCA6 and Bar genes, reverse transcriptase-PCR (RT-PCR) was conducted with RNAs extracted from those eight transgenic plants (Fig. 2). The transformed AtYUCCA6 and Bar genes were expressed in all transgenic lines as expected, while those were not detected in non-transgenic (NT) plants.
Confirmation of introduced genes from AtYUCCA6 transgenic soybean plants (T0) using PCR. Genomic DNAs were extracted from transgenic plants (T0). aAtYUCCA6 gene. bBar gene. c The DNAs between Bar gene and left border (LB). d The DNAs between right border (RB) and AtYUCCA6 gene. NT, non-transgenic plant; #2, #3, #4, #5, #9, #10, #12, and #14, AtYUCCA6 transgenic lines (T0)
Transcript level of AtYUCCA6 and Bar genes in transgenic plants (T0) using reverse transcriptase-PCR (RT-PCR). Total RNAs were extracted from AtYUCCA6 transgenic plants (T0), and RT-PCR was used to confirm gene expressions. TUB gene was used as a quantitative control. NT, non-transgenic plant; #2, #3, #4, #5, #9, #10, #12, and #14, AtYUCCA6 transgenic lines (T0)
To investigate the number of transgene insertions in the selected eight transgenic lines, genomic Southern blot analysis was carried out using leaf samples from T2 seedlings (Fig. 3). Genomic DNAs from NT and transgenic plants were digested with HindIII and hybridized with Bar probe. All eight transgenic lines (#2, #3, #4, #5, #9, #10, #12, and #14) showed multiple insertion events. Lines #4 and #5 showed similar patterns in the result; they seem to be clones generated from the same shoot pad.
Genomic Southern blot analysis of AtYUCCA6 transgenic soybean. Twenty micrograms of genomic DNAs were digested with HindIII and hybridized with Bar probe. The approximate DNA size markers are indicated on the right. NT, non-transgenic plant; #2, #3, #4, #5, #9, #10, #12, and #14, AtYUCCA6 transgenic lines (T2)
Drought tolerance of YUCAA6 transgenic soybean plants
Drought tolerance tests were done with transgenic lines #2, #3, and #5, from which we were able to harvest T2 seeds relatively quickly out of the eight transgenic lines that underwent proliferation in the greenhouse. We investigated the response to drought stress in these three lines and compared them with NT plants. All plants were drought-treated for 14 days under the same conditions. Among them, NT plants started to lose vigor on the 11th day of drought, and all had withered completely at the end of the 14-day drought treatment. On the other hand, transgenic lines #2, #3, and #5 were less affected by drought treatment and survived in the water-deficit conditions. When re-watered for 3 days, NT plants did not survive and eventually died, whereas the transgenic lines #2, #3, and #5 fully recovered and continued to grow (Fig. 4a). And then, we confirmed the relative expression level of AtYUCCA6 at 14 days after drought stress by qRT-PCR and found that the expression was confirmed in all transgenic lines #2, #3, and #5 (Fig. 4b).
Drought tolerance of AtYUCCA6 soybean transgenic plants (T2). a Analysis of drought tolerance of AtYUCCA6 transgenic plants compared with NT plants. Plants were grown on soil until leaves were fully expanded on two nodes under the same conditions. Plants were deprived of water for 14 days, and then re-watered for 3 days (n = 12 each). The photographs were taken 7, 11, and 14 days after drought stress and 3 days after re-watering. bAtYUCCA6 gene expression with detached leaves at 14 days after drought stress using real-time PCR (qRT-PCR). c, d Calculation of chlorophyll content and ion leakage at the indicated days after drought treatment from two-node leaves (n = 6 each). NT, non-transgenic plant; #2, #3, #4, #5, #9, #10, #12, and #14, AtYUCCA6 transgenic lines (T2). Error bars indicate mean ± standard deviation. Asterisks indicate significant changes compared with NT (*p < 0.05; **p < 0.01)
The physiological aspects of transgenic lines were investigated by measuring changes in chlorophyll content and ion leakage during water-deficit conditions. The chlorophyll contents of transgenic lines #2, #3, and #5 were significantly higher than those of NT plants after 14 days of drought treatment (p < 0.01); the increased drought tolerance may have resulted from the maintenance of chlorophyll content (Fig. 4c). Moreover, ion leakage was increased in transgenic lines #2, #3, and #5, but significantly less (p < 0.01) than in NT plants; these transgenic lines were less affected by drought treatment because of the prevention of cell- membrane damage (Fig. 4d).
The water loss by transpiration was investigated by weighing detached leaves from transgenic and NT plants for 200 min at intervals of 40 min (Fig. 5). The leaves in NT started to curl up as a withered phenotype from 120 min after detached, while those in transgenic lines did not (Fig. 5a). After 200 min of drought treatment, the water content of NT plant leaves was only 47% of the initial leaf weight (Fig. 5b). In contrast, those of transgenic lines #2, #3, and #5 were about 5 to11% greater than that of the NT plants (p < 0.05 in lines #3 and #5). This result suggests that the AtYUCCA6 gene appeared to have a positive effect on drought tolerance by slowing water loss.
The water loss content of AtYUCCA6 transgenic plants (T2). Plants were grown on soil until leaves were fully expanded on two nodes; leaves were detached (n = 6 each) and weighed at the indicated times after drought treatment (a, b). NT, non-transgenic plant; #2, #3, #4, #5, #9, #10, #12, and #14, AtYUCCA6 transgenic lines (T2). Error bars indicate mean ± standard deviation. Asterisks indicate significant changes compared with NT (*p < 0.05; **p < 0.01)
Low ROS accumulation of AtYUCCA6 transgenic soybean plants
ROS is known to accumulate under drought stress, and transgenic leaves of Arabidopsis and potato overexpressing the AtYUCCA6 gene have been reported to have levels of ROS lower than those in leaves of the wild-type plants (Park et al. 2013; Cha et al. 2015; Chan et al. 2016). Thus, we analyzed ROS accumulation using DAB staining of detached leaves from transgenic soybean plants under water-deficit conditions (Fig. 6). By observing DAB stained leaves under the stereomicroscope, ROS accumulation could be visualized by the brown color in leaves of NT plants at the 11th day of drought treatment, and severe dark browning shown after 14 days of drought treatment. However, transgenic lines #2, #3, and #5 did not show significant browning until day 14 of drought treatment (Fig. 6a). To confirm the intensity of DAB staining in drought-treated transgenic plants, ROS accumulation was quantified by the image analyzing program. In transgenic lines #2, #3, and #5, accumulation of ROS was significantly less than NT plants after 14 days of drought treatment (p < 0.05 in lines #2 and #3) (Fig. 6b).
Low ROS accumulation in AtYUCCA6 soybean transgenic plant leaves (T2). a ROS accumulation visualized by DAB staining. Plants were grown on soil until leaves were fully expanded on two nodes and deprived of water for 14 days. Detached leaves subjected to drought stress were stained with DAB (1 mg ml−1, pH 3.8) to examine ROS accumulation. b Quantitative analysis of DAB staining. ROS accumulation in detached leaves shown in a were quantified by analyzing the image analyzer. Data represent that mean ± SE from two biological replicates. NT, non-transgenic plant; #2, #3, #4, #5, #9, #10, #12, and #14, AtYUCCA6 transgenic lines (T2). DAD, day after drought treatment. Asterisks indicate significant changes compared with NT plants (*p < 0.05; **p < 0.01)
Discussion
Auxin, an essential regulator of plant growth and development, plays an important role in responses to environmental stress, such as drought, salinity, and pathogens. ROS is known to be accumulated under drought stress in plants (Miller et al. 2010). YUCCA6 as a flavin monooxygenase enzyme converts IPA to auxin which is a downstream step of Trp aminotransferase (TAA1/TAR1/TAR2) converting Trp to IPA (Zhao 2012, 2014). Overexpression of the AtYUCCA6 gene in Arabidopsis and potato plants exhibited auxin overproduction and drought tolerance. These transgenic plants also showed less ROS than did wild-type plants under drought stress conditions (Park et al. 2013; Cha et al. 2015; Chan et al. 2016).
Our stable Agrobacterium-mediated soybean transformation has been established based on the half-seed (Paz et al. 2006) and cotyledonary-node (Hinchee et al. 1988) methods. This method included a mixture of thiol compounds, such as l-cysteine, sodium thiosulfate, and dithiothreitol, in a co-cultivation medium, which inhibited the activity of enzymatic browning and cell death in the wounded area. These additional treatments resulted in significantly increased T-DNA delivery into cotyledonary cells (Olhoft and Somer 2001; Olhoft et al. 2003). Thus, our modified transformation protocol enabled us to produce stable transgenic soybean plants with agronomically important genes for practical use in agriculture. Based on the evidence from previous studies, the AtYUCCA6 gene was transformed into soybean via Agrobacterium-mediated transformation to identify increased drought tolerance. Transgenic soybean plants showed normal growth under drought stress, whereas NT plants withered and died eventually. Drought stress resulted in a significant increase in ion leakage and low chlorophyll contents from leaves of NT plants. However, the phenotypic changes of transgenic plants were less affected by drought conditions, their cell membranes were protected, and chlorophyll content was also maintained. These physiological responses showed that drought tolerance was significantly increased by introducing the AtYUCCA6 gene into transgenic soybeans.
A correlation between AtYUCCA6 overexpression and low ROS accumulation was also investigated in soybean leaves. ROS, the result of the partial reduction of atmospheric O2, is continuously produced in plants under normal conditions with a relatively low level in organelles, such as chloroplasts, mitochondria, and peroxisomes, that function as components of a stress-signaling pathway. When plants are exposed to environmental stresses, such as drought, the production of ROS is significantly increased. A certain level of ROS toxicity results in oxidative damage to cellular membranes and other cellular components, including membrane lipids, chlorophyll, nucleic acid, and proteins. ROS can cause membrane lipid peroxidation, ion leakage, and chlorophyll loss that lead to cell death. Chloroplasts are particularly susceptible to ROS because of their damage in the photosynthetic electron-transfer system. In addition, when the concentration of ROS exceeds a certain range, it activates a programmed cell-death response in cells (Wang et al. 2005; Cruz de Carvalho 2008; Yasar et al. 2008; Gill and Tuteja 2010; Miller et al. 2010; Wu et al. 2008). When ROS accumulation was visualized by DAB staining in soybean leaves subjected to drought stress, the severe brown color related to ROS accumulation was detected in non-transgenic leaves. In contrast, lower ROS accumulation was exhibited by transgenic leaves. Among the transgenic plants (lines #2, #3, and #5), line #5 showed the greatest drought tolerance and the lowest ROS accumulation, which seemed to be associated with the highest level of AtYUCCA6 expression. Our results suggest that the level of AtYUCCA6 expression was correlated with the change of ROS accumulation. Low ROS accumulation resulted in the protection of cell membranes and the maintenance of chlorophyll content, and finally increased drought tolerance in AtYUCCA6 transgenic plants under drought treatment.
In our previous study, soybean transgenic plants overexpressing the AtABF3 gene reduced stomatal opening and increased drought tolerance under water-stress conditions (Kim et al. 2018). However, closure of stomata had a negative effect on plant height. AtYUCCA6 overexpressing soybean transgenic plants are not likely to have a negative impact on plant growth, since they are associated with ROS scavenging instead of stomatal closure. Under the natural condition, a field test of AtYUCCA6 transgenic soybeans will be carried out in the near future.
Soybean, a drought-sensitive crop, is severely affected by drought stress, which is the major yield-limiting factor in crop plants. In this study, we introduced the AtYUCCA6 gene from Arabidopsis into soybean using Agrobacterium-mediated transformation to increase drought tolerance with low ROS accumulation. As always, the modest significance of our work is to test any available gene from a model plant in field-crop soybeans to evaluate its potential. The valuating gene via genetic transformation is urgently needed in the era of outpouring of genomics.
References
Blomster T, Salojärvi J, Sipari N, Brosché M, Ahlfors R, Keinänen M, Overmyer K, Kangasjärvi J (2011) Apoplastic reactive oxygen species transiently decrease auxin signaling and cause stress-induced morphogenic response in Arabidopsis. Plant Physiol 157:1866–1883
Cha JY, Kim WY, Kang SB, Kim JI, Baek D, Jung IJ, Kim MR, Li N, Kim HJ, Nakajima M, Asami T, Sabir JSM, Park HC, Lee SY, Bohnert HJ, Bressan RA, Pardo JM, Yun DJ (2015) A novel thiol-reductase activity of Arabidopsis YUC6 confers drought tolerance independently of auxin biosynthesis. Nat Commun 6:8041
Chan Z, Yokawa K, Kim WY, Song CP (2016) Editorial: ROS regulation during plant abiotic stress responses. Front Plant Sci 7:1536
Cheng Y, Dai X, Zhao Y (2015) Auxin biosynthesis by the YUCCA Flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20:1790–1799
Cruz de Carvalho MH (2008) Drought stress and reactive oxygen species: Production, scavenging and signaling. Plant Signal Behav 3(3):156–165
Dai X, Mashiguchi K, Chen Q, Kasahara H, Kamiya Y, Ojha S, DuBois J, Ballou D, Zhao Y (2013) The Biochemical mechanism of auxin biosynthesis by an Arabidopsis YUCCA flavin-containing monooxygenase. J Biol Chem 288(3):1448–1457
Dan Y (2008) Biological functions of antioxidants in plant transformation. In Vitro Cell Dev Biol Plant 44(3):149–161
Di R, Purcell V, Collins GB, Ghabrial SA (1996) Production of transgenic lines expressing the bean pod mottle virus coat protein precursor gene. Plant Cell Rep 15:746–750
Du H, Zeng X, Zhao M, Cui X, Wang Q, Yang H (2016) Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. J Biotechnol 217:90–97
Fan L, Zheng S, Wang X (1997) Antisense suppression of phospholipase Dα retards abscisic acid- and ethylene-promoted senescence of postharvest Arabidopsis leaves. Plant Cell 9:2183–2196
Gill SS, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48:909–930
Hinchee MAW, Connor-Ward DV, Newell CA, McDonnell RE, Sato SJ, Gasser CS, Fischhoff DA, Re DB, Fraley RT, Horsch RB (1988) Production of transgenic soybean plants using Agrobacterium-mediated DNA transfer. Nat Biotechnol 6:915–922
Iglesias MJ, Terrile MC, Bartoli CG, D’Ippólito S, Casalongué CA (2010) Auxin signaling participates in the adaptative response against oxidative stress and salinity by interacting with redox metabolism in Arabidopsis. Plant Mol Biol 74:215–222
Ke Q, Wang Z, Ji CY, Jeong JC, Lee HS, Li H, Xu B, Deng X, Kwak SS (2015) Transgenic poplar expressing Arabidopsis YUCCA6 exhibits auxin-overproduction phenotypes and increased tolerance to abiotic stress. Plant Physiol Biochem 94:19–27
Kim JI, Sharkhuu A, Jin JB, Li P, Jeong JC, Baek D, Lee SY, Blakeslee JJ, Murphy AS, Bohnert HJ, Hasegawa PM, Yun DJ, Bressan R (2007) yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes. Plant Physiol 145:722–735
Kim JI, Murphy AS, Baek D, Lee SW, Yun DJ, Bressan RA, Narasimhan ML (2011) YUCCA6 over-expression demonstrates auxin function in delaying leaf senescence in Arabidopsis thaliana. J Exp Bot 62(11):3981–3992
Kim MJ, Kim JK, Kim HJ, Pak JH, Lee JH, Kim DH, Choi HK, Jung HW, Lee JD, Chung YS, Ha SH (2012) Genetic modification of the soybean to enhance the β-carotene content through seed-specific expression. PLoS One 7(10):e48287
Kim HJ, Kim MJ, Pak JH, Jung HW, Choi HK, Lee YH, Baek IY, Ko JM, Jeong SC, Pack IS, Ryu KH, Chung YS (2013a) Characterization of SMV resistance of soybean produced by genetic transformation of SMV-CP gene in RNAi. Plant Biotechnol Rep 7:425–433
Kim JI, Baek D, Park HC, Chun HJ, Oh DH, Lee MK, Cha JY, Kim WY, Kim MC, Chung WS, Bohnert HJ, Lee SY, Bressan RA, Lee SW, Yun DJ (2013b) Overexpression of Arabidopsis YUCCA6 in potato results in high-auxin developmental phenotypes and enhanced resistance to water deficit. Mol Plant 6(2):337–349
Kim HJ, Kim MJ, Pak JH, Im HH, Lee DH, Kim KH, Lee JH, Kim DH, Choi HK, Jung HW, Chung YS (2016) RNAi-mediated soybean mosaic virus (SMV) resistance of a Korean soybean cultivar. Plant Biotechnol Rep 10:257–267
Kim HJ, Cho HS, Pak JH, Kim KJ, Lee DH, Chung YS (2017a) Overexpression of a chromatin architecture-controlling ATPG7 has positive effect on yield components in transgenic soybean. Plant Breed Biotechnol 5(3):237–242
Kim MJ, Kim HJ, Pak JH, Cho HS, Choi HK, Lee DH, Chung YS (2017b) Overexpression of AtSZF2 from Arabidopsis showed enhanced tolerance to salt stress in soybean. Plant Breed Biotechnol 5(1):1–15
Kim HJ, Cho HS, Pak HJ, Kwon T, Lee JH, Kim DH, Lee DH, Kim CG, Chung YS (2018) Confirmation of drought tolerance of ectopically expressed AtABF3 gene in soybean. Mol Cells 41(5):413–422
Li S, Cong Y, Liu Y, Wang T, Shuai Q, Chen N, Gai J, Li Y (2017) Optimization of Agrobacterium-mediated transformation in soybean. Front Plant Sci 8:246
Manavalan LP, Guttikonda SK, Tran LSP, Nguyen HT (2009) Physiological and molecular approaches to improve drought resistance in soybean. Plant Cell Physiol 50(7):1260–1276
Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R (2010) Reactive oxygen species homeostasis and signaling during drought and salinity stresses. Plant Cell Environ 33:453–467
Olhoft PM, Somer DA (2001) l-Cysteine increases Agrobacterium-mediated T-DNA delivery into soybean cotyledonary-node cells. Plant Cell Rep 20:706–711
Olhoft PM, Flagel LE, Donovan CM, Somers DA (2003) Efficient soybean transformation using hygromycin B selection in the cotyledonary-node method. Planta 216:723–735
Park HC, Cha JY, Yun DJ (2013) Roles of YUCCAs in auxin biosynthesis and drought stress responses in plants. Plant Signal Behav 8(6):e24495
Paz MM, Martinez JC, Kalvig AB, Fonger TM, Wang K (2006) Improved cotyledonary node method using an alternative explant derived from mature seed for efficient Agrobacterium-mediated soybean transformation. Plant Cell Rep 25:206–213
Santner A, Estelle M (2009) Recent advances and emerging trends in plant hormone signalling. Nature 459:25
Sekulska-Nalewajko J, Goclawski J, Chojak-Koźniewska J, Kuźniak E (2016) Automated image analysis for quantification of reactive oxygen species in plant leaves. Methods 109:114–122
Shi H, Chen L, Ye T, Liu X, Ding K, Chan Z (2014) Modulation of auxin content in Arabidopsis confers improved drought stress resistance. Plant Physiol Biochem 82:209–217
Verma K, Saini R, Rani A (2014) Recent advances in the regeneration and genetic transformation of soybean. J Innov Biol 1(1):15–26
Wang FZ, Wang QB, Kwon SY, Kwak SS, Su WA (2005) Enhanced drought tolerance of transgenic rice plants expressing a pea manganese superoxide dismutase. J Plant Physiol 162:465–472
Wu C, Niu Z, Tang Q, Huang W (2008) Estimating chlorophyll content from hyperspectral vegetation indices: modeling and validation. Agric For Meteorol 148:1230–1241
Yasar F, Ellialtioglu S, Yildiz K (2008) Effect of salt stress on antioxidant defense systems, lipid peroxidation, and chlorophyll content in green bean. Russ J Plant Physiol 55(6):782–786
Zhang ZY, Xing AQ, Staswick P, Clemente TE (1999) The use of glufosinate as a selective agent in Agrobacterium-mediated transformation of soybean. Plant Cell Tissue Org 56:37–46
Zhang H, Liu W, Wan L, Li F, Dai L, Li D, Zhang Z, Huang R (2010) Functional analyses of ethylene response factor JERF3 with the aim of improving tolerance to drought and osmotic stress in transgenic rice. Transgenic Res 19:809–818
Zhao Y (2012) Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol Plant 5:334–338
Zhao Y (2014) Auxin biosynthesis. Arabidopsis Book 12:e0173
Acknowledgements
This work was supported by the Next-Generation BioGreen 21 Program, Rural Development Administration (PJ01366501 granted to Y. S. Chung), and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2017R1A6A3A11028883 granted to H. J. Kim) in the Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11816_2019_527_MOESM1_ESM.tif
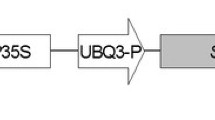
Supplementary material 1 Fig. S1. Vector used for soybean transformation. Amplified AtYUCCA6 (1,281 bp size) was subcloned into pPZP-3’PinII-Bar vector for soybean transformation. LB/RB, left/right T-DNA border; p35S/T35S, CaMV (cauliflower mosaic virus) 35S promoter/terminator; 3’PinII/3’nos, terminator; Bar, coding region of the DL-phosphinothricin resistance gene. HindIII and PstI restriction enzyme sites are marked (TIF 63 KB)
11816_2019_527_MOESM2_ESM.tif
Supplementary material 2 Fig. S2. AtYUCCA6overexpression soybean transgenic plants. (a) Co-cultivation of half-seed explants after inoculation (left) and at 5 days after inoculation (right). (b) Shoot induction on SIM without PPT for 14 days. (c) Shoot induction on SIM with 10 mg l-1 PPT for another 14 days. (d) Shoot elongation on SEM with 5 mg l-1 PPT. (e) Root formation (f) Acclimation of putative transgenic plant in a small pot. (g) Transgenic plant (T0) grown in a large pot in the greenhouse. (h) Herbicide (100 mg l-1 PPT) painting showing sensitivity in NT plant leaves (left) and resistance in transgenic plant leaves (right) (TIF 1279 KB)
Rights and permissions
About this article
Cite this article
Park, J.S., Kim, H.J., Cho, H.S. et al. Overexpression of AtYUCCA6 in soybean crop results in reduced ROS production and increased drought tolerance. Plant Biotechnol Rep 13, 161–168 (2019). https://doi.org/10.1007/s11816-019-00527-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-019-00527-2