Abstract
Aims
Pseudomonas koreensis, a subgroup of the P. fluorescens complex, is a potential plant growth-promoting rhizobacterium. This study explored the mechanisms of plant growth promotion by P. koreensis and its potential to induce stress tolerance in tomato.
Methods
Tomato plants in pots were inoculated with P. koreensis GS and cultured for 60 days. RNA sequencing and gas chromatography-mass spectrometry were used to detect global transcriptomic and metabolomic changes in tomato leaves. Liquid chromatography-mass spectrometry was used for quantification of plant hormones.
Results
The inoculated plants showed more vigorous growth, with higher leaf chlorophyll content and shoot biomass compared with uninoculated controls. The activities of several defense enzymes (e.g., phenylalanine ammonia-lyase) were enhanced in the leaves of inoculated plants. A total of 737 differentially expressed genes were identified, which were related to plant hormone biosynthesis, MAPK signaling transduction, and polyphenol biosynthesis. The contents of specific metabolites related to plant growth and stress tolerance, including polyphenols (phenylpropanoids) and amino acids (tryptophanand and proline), were increased after inoculation. Plant hormones such as indole-3-acetic acid participated in plant growth promotion by P. koreensis.
Conclusions
P. koreensis promoted plant growth and showed potential to induce stress tolerance in tomato by enhancing auxin and polyphenol-related pathways.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
China is the world’s leading producer of tomato, accounting for approximately 20.9 % (999,312 ha) of global harvested area and 31.8 % (5.631 × 1010 kg) of global production in 2016. However, the average yield per hectare of tomato in China (56,350 kg) is still lower than the global average (Wang et al. 2018). At least 10 diseases cause significant tomato yield reduction in China (Li 2011). In recent years, the application of beneficial microbes as biocontrol agents has received much attention because they are environmentally friendly, low in cost, and easy to use.
Plant growth-promoting rhizobacteria (PGPR) are naturally occurring soil bacteria that benefit plants by improving plant productivity and immunity (Porcel et al. 2014). Pseudomonas, Bacillus, and Streptomyces spp. are the commonest PGPR, extensively used for plant growth promotion and disease control (Fahimi et al. 2014; Li et al. 2019; Lim and Kim 2009). Widely accepted mechanisms for plant growth promotion by PGPR include: (1) modulation of the biosynthesis of plant growth regulators such as indole-3-acetic acid (IAA) (Acuna et al. 2011) and gibberellic acid (GA) (Ahmed et al. 2014); (2) enhancement of the solubilization of mineral phosphates and other nutrients with low bioavailability in soil (Hayat et al. 2010); and (3) direct participation in atmospheric nitrogen fixation or indirect contribution to nitrogen fixation efficiency of rhizobia (Figueiredo et al. 2008; Habibi et al. 2014).
Evidence suggests that PGPR can control plant diseases through direct mechanisms—antagonistic activity against pathogens via competition for nutrients and production of antibiotics or lytic enzymes, and indirect mechanisms—induction of plant resistance to pathogens and regulation of mitogen-activated protein kinase (MAPK) cascades in plants (Pieterse et al. 2014). When plants suffer abiotic stresses such as drought, salinity, and heavy metals, PGPR can be involved in the upregulation of plant secondary metabolic pathways such as polyphenol synthesis (Kumar et al. 2016). Polyphenols including flavonoids, phenylpropanoids, anthocyanins, and lignins can help plants withstand various biotic and abiotic stresses (Martins et al. 2013; Moura et al. 2010; Trinh et al. 2018; Wang et al. 2016).
Pseudomonas represents a major group of PGPR and members such as P. fluorescens have been reported to be beneficial microbes that mediate plant growth promotion and/or disease suppression (Lopes et al. 2018). P. koreensis is a subgroup of closely related strains within the P. fluorescens complex (Rangel et al. 2016; Rafikova et al. 2016) reported that in a field test that P. koreensis strain IB-4 had a positive effect on potato yield and resistance to plant pathogens through its antifungal activity, nitrogenase activity, and ability to synthesize IAA and cytokinin-like compounds. Hultberg et al. (2010) showed that a biosurfactant produced by P. koreensis strain 2.74 suppressed disease in tomato plants infected by Pythium ultimum in greenhouse trials. These studies suggest that P. koreensis is a potential PGPR species. Our recent study showed that the addition of cell-free filtrate from P. koreensis strain GS promoted the growth of tomato seedlings under laboratory conditions (Guo et al. 2020). However, there is a lack of systematic research to clarify the mechanisms of plant growth promotion by strain GS and the global transcriptomic and metabolomic changes in plants elicited by P. koreensis.
In the present study, we investigated the effects of P. koreensis GS on plant growth and defense enzyme activity in tomato. RNA sequencing (RNA-Seq) and metabolome analysis were used to explore the possible mechanisms underpinning plant growth promotion by P. koreensis and its potential to induce stress tolerance in plants. We hypothesized that: (1) P. koreensis promotes the growth of tomato plants by increasing the expression of functional genes and specific metabolites related to plant growth; and (2) P. koreensis shows potential to induce stress tolerance in tomato plants via enhancing defense enzyme activities and the biosynthesis of defense metabolites such as polyphenols and amino acids. This study presents the first evidence of transcriptomic and metabolic changes that occur in tomato plants treated with P. koreensis.
Materials and methods
Strain, seeds, and soil
Pseudomonas koreensis strain GS (PRJNA517377) was previously isolated from the rhizosphere of ginseng (Panax ginseng C.A. Mey) in a brown loam soil in the southern piedmont of the Lesser Khingan Mountains, China (46°57’ N, 128°16’ E). Strain GS was preserved by the Resource and Environmental Biology Laboratory in the College of Resources & Environment, Northwest A&F University (Yangling, Shaanxi, China). Seeds of tomato (Lycopersicon esculentum Mill.) cultivar ‘Jinpeng 1’ were purchased from Xi’an Jinpeng Seedling Co., Ltd. (Xi’an, Shaanxi Province, China). Fertile field soil was collected from farmland in Yangling. The soil type was classified as Eum-Orthric Anthrosols (Staff 2010). The soil had a pH of 7.67 and contained total organic carbon of 12 g kg− 1, total nitrogen of 1.66 g kg− 1, available phosphorous of 26.3 mg kg− 1, and available potassium of 155.9 mg kg− 1. The soil was passed through a 2.0-mm sieve and mixed thoroughly with compound fertilizer (N:P:K, 15:6.5:12.4) at a rate of 3 ‰ before use.
Pot experiments
Tomato seeds (three seeds per pot) were drilled into 60 pots (diameter × height, 15 × 12 cm) each containing 1.2 kg of fresh soil. All pots were placed in a greenhouse at 28 °C, with sunlight for 12 hours per day on average. The pots were watered once a day with 120 mL of sterilized distilled water per pot. When the first true leaves appeared, one seedling with uniform growth was selected and retained in each pot. The experimental design was a randomized complete block design with three blocks, and each block contained 20 pots. The pots in each block were divided randomly and equally into a control group and treatment group, with 10 plants per group.
Strain GS was cultured in beef extract peptone broth at 37 °C with oscillation at 180 rpm. The 72-hour-old culture was diluted with sterile phosphate-buffered saline. After the first true leaves had just emerged, one group of tomato seedlings was watered with 100 mL of diluted cell culture (1 × 107 mL− 1 colony-forming units) per pot (treatment group), and the other group received 100 mL of sterile phosphate-buffered saline per pot (control group). No pesticides or fungicides were applied, and the plants did not show any symptoms of disease during the experiment.
Plant growth measurements and leaf sampling
Plant height and stem diameter were measured 15, 30, 45, and 60 days post-inoculation (dpi). Leaf chlorophyll content (SPAD value) was measured at 30 and 60 dpi using a SPAD-502plus (Konica Minolta, Tokyo, Japan). Fresh weight of shoot biomass was measured at 60 dpi.
Leaf samples were taken at 30 dpi for physiological and molecular analyses. For enzyme activity assays, three biological samples (replicates) were taken from each group, and a composite sample of biological replicates was obtained from a mixture of the 4th and 5th leaves five of plants randomly collected in each block. For transcriptome analysis and quantitative validation of functional gene expression, three biological samples (replicates) were taken from each group, and a composite sample of biological replicates was obtained from a mixture of the 3rd leaf of five plants randomly collected in each block. For metabolome and hormone analyses, seven biological samples (replicates) were taken from each group, and each biological replicate was obtained from the 2nd leaf of one plant randomly selected in three blocks. Leaf samples used for enzyme activity assays were directly placed into an ice-box after collection. The remaining leaf samples were washed with distilled water and dried on blotting paper, then immediately frozen in liquid nitrogen. All samples were taken to the laboratory and stored at − 80 °C until use.
Defense enzyme activity assays
Polyphenol oxidase (PPO) activity was assayed using the method of Li (1998). An absorbance change of 0.001 units per min at 398 nm was defined as one unit of PPO activity (unit: U min− 1). Catalase (CAT) activity was assayed using the method of Gao (2006). An absorbance change of 0.01 units per min at 240 nm was defined as one unit of CAT activity (unit: U min− 1). Phenylalanine ammonia-lyase (PAL) and peroxidase (POD) activities were assayed using the method of Silva et al. (2004). An absorbance change of 0.01 units per min at 290 nm (or 470 nm) was defined as one unit of PAL (or POD) activity (unit: U min− 1).
Transcriptome analysis
RNA isolation and sequencing
Leaf samples were sent to BGI (Shenzhen, China) for RNA isolation and transcriptome sequencing. Total RNA was isolated from leaf samples (each 0.1 g) using an RNeasy mini kit (Qiagen, Hilden, Germany). RNA purity was determined using a NanoPhotometer® (IMPLEN, Westlake Village, USA). RNA concentration was measured using a Qubit® RNA Assay Kit in a Qubit® 2.0 Fluorometer (Life Technologies, Carlsbad, USA). The RNA integrity number (RIN) was assessed using an RNA Nano 6000 Assay Kit on the Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, USA). The samples were considered acceptable if 2.2 ≥ A260/A280 ≥ 2.0, 28 S/JZ18 S ≥ 1.5, and RIN ≥ 7.5.
Library preparations were sequenced on the BGISEQ-500 platform (BGI, Shenzhen, China), and 50-bp single-end reads were generated. Raw reads were first processed using SOAPnuke (parameters: −l 5 − q 0.5 − n 0.1) and then filtered using Trimmomatic (parameters: ILLUMINACLIP:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:50). The obtained clean reads were aligned to the tomato reference genome using HISAT and then mapped to the reference sequence using Bowtie2 (Johns Hopkins University, Baltimore, MD, USA). The gene and transcript expression levels of each sample were calculated using RSEM (http://deweylab.biostat.wisc.edu/rsem/).
The open reading frame (ORF) of each unigene was detected using getorf (http://emboss.sourceforge.net/apps/cvs/emboss/apps/getorf.html). The ORFs were aligned to transcription factor domains from the Plant Transcription Factor database (PlnTFDB; http://plntfdb.bio.uni-potsdam.de/) using hmmsearch (http://hmmer.org/). The function of unigenes was identified based on the characteristics of transcription factor families described by PlnTFDB (http://plntfdb.bio.uni-potsdam.de/v3.0/).
Identification of differentially expressed genes (DEGs)
Differential gene expression analysis was performed using the method of Zhao et al. (2018). High-quality clean reads were mapped to the reference transcriptome, and the transcript levels of unigenes were analyzed using TopHat (http://ccb.jhu.edu/software/tophat/index.shtml) and Cufflinks (http://cole-trapnelllab.github.io/cufflinks/). The transcript levels were normalized to Fragments Per Kilobase of exon model per Million mapped reads (FPKM) (Trapnell et al. 2010) as follows: FPKM = 106C/NL/103, where C denotes the number of fragments aligned to a particular gene, N denotes the total number of fragments aligned to all genes, and L denotes the number of bases in the particular gene. Sequencing was used to identify genes that were differentially expressed in different groups. False discovery rates (FDRs) were used in multiple hypothesis testing to correct p-values. The FDRs were statistically preset to values < 0.05, and differential gene expression was calculated based on FPKM values (Zenoni et al. 2010). An absolute value of |fold-change| > 2 and p < 0.001 were used as the thresholds to screen DEGs.
Gene annotation and functional enrichment analysis
Gene Ontology (GO) analysis of DEGs was performed using the PANTHER (Protein Annotation Through Evolutionary Relationship) classification system (http://www.pantherdb.org/data/). Pathway enrichment analysis was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.kegg.jp/). The Basic Local Alignment and Search Tool (BLASTn) was used to identify homologues in the tomato genome (http://solgenomics.net/).
Validation of transcriptome data by real‐time PCR
Reverse transcription quantitative real-time PCR (RT-qPCR) was used to validate the RNA-Seq data for selected DEGs. The RNA used for quantitative validation of functional gene expression was derived from RNA extracted for transcriptome analysis, and random primers (Table S1) were used for reverse transcription. Total RNA (500 ng) was reverse-transcribed into cDNA using the following program: 25 °C for 10 min, 42 °C for 20 min, 85 °C for 5 s, and holding at 12 °C. Then, 1 µL of cDNA was used as template for RT-qPCR in the following conditions: 95 °C for 180 s, followed by 40 cycles of 95 °C for 10 s, 55 °C for 30 s, and 72 °C for 30 s. Data acquisition and analysis were performed using Bio-Rad iQ5 software (V.2.1.97; Bio-Rad Laboratories, Hercules, USA), with the number of cycles required for the fluorescent signal to cross the threshold in RT-qPCR as the cycle threshold (CT) (Ali-Benali et al. 2005). Actin served as an internal reference. The relative expression level of target genes was calculated using the 2−ΔΔCT method (Schmittgen and Livak 2008).
Metabolome and hormone analyses
Gas chromatography‐mass spectrometry (GC-MS) of metabolites
Leaf samples (each 100 mg) and five steel beads were transferred into 5-mL centrifuge tubes. Following pretreatment with liquid nitrogen (5 min), the tubes were placed in a high-flux tissue grinding apparatus (SCIENTZ-48; Xinzhi, Ningbo, China) at 70 Hz for 1 min. Then, 400 µL of methanol (precooled to − 20 °C) were added to each tube, followed by the addition of 60 µL of ribitol (0.2 mg mL− 1 stock in methanol) as an internal quantitative standard. After vortexing, the mixtures were sonicated at room temperature for 30 min. Then, 750 µL of chloroform (precooled to − 20 °C) and 1,400 µL of deionized water (4 °C) were added to each tube. The mixtures were vortexed for 60 s and then centrifuged at 18,756 x g for 10 min at 4 °C.
Aliquots (each 1 mL) of the supernatants were transferred into fresh centrifuge tubes and dried by vacuum concentration. Afterward, 60 µL of methoxyamine pyridine solution (15 mg mL− 1) was added to each tube. The mixtures were vortexed for 30 s and allowed to react for 120 min at 37 °C. Then, 60 µL of N, O-bis (trimethylsilyl) trifluoroacetamide reagent (containing 1 % trimethyl chlorosilane) was added and the mixtures were allowed to react for another 90 min at 37 °C. Following reaction, the tubes were centrifuged at 13,780 x g for 10 min at 4 °C and the supernatants were collected for use. For quality control, 20 µL of each prepared sample was pooled to determine deviations of the analytical results, and the deviations from pooled mixtures were then compared to analytical errors. The remaining samples were used for GC-MS analysis.
To separate the derivatives, GC was performed on an HP-5MS capillary column (30 m × 250 µm i.d., 0.25 µm film thickness; Agilent J 7980A; Agilent J & W Scientific, Folsom, USA) coated with 5 % phenyl and 95 % methylpolysiloxane. Helium was used as the carrier gas at a constant flow rate of 1 mL min− 1. Sample (each 1 µL) was injected in split mode with a 20:1 split ratio using an autosampler. The injection temperature was maintained at 280 °C, with the interface set to 150 °C and the ion source adjusted to 230 °C. The temperature ramping program was as follows: start at 60 °C for 2 min, ramp to 300 °C at 10 °C min− 1, hold at 300 °C for 5 min. MS was performed using the full-scan mode over the mass-to-charge ratio range from m/z 35 to 750 (Agilent J 5975C; Agilent J & W Scientific, Folsom, USA).
To classify and annotate metabolites, the raw data were converted into netCDF format using G1701 MSD ChemStation (E.02.00.493) and then preprocessed using the XCMS package of R (v3.3.2) for peaks identification, peaks filtration, and peaks alignment. The main parameters were as follows: bw = 2, minfrac = 0.3, fwhm = 3, snthresh = 0, step = 0.1, steps = 2, mzdiff = 0.5, and profmethod = binlin. The data matrix including the mass-to-charge ratio (m/z), retention time (RT), and peak area (intensity) was obtained. After that, annotation of the metabolites was performed using the AMDIS program combined with the National Institute of Standards and Technology (NIST) commercial database (http://www.nist.gov/) and Wiley registry metabolome database (http://sciencesolutions.wiley.com/.). Among them, the metabolite alkane retention index was used for further substance identification according to the retention index provided by The Golm Metabolome Database (GMD; http://gmd.mpimp-golm.mpg.de). To compare the data of different magnitudes, the peak area of the data was normalized by the internal standard. Finally, the metabolites obtained by annotation were divided into nine categories, amino acids, sugars, nucleotides, organic acids, fatty acids, phosphoric acids, amines, polyols, and others. The screening conditions of differential metabolites were as follows: variable importance in the project (VIP) ≥ 1 and p-value ≤ 0.05.
Liquid chromatography‐mass spectrometry (LC-MS) of plant hormones
Leaf samples (each 0.3 g) were extracted with 10 mL of acetone–methanol solution (4:1, v/v) by shaking for 1 min and incubating in a 55 °C water bath for 1 h. After centrifugation (13,780 x g, 4 °C), the supernatants were collected into fresh tubes with 5 mL of acetone–methanol solution, followed by shaking for 1 min and incubation at 55 °C for 30 min. After centrifugation (13,780 x g, 4 °C) and concentration, the materials were redissolved with 240 µL of 80 % methanol and filtered through a 0.22-µm microporous filter. The filtrates were analyzed by LC-MS. The samples were protected from light and kept at low temperature during preparation and analysis.
LC was conducted using an ACQU ITY UPLC® BEH C18 column (2.1 × 100 mm, 1.7 µm; Waters, Milford, USA). The injection volume was 5 µL and the column temperature was maintained at 40 °C. The mobile phase consisted of 0.1 % acetic acid (A) and acetonitrile (B). The gradient elution conditions were: 0–2 min, 90–75 % A; 2–3 min, 75 % A; 3–4 min, 75–10 % A; 4–5 min, 10 % A; 5–6 min, 10–90 % A; and 6–8 min, 90 % A. MS was conducted using an electrospray ionization source in both positive and negative ionization modes, with an ion source temperature of 500 °C and ion source voltage of 5000 V/−4500 V. Collision gas was maintained at 6 psi, with air curtain gas at 30 psi, and atomization and auxiliary gases at 50 psi. Multipath monitoring was used for scanning (Bajad et al. 2006). The linearity of the measurement was evaluated by analyzing different concentrations of standard solutions of IAA, abscisic acid (ABA), salicylic acid (SA), and jasmonic acid (JA). Calibration standards were prepared by diluting stock solutions to give the concentrations indicated in Table S2.
Statistical analysis
Data are expressed as means ± standard deviation (SD). The comparison of group means was performed using Student’s t-test with DPS v4.0 (Data Processing System; http://www.chinadps.net). A P-value of < 0.05 was considered to indicate a significant difference. Enrichment of DEGs in biotic stress pathways was analyzed using MAMPAN (Thimm et al. 2004). Hierarchical cluster of differential metabolites was constructed by the pheatmap package in R v3.3.2 (http://www.r-project.org/). The correlations between all DEGs (fold-change data) and differential metabolites (normalized peak area data) involved in the same metabolic pathway were determined using the package psych in R (http://personality-project.org/r/psych) based on metabolic pathways recalculated at the KEGG website (http://www.kegg.jp/). Pearson’s correlation coefficient was used to evaluate the consistency between RNA-Seq and RT-qPCR data.
Results
Plant growth promotion by P. koreensis GS
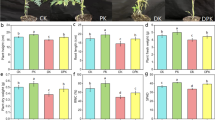
After treatment with P. koreensis GS, the growth and physiological parameters of tomato plants generally increased compared with those in the control group (Fig. 1). Plant height (Fig. 1b) and stem diameter (Fig. 1c) in the treatment group were respectively 6.6–19.5 % and 14.6–22.3 % higher than those in the control group from 30 to 60 dpi. Leaves from the treated plants appeared to be greener and their SPAD values increased significantly, by 15.1 % (P < 0.05; 30 dpi) and 8.3 % (P < 0.05; 60 dpi), compared with values for the control group (Fig. 1d). Significant increases (P < 0.05) also occurred in fresh weight of the treatment group at 60 dpi (61.5 %; P < 0.05) compared with the control group (Fig. 1e). The growth-promoting effect of P. koreensis GS was verified in terms of plant dry weight in an independent experiment (Note S1; Fig. S1).
Plant growth of tomato in the presence and absence of P. koreensis GS. (a) A photograph of potted plants 60 days post-inoculation; (b) plant height; (c) stem diameter; (d) leaf chlorophyll content (SPAD value); and (e) shoot biomass (fresh weight). Data are the means of 30 biological replicates (3 × 10). * P < 0.05 compared with control group (Student’s t-test)
Enhancement of defense enzyme activities by P. koreensis GS
Several defense enzymes showed higher activities in tomato leaves from the treatment group than in those from the control group (Fig. 2). Leaf PPO (Fig. 2a), CAT (Fig. 2b), and PAL (Fig. 2c) activities in the treatment group increased at 30 and 60 dpi, by 18.8–27.3 %, 38.8–25.3 %, and 11.3–22.4 % respectively, compared with levels in the control group. With regard to leaf POD activity, the treatment group showed a slight, but not significant, decrease at 30 dpi compared with the control group, whereas a 39.8 % increase occurred at 60 dpi (Fig. 2d).
Defense enzyme activities in the leaves of tomato plants inoculated with and without P. koreensis GS. (a) Polyphenol oxidase (PPO) activity; (b) catalase (CAT) activity; (c) phenylalanine ammonia-lyase (PAL) activity; and (d) peroxidase (POD) activity. Data are the means of three biological replicates. *P < 0.05 compared with the control group (Student’s t-test)
Global transcriptomic changes induced by P. koreensis GS
Overall changes in gene expression
A total of 23,252 transcripts, amounting to ~ 68 % of the reference transcripts (n = 33,810), were detected in tomato leaves. From these, 737 genes (3.2 %) were identified as DEGs; among them, 571 were upregulated and 166 were downregulated in plants treated with strain GS relative to the control group. Although most of the DEGs have been functionally annotated, 35 (4.7 %) remain of unknown function, and 22 (3.0 %) encode either hypothetical proteins or proteins with putative functions. The transcriptome data were deposited in the Sequence Read Archive with accession number PRJNA544688.
GO functional enrichment analysis of DEGs
Functional enrichment analysis revealed that the 737 DEGs were enriched in 1407 GO terms (Table S3). Significant expression differences were found in 196 GO terms in the categories biological processes (110, 56.1 %), cellular components (24, 12.2 %), and molecular functions (62, 31.6 %). In biological processes, 51, 46, 41, 41, 33, 29, and 28 DEGs were associated with catabolic process, cellular catabolic process, carbohydrate metabolic process, organic substance catabolic process, cell wall organization or biogenesis, lipid metabolic process, and secondary metabolic process, respectively. In cellular components, 194 and 178 DEGs were related to membrane and membrane part, respectively. In molecular functions, 116 and 116 DEGs were related to metal ion binding and cation binding, respectively.
KEGG pathway enrichment analysis of DEGs
The 737 DEGs were mapped to the KEGG database and 95 enriched pathways were identified. Table 1 lists the top 11 significantly enriched pathways, namely flavonoid biosynthesis, photosynthesis-antenna proteins, phenylpropanoid biosynthesis, flavone and flavonol biosynthesis, stilbenoid, diarylheptanoid and gingerol biosynthesis, anthocyanin biosynthesis, brassinosteroid biosynthesis, and MAPK signaling pathway-plant. Unigenes assigned to these pathways are summarized in Table S4.
Differential gene expression in functional pathways
Growth regulation KEGG analysis revealed that two DEGs involved in GA signal transduction were upregulated in the treatment group compared with the control group (Fig. 3). GA receptor genes GID2 (GA-insensitive dwarf 2) and PIF3 (phytochrome-interacting factor 3) were upregulated by 5.2- and 2.2-fold, respectively. In auxin signal transduction, one early auxin response genes Aux/IAA (auxin-responsive protein/IAA) and two GH3 (Gretchen Hagen 3) were upregulated by 2.2-, 2.5-, and 2.9-fold, respectively, while SAUR (small auxin-up RNA) was downregulated by 20.8-fold. All five DEGs associated with brassinosteroid biosynthesis were upregulated. Specifically, BAK1 (brassinosteroid insensitive 1-associated receptor kinase 1) was upregulated by 4.0-fold, and four TCH4 (xyloglucan: xyloglucosyl transferase) genes were upregulated by 2.0- to 3.5-fold.
Regulation of plant hormone biosynthesis and response pathways in tomato leaves by P. koreensis GS. The red box represents upregulation and the green box represents downregulation. Hormone signal transduction: KEGG PATHWAY Map04075 (http://www.genome.jp/dbget-bin/www_bget?map04075). (For interpretation of color in this figure, please refer to the online version of this article)
MAPK signaling pathway-plant
KEGG analysis showed that there were 34 DEGs involved in the pathway MAPK signaling pathway-plant, 27 (79.4 %) of which were upregulated. Six DEGs were identified as leucine-rich repeat receptor-like serine/threonine-protein kinase genes, FLS2 (flagellin-sensing 2), four of which were upregulated by 2.2–26.8-fold. In addition, the leucine-rich repeat receptor-like serine/threonine-protein kinase gene ER (ERECTA) was upregulated by 4.1-fold.
A number of genes downstream of the MAPK pathway were also upregulated by 2.1–3.5-fold, including MKK4_5 (mitogen-activated protein kinase kinase 4/5), MKS1 (MAP kinase substrate 1), WRKY33 (WRKY transcription factor 33), MYF6 (myogenic factor 6), ATF-4 (cyclic AMP-dependent transcription factor ATF-4), SHOC2 (leucine-rich repeat protein), and ACS6 (1-aminocyclopropane-1-carboxylate synthase). With regard to pathogenesis-related (PR) proteins, expression of PR1 genes was upregulated by 22.2-, 5.5-, and 195.4-fold, while one PR1 gene was downregulated by 2-fold.
With respect to ethylene and JA signal transduction, three genes including E3.2.1.14 (chitinase), VSP2 (vegetative storage protein 2), and GLCAK (glucuronokinase) were upregulated by 2.1-, 3.0-, and 2.4-fold, respectively. Considering ABA signal transduction, three genes including PYL (ABA receptor) and PRF (disease resistance protein) were upregulated by 2.1–9.6-fold. In addition, MAPKKK17_18 (mitogen-activated protein kinase kinase kinase) was downregulated.
The 737 DEGs were introduced into the corresponding biotic stress pathway in Mapman, 308 (41.8 %) of which were enriched in this pathway. Among them, 62 of the 70 (88.6 %) genes related to PR protein, auxin, and cell wall were upregulated, and 39 of the 53 (73.6 %) genes related to signaling and MAPK were also upregulated (Fig. 4). The results of biotic stress pathway analysis were partially consistent with the results of KEGG analysis (Table 1).
Biotic stress pathway analysis of differently expressed genes (DEGs) in leaves of tomato plants treated with P. koreensis GS. Enrichment of DEGs in the biotic stress pathway was analyzed using MAPMAN, with upregulated (red) and downregulated (green) genes from the treated plants compared with the control plants
Polyphenol biosynthesis
There were five enriched KEGG pathways related to the biosynthesis of polyphenols. Twenty-five of the 32 (78.1 %) DEGs involved in phenylpropanoid biosynthesis were upregulated by 2.0–13.1-fold. These genes encode a total of 16 key enzymes, including POD, coniferyl-alcohol glucosyltransferase, and PAL (Fig. S2). Seventeen of the 21 (81.0 %) DEGs involved in flavonoid biosynthesis were upregulated by 2.1–4.4-fold. These genes encode a total of 13 key enzymes, including flavanone 3-dioxygenase, flavonoid 3’-monooxygenase, and naringenin 3-dioxygenase (Fig. S3). All five DEGs involved in flavone and flavonol biosynthesis were upregulated, and they encode flavonoid 3’,5’-hydroxylase, flavonol-3-O-L-rhamnoside-7-O-glucosyltransferase, caffeoyl-CoA O-methyltransferase, flavonol 3-O-glucosyltransferase, and flavonoid 3’-monooxygenase, respectively (Fig. S4). Table S5 summarizes the details (gene number, expression change, and protein function) of genes related to polyphenol biosynthesis pathways.
Moreover, seven of 10 (70 %) DEGs involved in stilbenoid, diarylheptanoid, and gingerol biosynthesis were upregulated by 2.1–4.4-fold, and they encode four key enzymes, namely shikimate O-hydroxycinnamoyltransferase, caffeoyl-CoA O-methyltransferase, coumaroylquinate (coumaroylshikimate) 3’-monooxygenase, and linoleate 9S-lipoxygenase (Fig. S5). All three DEGs involved in anthocyanin biosynthesis were upregulated, and these genes encode solute carrier family 39 (zinc transporter), member 1/2/3, and flavonol 3-O-glucosyltransferase (Fig. S6).
PCR validation of RNA-Seq results
Eight upregulated DEGs involved in growth regulation, MAPK signaling, and polyphenol biosynthesis were selected for validation by RT-qPCR (Fig. 5). The RT-qPCR data showed that these DEGs exhibited 2.2- to 5.4-fold changes in their expression levels after treatment with strain GS relative to the control group (P < 0.001). The expression changes of each gene revealed by RT-qPCR were consistent with those obtained from the RNA-Seq data (Pearson’s correlation coefficient r = 0.707). For example, the expression changes of WRKY33, E1.11.1.7 (peroxidase), and PR1 in RT-qPCR analysis were 1.4-, 6.6-, and 4.1-fold (P < 0.001), while the corresponding expression changes determined in RNA-Seq analysis were 2.3-, 5.3-, and 5.4-fold, respectively (P < 0.01). There were no significant differences in the gene expression changes between RT-qPCR and RNA-seq data (t = 0.389, P = 0.709). These results indicate that the RNA-Seq data were reliable.
Expression changes of eight differentially expressed genes involved in plant growth regulation and defense response in tomato leaves for validation of RNA sequencing (RNA-Seq) data by reverse transcription quantitative real-time PCR (RT-qPCR). There were no significant differences in the gene expression changes between RT-qPCR and RNA-seq data (t = 0.389, P = 0.709; by Student t test)
Global metabolomic changes induced by P. koreensis GS
Seventy-nine compounds were identified in tomato leaves by GC-MS. There were 25 organic acids, 21 amino acids, one amine, ten sugars, three fatty acids, one nucleotide, four phosphoric acids, eight polyols, and six compounds of other categories. Comprehensive analysis showed that the contents of 30 compounds (38.0 %) changed significantly in the treatment group relative to the control group (Fig. 6a), and the details (VIP value, p value, and fold-change) of differential metabolites are listed in Table S6. Among these, 10 organic acids showed significant changes, seven of which were increased. All seven amino acids with significant changes were increased (Fig. 6b).
Metabolomic profiles (a) and hierarchical clustering (b) of metabolites in the leaves of tomato plants treated with P. koreensis GS. Data from seven biological replicates per group are shown. (a) Red and blue colors indicate high and low levels, respectively. (b) Red color represents significant upregulation, blue color represents significant downregulation, and gray color represents no significant change
Links between transcriptome and metabolome
Transcriptome–metabolome co-analysis (Table 2) showed that all the metabolites involved in the phenylpropanoid biosynthesis pathway (which contained 32 DEGs, including 25 upregulated) were enriched after treatment with P. koreensis GS. In particular, phenylalanine, tyrosine, and trans-ferulic acid contents were increased by 1.8-, 1.3-, and 1.4-fold in the treatment group compared with the control group, respectively. Moreover, significant changes occurred in one, four, five, and six compounds related to tryptophan metabolism (five DEGs), ABC transporters (three DEGs), 2-oxocarboxylic acid metabolism (seven DEG), and aminoacyl-tRNA biosynthesis pathways (one DEG). The metadata for transcriptome-metabolome co-analysis are summarized in Table S7.
Changes in plant hormones in response to P. koreensis GS
Plant hormone analysis showed that mean ABA content in tomato leaves from the control group (113.46 ng g− 1 fresh weight) was 1.64-fold that in the treatment group (69.34 ng g− 1 fresh weight). There was no significant difference in SA content between the two groups (355.73 ng g− 1 vs. 366.69 ng g− 1 fresh weight). JA was not detected in any group. IAA was detected in the treatment group (9.35 ng g− fresh weight), but not in the control group (below the limit of detection; Fig. 7).
Discussion
PGPR such as P. koreensis are soil microbes that colonize plant roots and acquire nutrients from root exudates, which, in turn, benefit plants by increasing tissue growth and/or suppressing disease development (Rafikova et al. 2016). Our recent research showed that the addition of cell-free filtrate of P. koreensis GS increased hypocotyl and radicle length of tomato seedlings grown in Petri dishes, while the expression of plant growth-related genes (i.e., expansin genes, LeEXP2 and LeEXP18) in tomato leaves was also upregulated (Guo et al. 2020). In the present study, we verified the effect of P. koreensis GS on plant growth promotion using a dilute cell suspension, which effectively increased plant height, stem diameter, and shoot biomass of tomato seedlings grown in pots. Both the culture filtrate and diluted culture of P. koreensis GS showed the ability to promote the growth of tomato plants. However, transcriptome analysis revealed that the expression of expansin genes (LeEXP2 and LeEXP18) was not significantly regulated by the diluted culture treatment compared with the control group in the present study. We infer that this discrepancy may be due to the differences of effective bacterial components and plant gene expression patterns over different growth stages of tomato (7 days in the previous study vs. 30 days in the present study). Further, we demonstrated via transcriptome and metabolome analyses that tomato plants responded to P. koreensis GS by modulating multiple growth and defense factors and enhancing biosynthesis of defense metabolites. The results provide insight into the mechanisms underpinning PGPR–plant interactions, and will be useful for field application of potential PGPR such as P. koreensis GS in increasing plant stress tolerance.
IAA is a phytohormone that plays a central role in plant growth and development. Our previous research found that P. koreensis GS produced IAA in laboratory cultures (Guo et al. 2020). In addition, a preliminary colonization assay indicated that P. koreensis GS effectively colonized the root zone, root surface, and root endosphere of tomato plants, with the root surface being an ideal habitat for this bacterium (Note S2; Table S8). In the present study, hormone analysis by LC-MS revealed that following application of P. koreensis GS, the IAA content in tomato leaves was much higher than that in the control group. Taken together, higher concentrations of tryptophan in the leaves and upregulation of auxin early response genes in inoculated plants suggest that the IAA is of plant origin and its biosynthesis is stimulated by the presence of P. koreensis GS, or the IAA may be produced by a combination of the bacterium and tomato plants. Further experimentation should be conducted to verify these possibilities.
IAA biosynthesis is mainly divided into tryptophan-dependent and tryptophan-independent pathways (Cohen et al. 2003). However, most important catalytic enzymes and key regulatory genes related to IAA biosynthesis are found in the tryptophan-dependent pathway (Zhao 2010). Here, metabolome analysis by GC-MS indicated that the tryptophan level in tomato leaves was increased by application of P. koreensis GS, which might explain the increase of IAA content in plant tissues. Aux/IAA proteins such as IAA1 (Yang et al. 2004) and IAA19 (Kohno et al. 2012) are major components involved in the growth and morphogenesis of plant stems and leaves. In our study, transcriptome analysis by RNA-Seq revealed upregulation of the auxin early response genes Aux/IAA and GH3 in tomato leaves following application of P. koreensis GS. In a previous study, AtIAA19 overexpression in Arabidopsis was found to promote the growth of roots, stems, and leaves, and advance flowering (Kohno et al. 2012). Taken together, our results indicate that application of P. koreensis GS may promote the growth of tomato plants by facilitating the secretion and signal transduction of auxin.
Ryu et al. (2005) have indicated that GA, brassinosteroid, IAA, and SA signaling pathways are involved in elicitation of plant growth promotion by PGPR such as P. fluorescens 89B-61 and Bacillus subtilis GB03. In the present study, the transcriptome results revealed upregulation of GID2 and PIF3 expression in tomato leaves following application of P. koreensis GS. Like GID1 (a GA receptor protein) and DELLA (a negative regulator of GA signaling), GID2 (a protein associated with DELLA protein degradation) is an essential component of GA signaling (Sun 2010; Ueguchi-Tanaka et al. 2007). In addition, PIFs act as negative regulators in light signal transduction, and they not only mediate GA biosynthesis but also participate in GA signal transduction (de Lucas and Prat 2014). GAs can promote the growth and development of plant tissues, especially in key stages such as seed breaking dormancy (Seo et al. 2009), seedling maturation, and vegetative development to reproductive development (Wasilewska et al. 1987). DELLA protein inhibits plant growth in the absence of GA or its presence at low levels, whereas GA binds to the receptor GID1 when cells perceive high levels of GA; this drives GID1 to change conformation and interact with DELLA protein. The GID1–DELLA protein interaction induces DELLA ubiquitination and subsequent degradation, which in turn promotes an increase of free PIFs and their subsequent binding to the target gene promoter. Eventually, GA downstream signaling regulation is activated, thereby facilitating plant growth and development (Hirano et al. 2008; Sun 2010). Moreover, brassinosteroids play a role in vital plant growth and development processes (Li et al. 2002). In this study, application of P. koreensis GS induced upregulation of BAK1 and TCH4, genes involved in brassinosteroid biosynthesis. BAK1 is a serine/threonine protein kinase participating in the signal transmission process of brassinosteroids (Li et al. 2002), and TCH4 acts on the major hemicellulose of the plant cell wall (Xu et al. 1995). In summary, our results indicate that the promotion of tomato plant growth by P. koreensis GS may be related to the upregulation of GA and brassinosteroid signaling. In support of our first hypothesis, P. koreensis could promote tomato plant growth by facilitating the secretion and signal transduction of specific hormones.
MAPK is a serine/threonine protein kinase involved in plant hormone production, defense gene activation, phytoalexin biosynthesis, and cell wall strengthening (Meng and Zhang 2013). In the present study, five of the eight DEGs involved in FLS2 and BAK1 synthesis were upregulated in tomato leaves after treatment with P. koreensis GS. A previous study has shown that BAK1 can phosphorylate FLS2, activating their respective kinase activities (Schulze et al. 2010). Following their activation, FLS2 and BAK1 transmit extracellular signals to plant cells. Then, WRKY33 and PR1 are activated respectively by MKS1 and VIP1 (VirE2-interacting protein 1) to participate in plant disease resistance (Andreasson et al. 2005; Pitzschke et al. 2009). WRKY33 can promote the production of phytoalexin camalexin (Mao et al. 2011), and PR1 is a component of the plant defense system that is closely related to disease resistance. Here, our results showed that 11 of the 12 DEGs associated with downstream components of the MAPK signaling pathway (MKK4.5, VIP1, MKS1, WRKY22.29, WRKY33, and PR1) were upregulated in tomato leaves following application of P. koreensis GS. Similarly, Xie et al. (2017) observed that B. amyloliquefaciens FZB42 induced expression of several disease resistance genes, including BAK1, FLS2, PR1, and WRKY29, in rice leaves. The results indicate that application of P. koreensis GS can upregulate the expression of key genes in the MAPK signaling pathway, potentially improving disease resistance in tomato plants.
Polyphenols such as flavonoids, phenylpropanoids, flavones, flavonols, anthocyanins, and lignin are metabolites involved in plant response to biotic and abiotic stresses (Atkinson et al. 2011; Cuong et al. 2018; Moura et al. 2010; Van den Ende and El-Esawe 2014). Polyphenol biosynthesis in vegetative tissues can be induced by specific physiological and environmental factors, thus protecting plants against pathogens, UV radiation (Manuel et al. 2017), low temperature, drought, and high salinity (Catala et al. 2011; Nakabayashi et al. 2014; Shi and Xie 2014). In the present study, the metabolome results showed that application of P. koreensis GS induced an increase in the phenylalanine content in tomato leaves. The production of phenolics is mainly through the phenylpropanoid metabolic pathway, which begins with phenylalanine (Cuong et al. 2018). In addition, our analysis of transcriptome results demonstrated that flavonoid biosynthesis, phenylpropanoid biosynthesis, flavone and flavonol biosynthesis, stilbenoid, diarylheptanoid and gingerol biosynthesis, and anthocyanin biosynthesis pathways were all enriched following application of P. koreensis GS. In particular, DEGs related to flavone and flavonol biosynthesis and anthocyanin biosynthesis were upregulated in tomato leaves. Trinh et al. (2018) also showed that in the absence of stress conditions, Paenibacillus pabuli strain P7S increased the anthocyanin content and biomass of (A) thaliana seedlings, along with higher expression of key genes regulating anthocyanin and flavonoid biosynthesis pathways. Moreover, we found that some of the key enzyme-encoding genes in the flavonoid biosynthesis pathway, such as CHS, CHI, and F3H, were upregulated in tomato leaves in the presence of P. koreensis GS. The expression of PAL and POD genes involved in phenylpropanoid biosynthesis was increased, which is in agreement with the enhancement of PAL and POD activities we observed in tomato leaves. Martins et al. (2013) reported that without pathogen inoculation, (B) subtilis strains UFLA285 and ALB629 increased phenylalanine ammonia lyase activity and phenolic content, while strain ALB629 also facilitated lignin accumulation in bean (Phaseolus vulgaris) compared with uninoculated controls. Further, our co-analysis of plant transcriptome and metabolome data showed consistent expression patterns of genes related to the biosynthesis of phenylpropane and associated metabolites. In particular, the metabolites phenylalanine, tyrosine, and trans-ferulic acid were enriched in tomato leaves by application of P. koreensis GS. Hence, we found support for our second hypothesis that P. koreensis shows potential to induce stress tolerance in tomato plants by enhancing the secretion of polyphenols, and this effect will not affect normal plant growth under non-stress conditions.
Proline can regulate osmotic potential in plant cells and maintain their normal physiological function. Here, the metabolome results showed that the content of eight amino acids, including proline, increased in tomato leaves after treatment with P. koreensis GS. Ghosh et al. (2017) also found a PGPR-mediated enhancement of proline turnover in (A) thaliana in dehydration conditions. In addition, wheat plants showed an increase in dry biomass and proline content under different salinity regimes when co-inoculated with two PGPR strains, (B) subtilis SU47 and Arthrobacter sp. SU18 (Upadhyay et al. 2012). These findings lead us to conclude that P. koreensis GS could mitigate drought-induced osmotic stress in tomato plants. Moreover, organic acids secreted by plant roots are often considered to play an essential role in plant tolerance to stresses such as heavy metals, salinity, and drought (Incerti et al. 2007; Fu et al. 2010). In the present study, the contents of six organic acids such as oxalic acid and succinic acid were substantially increased in tomato plants after application of P. koreensis GS. The increase of low-molecular-weight organic acids might indirectly enhance plant tolerance to adverse environmental conditions, because they could bind to heavy metals and thereby reduce metal toxicity to the plants (Chen et al. 2015). In addition, plant-derived organic acids play a role in the activation and absorption of soil nutrients. de Werra et al. (2009) found that P. fluorescens strain CHA0 secreted gluconate to dissolve mineral phosphate in soil. In summary, the increase of organic acid biosynthesis modulated by PGPR such as P. koreensis GS may be beneficial for enhancing plant stress tolerance and improving soil nutrient availability, thereby facilitating plant growth.
Conclusions
This study elaborated complex mechanisms underpinning PGPR–plant interactions based on pot experiments. In tomato, hormone biosynthesis and signal transduction, MAPK signaling, and polyphenol biosynthesis pathways were regulated by the presence of P. koreensis strain GS. In addition, metabolites related to plant growth and defense responses (particularly tryptophan, proline, and phenylalanine—the precursor of polyphenols) were enriched by application of the bacterium. This study provides insight into the molecular mechanisms of plant growth promotion by P. koreensis and its potential to induce stress tolerance, which may be useful for field application of the microbial agent in tomato production. However, because the present study was conducted under non-stress conditions, the potential for P. koreensis to induce plant stress tolerance needs to be verified by further testing under stress conditions.
Abbreviations
- ABA:
-
Abscisic acid
- BAK1:
-
Brassinosteroid insensitive 1-associated receptor kinase 1
- DEGs:
-
Differentially expressed gene
- GA:
-
Gibberellic acid
- GID2:
-
GA-insensitive dwarf 2
- JA:
-
Jasmonic acid
- PAL:
-
Phenylalanine ammonia-lyase
- PIF3:
-
Phytochrome-interacting factor 3
- PPO:
-
Polyphenol oxidase
- RNA-Seq:
-
RNA sequencing
- TCH4:
-
xyloglucan:xyloglucosyl transferase TCH4
- WRKY33:
-
WRKY transcription factor 33
- Aux/IAA:
-
Auxin-responsive protein/IAA
- CAT:
-
Catalase
- FLS2:
-
Flagellin-sensing 2
- GH3:
-
Gretchen Hagen 3
- IAA:
-
Indole-3-acetic acid
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- PGPR:
-
Plant growth-promoting rhizobacterium
- POD:
-
Peroxidase
- PR1:
-
Pathogenesis-related protein1
- SA:
-
Salicylic acid
- VIP1:
-
VirE2-interacting protein 1
References
Acuna JJ, Jorquera MA, Martinez OA, Menezes-Blackburn D, Fernandez MT, Marschner P, Greiner R, Mora ML (2011) Indole acetic acid and phytase activity produced by rhizosphere bacilli as affected by pH and metals. J Plant Nutr Soil Sci 11:1–12. https://doi.org/10.4067/s0718-95162011000300001
Ahmed EA, Hassan EA, Tobgy KMKE, Ramadan EM (2014) Evaluation of rhizobacteria of some medicinal plants for plant growth promotion and biological control. Ann Agric Sci 59:273–280. https://doi.org/10.1016/j.aoas.2014.11.016
Ali-Benali MA, Alary R, Joudrier P, Gautier MF (2005) Comparative expression of five lea genes during wheat seed development and in response to abiotic stresses by real-time quantitative RT-PCR. Biochim Biophys Acta 1730:56–65. https://doi.org/10.1016/j.bbaexp.2005.05.011
Andreasson E et al (2005) The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J 24:2579–2589. https://doi.org/10.1038/sj.emboj.7600737
Atkinson NJ, Dew TP, Orfila C, Urwin PE (2011) Influence of combined biotic and abiotic stress on nutritional quality parameters in tomato (Solanum lycopersicum). J Agr Food Chem 59:9673–9682. https://doi.org/10.1021/jf202081t
Bajad SU, Lu WY, Kimball EH, Yuan J, Peterson C, Rabinowitz JD (2006) Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J Chromatogr A 1125:76–88. https://doi.org/10.1016/j.chroma.2006.05.019
Catala R, Medina J, Salinas J (2011) Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc Natl Acad Sci U S A 108:16475–16480. https://doi.org/10.1073/pnas.1107161108
Chen MJ, Liu QW, Tan JY, Xue-Mei LI (2015) Research advance on effects of heavy metal stress on plant organic acid metabolism. Guangdong Agric Sci 24:86–91. https://doi.org/10.16768/j.issn.1004-874x.2015.24.059
Cohen JD, Slovin JP, Hendrickson AM (2003) Two genetically discrete pathways convert tryptophan to auxin: more redundancy in auxin biosynthesis. Trends Plant Sci 8:0–199. https://doi.org/10.1016/S1360-1385(03)00058-X
Cuong DM, Kwon SJ, Jeon J, Park YJ, Park JS, Park SU (2018) Identification and characterization of phenylpropanoid biosynthetic genes and their accumulation in bitter melon (Momordica charantia). Molecules 23. https://doi.org/10.3390/molecules23020469
de Lucas M, Prat S (2014) PIFs get BRright: Phytochrome interacting factors as integrators of light and hormonal signals. New Phytol 202:1126–1141. https://doi.org/10.1111/nph.12725
de Werra P, Pechy-Tarr M, Keel C, Maurhofer M (2009) Role of gluconic acid production in the regulation of biocontrol traits of Pseudomonas fluorescens CHA0. Appl Environ Microbiol 75:4162–4174. https://doi.org/10.1128/aem.00295-09
Fahimi A, Ashouri A, Ahmadzadeh M, Naveh VH, Asgharzadeh A, Maleki F, Felton GW (2014) Effect of PGPR on population growth parameters of cotton aphid. Arch Phytopathol Plant Prot 47:1274–1285. https://doi.org/10.1080/03235408.2013.840099
Figueiredo MVB, Martinez CR, Burity HA, Chanway CP (2008) Plant growth-promoting rhizobacteria for improving nodulation and nitrogen fixation in the common bean (Phaseolus vulgaris L.). World J Microb Biot 24:1187–1193. https://doi.org/10.1007/s11274-007-9591-4
Fu XP, Dou CM, Hu SP, Chen XC, Shi JY, Chen YX (2010) A review of progress in roles of organic acids on heavy metal resistance and detoxification in plants. Chin J Plant Ecol 34:1354–1358. https://doi.org/10.3773/j.issn.1005-264x.2010.11.013
Gao JF (2006) Experimental techniques of plant physiology. Higher Education, Beijing, pp 214–215
Ghosh D, Sen S, Mohapatra S (2017) Modulation of proline metabolic gene expression in Arabidopsis thaliana under water-stressed conditions by a drought-mitigating Pseudomonas putida strain. Ann Microbiol 67:655–668. https://doi.org/10.1007/s13213-017-1294-y
Guo Q, Shi MD, Chen L, Zhou JH, Zhang LX, Li YL, Xue QH, Lai HX (2020) The biocontrol agent Streptomyces pactum increases Pseudomonas koreensis populations in the rhizosphere by enhancing chemotaxis and biofilm formation. Soil Biol Biochem 144:107755. https://doi.org/10.1016/j.soilbio.2020.107755
Habibi S, Djedidi S, Prongjunthuek K, Mortuza MF, Ohkama-Ohtsu N, Sekimoto H, Yokoyoma T (2014) Physiological and genetic characterization of rice nitrogen fixer PGPR isolated from rhizosphere soils of different crops. Plant Soil 379:51–66. https://doi.org/10.1007/s11104-014-2035-7
Hayat R, Ali S, Amara U, Khalid R, Ahmed I (2010) Soil beneficial bacteria and their role in plant growth promotion: a review. Ann Microbiol 60:579–598. https://doi.org/10.1007/s13213-010-0117-1
Hirano K, Aya K, Hobo T, Sakakibara H, Kojima M, Shim RA, Hasegawa Y, Ueguchi-Tanaka M, Matsuoka M (2008) Comprehensive transcriptome analysis of phytohormone biosynthesis and signaling genes in microspore/pollen and tapetum of rice. Plant Cell Physiol 49:1429–1450. https://doi.org/10.1093/pcp/pcn123
Hultberg M, Alsberg T, Khalil S, Alsanius B (2010) Suppression of disease in tomato infected by Pythium ultimum with a biosurfactant produced by Pseudomonas koreensis. Biocontrol 55:435–444. https://doi.org/10.1007/s10526-009-9261-6
Incerti A, Navari-Izzo F, Pardossi A, Mensuali A, Izzo R (2007) Effect of sea water on biochemical properties of fruit of tomato (Lycopersicon esculentum Mill.) genotypes differing for ethylene production. J Sci Food Agric 87:2528–2537. https://doi.org/10.1002/jsfa.3020
Kohno M, Takato H, Horiuchi H, Fujita K, Suzuki S (2012) Auxin-nonresponsive grape Aux/IAA19 is a positive regulator of plant growth. Mol Biol Rep 39:911–917. https://doi.org/10.1007/s11033-011-0816-0
Kumar S et al (2016) Paenibacillus lentimorbus inoculation enhances tobacco growth and extenuates the virulence of cucumber mosaic virus. PLos One 11. https://doi.org/10.1371/journal.pone.0149980
Li B (1998) Changes in activities and eletrophretic patterns of peroxidase and polyphenoloxidase in cucumbers during infection with Cladosporium cucumerinum. Sci Agric Sin 31:86–88
Li J (2011) Chinese tomato breeding. China Ariculture, Beijing, p 275
Li J, Wen JQ, Lease KA, Doke JT, Tax FE, Walker JC (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110:213–222. https://doi.org/10.1016/s0092-8674(02)00812-7
Li YL, Guo Q, Li YZ, Sun YF, Xue QH, Lai HX (2019) Streptomyces pactum Act12 controls tomato yellow leaf curl virus disease and alters rhizosphere microbial communities. Biol Fertil Soils 55:149–169. https://doi.org/10.1007/s00374-019-01339-w
Lim JH, Kim SD (2009) Synergistic plant growth promotion by the indigenous auxins-producing PGPR Bacillus subtilis AH18 and Bacillus licheniforims K11. J Korean Soc Appl Bi 52:531–538. https://doi.org/10.3839/jksabc.2009.090
Lopes LD et al (2018) Genome variations between rhizosphere and bulk soil ecotypes of a Pseudomonas koreensis population. Environ Microbiol 20:4401–4414. https://doi.org/10.1111/1462-2920.14363
Manuel SP, Mayra AM, Sixta LP (2017) Biodegradable polymer hydrogels based in sorbitol and citric acid for controlled release of bioactive substances from plants (polyphenols). Curr Chem Biol 11:36–43. https://doi.org/10.2174/2212796810666161028114432
Mao GH, Meng XZ, Liu YD, Zheng ZY, Chen ZX, Zhang SQ (2011) Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23:1639–1653. https://doi.org/10.1105/tpc.111.084996
Martins SJ, de Medeiros FHV, de Souza RM, de Resende MLV, Ribeiro PM (2013) Biological control of bacterial wilt of common bean by plant growth-promoting rhizobacteria. Biol Control 66:65–71. https://doi.org/10.1016/j.biocontrol.2013.03.009
Meng XZ, Zhang SQ (2013) MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol 51:245–266. https://doi.org/10.1146/annurev-phyto-082712-102314
Moura JC, Bonine CA, de Oliveira Fernandes Viana J, Dornelas MC, Mazzafera P (2010) Abiotic and biotic stresses and changes in the lignin content and composition in plants. J Integr Plant Biol 52:360–376. https://doi.org/10.1111/j.1744-7909.2010.00892.x
Nakabayashi R et al (2014) Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J 77:367–379. https://doi.org/10.1111/tpj.12388
Pieterse CMJ, Zamioudis C, Berendsen RL, Weller DM, Van Wees SCM, Bakker PAHM (2014) Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol 52:347–375. https://doi.org/10.1146/annurev-phyto-082712-102340
Pitzschke A, Djamei A, Teige M, Hirt H (2009) VIP1 response elements mediate mitogen-activated protein kinase 3-induced stress gene expression. Proc Natl Acad Sci U S A 106:18414–18419. https://doi.org/10.1073/pnas.0905599106
Porcel R, Zamarreno AM, Garcia-Mina JM, Aroca R (2014) Involvement of plant endogenous ABA in Bacillus megaterium PGPR activity in tomato plants. Bmc Plant Biol 14:36. https://doi.org/10.1186/1471-2229-14-36
Rafikova GF, Korshunova TY, Minnebaev LF, Chetverikov SP, Loginov ON (2016) A new bacterial strain, Pseudomonas koreensis IB-4, as a promising agent for plant pathogen biological control. Microbiology 85:333–341. https://doi.org/10.1134/s0026261716030115
Rangel LI, Henkels MD, Shaffer BT, Walker FL, Davis EW, Stockwell VO, Bruck D, Taylor BJ, Loper JE (2016) Characterization of toxin complex gene clusters and insect toxicity of bacteria representing four subgroups of Pseudomonas fluorescens. PLoS One 11:e0161120. https://doi.org/10.1371/journal.pone.0161120
Ryu CM, Hu CH, Locy RD, Kloepper JW (2005) Study of mechanisms for plant growth promotion elicited by rhizobacteria in Arabidopsis thaliana. Plant Soil 268:285–292. https://doi.org/10.1007/s11104-004-0301-9
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C-T method. Nat Protoc 3:1101–1108. https://doi.org/10.1038/nprot.2008.73
Schulze B, Mentzel T, Jehle AK, Mueller K, Beeler S, Boller T, Felix G, Chinchilla D (2010) Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J Biol Chem 285:9444–9451. https://doi.org/10.1074/jbc.M109.096842
Seo M, Nambara E, Choi G, Yamaguchi S (2009) Interaction of light and hormone signals in germinating seeds. Plant Mol Biol 69:463–472. https://doi.org/10.1007/s11103-008-9429-y
Shi MZ, Xie DY (2014) Biosynthesis and metabolic engineering of anthocyanins in Arabidopsis thaliana. Recent Pat Biotechnol 8:47–60
Silva HSA, Romeiro RD, Macagnan D, Halfeld-Vieira BD, Pereira MCB, Mounteer A (2004) Rhizobacterial induction of systemic resistance in tomato plants: non-specific protection and increase in enzyme activities. Biol Control 29:288–295. https://doi.org/10.1016/s1049-9644(03)00163-4
Staff SS (2010) Keys to Soil Taxonomy, 11th edn. USDA-Natural Resources Conservation Service, Washington, DC
Sun TP (2010) Gibberellin-GID1-DELLA: a pivotal regulatory module for plant growth and development. Plant Physiol 154:567–570. https://doi.org/10.1104/pp.110.161554
Thimm O et al (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37:914–939. https://doi.org/10.1111/j.1365-313X.2004.02016.x
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–174. https://doi.org/10.1038/nbt.1621
Trinh CS, Jeong CY, Lee WJ, Truong HA, Chung N, Han J, Hong SW, Lee H (2018) Paenibacillus pabuli strain P7S promotes plant growth and induces anthocyanin accumulation in Arabidopsis thaliana. Plant Physiol Biochem 129:264–272. https://doi.org/10.1016/j.plaphy.2018.06.001
Ueguchi-Tanaka M et al (2007) Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 19:2140–2155. https://doi.org/10.1105/tpc.106.043729
Upadhyay SK, Singh JS, Saxena AK, Singh DP (2012) Impact of PGPR inoculation on growth and antioxidant status of wheat under saline conditions. Plant Biol (Stuttg) 14:605–611. https://doi.org/10.1111/j.1438-8677.2011.00533.x
Van den Ende W, El-Esawe SK (2014) Sucrose signaling pathways leading to fructan and anthocyanin accumulation: A dual function in abiotic and biotic stress responses? Environ Exp Bot 108:4–13. https://doi.org/10.1016/j.envexpbot.2013.09.017
Wang FB, Kong WL, Wong G, Fu LF, Peng RH, Li ZI, Yao QH (2016) AtMYB12 regulates flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis thaliana. Mol Genet Genomics 291:1545–1559. https://doi.org/10.1007/s00438-016-1203-2
Wang YQ, Zhang YX, Gao ZP, Yang WC (2018) Breeding for resistance to tomato bacterial diseases in China: challenges and prospects. Hortic Plant J 4:193–207. https://doi.org/10.1016/j.hpj.2018.08.004
Wasilewska LD, Bralczyk J, Szczegielniak J (1987) The role of gibberellin in regulation of dwarf plants development. Plant Sci 53:11–19. https://doi.org/10.1016/0168-9452(87)90172-5
Xie YL, Wu LM, Zhu BC, Wu HJ, Gu Q, Rajer FU, Gao XW (2017) Digital gene expression profiling of the pathogen-resistance mechanism of Oryza sativa 9311 in response to Bacillus amyloliquefaciens FZB42 induction. Biol Control 110:89–97. https://doi.org/10.1016/j.biocontrol.2017.04.009
Xu W, Purugganan MM, Polisensky DH, Antosiewicz DM, Fry SC, Braam J (1995) Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. Plant Cell 7:1555–1567. https://doi.org/10.1105/tpc.7.10.1555
Yang X et al (2004) The IAA1 protein is encoded by AXR5 and is a substrate of SCFTIR1. Plant J 40:772–782. https://doi.org/10.1111/j.1365-313X.2004.02254.x
Zenoni S, Ferrarini A, Giacomelli E, Xumerle L, Fasoli M, Malerba G, Bellin D, Pezzotti M, Delledonne M (2010) Characterization of transcriptional complexity during berry development in vitis vinifera using RNA-SEq. Plant Physiol 152:1787–1795. https://doi.org/10.1104/pp.109.149716
Zhao YD (2010) Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol 61:49–64. https://doi.org/10.1146/annurev-arplant-042809-112308
Zhao XM, Liu Y, Liu X, Jiang J (2018) Comparative transcriptome profiling of two tomato genotypes in response to potassium-deficiency stress. Int J Mol Sci 19. https://doi.org/10.3390/ijms19082402
Acknowledgements
We thank Dr. Chaofeng Lin (Transcend Envirotech Consulting. Co., Ltd., Qingdao, China) for improving the English. This work was supported by the Youth Fund of National Natural Science Foundation of China (31600407), the 2019 Frontier and Interdisciplinary Research Project of Basic Business Expenses in China (2452019177), and the Key R&D Program of Shaanxi Province, China (2019ZDLNY01-07-01).
Author information
Authors and Affiliations
Contributions
H.L. and Q.G. conceived the study and proposed the experimental approaches; Y.S., M.S., H.L., Y.L., and Q.G. conducted the experiments; Y.S., Q.G., X.H., and Y.J. contributed to data processing and analysis; and Q.G. and Y.S. wrote the manuscript. All authors read, commented on, and approved the final version of the manuscript.
Corresponding author
Ethics declarations
All authors are aware with the content of the manuscript and have agreed with the submission to Plant and Soil. The manuscript is not currently being considered for publication in another journal and the manuscript has not been published in whole or in part elsewhere.
Conflict of interest
The authors declare that they have no conflict of interest.
Human and animal rights
This article does not contain any studies with human or animal subjects.
Additional information
Responsible Editor: Luz E. de-Bashan.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 350 KB)
Rights and permissions
About this article
Cite this article
Guo, Q., Sun, Y., Shi, M. et al. Pseudomonas koreensis promotes tomato growth and shows potential to induce stress tolerance via auxin and polyphenol‐related pathways. Plant Soil 462, 141–158 (2021). https://doi.org/10.1007/s11104-021-04837-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-021-04837-9