Abstract
Key message
Two MYB genes have been identified which regulate apocarotenoid metabolism in Crocus sativus.
Abstract
Apocarotenoids like crocin, picrocrocin and safranal are restricted to genus Crocus and are synthesized by oxidative cleavage of zeaxanthin followed by glycosylation reactions. In Crocus sativus, these apocarotenoids are synthesized in stigma part of the flower in developmentally regulated manner. Most of the genes of apocarotenoid pathway are known, however, the mechanism that regulates its tissue and stage specific biosynthesis remains elusive. MYB family was identified as the largest transcription factor family from Crocus transciptome which indicated its possible role in apocarotenoid regulation besides regulating other metabolic pathways. Towards this, we started with identification of 150 MYB genes from Crocus transcriptome databases. The phylogenetic analysis of Crocus MYB genes divided them into 27 clusters. Domain analysis resulted in identification of four groups of MYBs depending upon the number of R repeats present. Expression profiling indicated that 12 MYBs are upregulated in stigma out of which expression of four genes CstMYB1, CstMYB14, CstMYB16 and CstMYB1R2 correlated with crocin accumulation. Transient overexpression of two nuclear localized MYB genes (CstMYB1 and CstMYB1R2) in Crocus confirmed their role in regulating carotenoid metabolism. Yeast-one-hybrid confirmed that CstMYB1 binds to carotenoid cleavage dioxygenase 2 (CCD2) promoter while CstMYB1R2 binds to phytoene synthase (PSY) and CCD2 promoters. Overall, our study established that CstMYB1 and CstMYB1R2 regulate apocarotenoid biosynthesis by directly binding to promoters of pathway genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Carotenoids are a subfamily of isoprenoids and form the most widespread of all natural secondary metabolites (Giuliano et al. 2008). In plants, they perform a variety of critical functions in light harvesting and photoreaction centers besides providing colors to many fruits and flowers (Lu and Li 2008). Carotenoids are important in their intact form and also constitute precursors for biosynthesis of bioactive compounds called apocarotenoids (Auldridge et al. 2006). Apocarotenoids are synthesized by oxidative cleavage of carotenoids in a number of plants including maize, tomato, Arabidopsis, Crocus etc. Crocus finds a special place because it is the only plant which produces crocin, picrocrocin and saffranal in significant quantities. These compounds impart color, flavor and aroma to saffron (dried Crocus stigmas) making it one of the world’s costliest spices (Ashraf et al. 2015). Crocus apocarotenoids have tremendous medicinal properties as well (Abdullaev and Espinosa-Aguirre 2004; Zhang et al. 2013). These properties make Crocus a very important crop to study.
Crocus sativus L. (Iridaceae) is a sterile triploid plant propagated vegetatively through corms (D'Agostino et al. 2007; Baba et al. 2017). Crocus synthesizes apocarotenoids in stigma of the flower and at specific developmental stages. Significant progress has been made in our understanding of carotenoid and apocarotenoid biosynthesis (Lu and Li 2008). In Crocus, apocarotenoids are synthesized by oxidative cleavage of zeaxanthin carried out by carotenoid cleavage dioxygenase 2 (CsCCD2) (Frusciante et al. 2014). However, the mechanism that regulates tissue and stage specific biosynthesis of apocarotenoids in Crocus still remains elusive with only a few reports (Ahrazem et al. 2018, 2019; Malik and Ashraf 2017; Ashraf et al. 2015). Therefore, more efforts need to be put in place to understand the regulatory mechanism.
In our previous study (Baba et al. 2015), we identified different transcription factor families from Crocus transcriptome. Our results indicated that MYB transcription factor family was the largest one and, therefore, prompted us to investigate its role in apocarotenoid regulation. MYB transcription factors represent one of the largest and most diverse families in plants (Dubos et al. 2010). They are known to regulate many cellular processes including primary and secondary metabolism. There are many reports where MYB genes are shown to play roles in regulation of secondary metabolites like flavonoids, anthocyanins, etc. (Stracke et al. 2007; Gonzalez et al. 2008). They also regulate biotic and abiotic stress responses in plants (Roy 2016). Recently several reports have demonstrated role of MYBs in regulating cartotenoid metabolism. For example, an R2R3 MYB transcription factor was shown to regulate carotenoid pigmentation in Mimulus lewisii flowers (Sagawa et al. 2016). In another study, an R2R3 MYB gene was reported to modulate carotenoid accumulation in kiwi fruit (Ampomah-Dwamena et al. 2019). Another MYB gene (WHITEPETAL1) was found to associate with a bHLH and a WD40 gene to regulate carotenoid dependent flower pigmentation in Medicago (Meng et al. 2019). Carotenoid metabolism in papaya is also regulated by MYB genes (Fu et al. 2020). Further, a SlMYB72 was reported to regulate carotenoid metabolism in tomato (Wu et al. 2020). These instances advocate possibility of involvement of MYB genes in regulating carotenoid and apocarotenoid biosynthesis in Crocus as well.
Here we report identificaton of 150 MYB genes from different Crocus transcriptome databases (Baba et al. 2015; Jain et al. 2016; Ahrazem et al. 2018, 2019; Tan et al. 2019). Phylogenetic analysis of Crocus MYB genes divided them into 27 clusters. Domain analysis classified them into four groups depending upon the number of R repeats. In silico tissue specific expression profile was available for 71 MYB genes which indicated that 12 MYBs are upregulated in stigma. This was further confirmed by performing qPCR. Correlation analysis between crocin accumulation and expression of different MYB genes identified four MYBs which showed positive relation between the two. Two genes (CstMYB1 and CstMYB1R2) were characterized further and were found to be transcriptionally active and nuclear localized. Transient overexpression of these two genes confirmed their role in regulating apocarotenoid biosynthesis in Crocus by enhancing expression of pathway genes. This resulted in enhanced production of carotenoids like lycopene, beta carotene and zeaxanthin and apocarotenoids like crocin. Yeast one hybrid demonstrated that these MYB genes directly bind to promoters of apocarotenoid pathway genes, thereby enhancing their expression.
Material and methods
Plant material collection and treatments
The plants of C. sativus used in the present study were grown in the experimental field at Indian Institute of Integrative Medicine (IIIM), Srinagar, India (longitude: 34°5′24′′N; latitude: 74°47′24′′ and altitude 1585 m.a.s.l). The Crocus plants were grown in raised beds. The corms were sown in the month of August at a depth of 8 cm–11 cm and inter-corm distance was about 7 cm–16 cm. Plant sample collection was performed during last week of October to second week of November in the subsequent year. The samples from different stages of stigma development i.e., orange, red (three days before anthesis) and three days after anthesis and different tissues i.e., corm, leaf, tepal, anther and stigma were collected, frozen in liquid nitrogen and stored at − 80 °C till further use.
For transient overexpression studies, corms were sown in pots in late August/early September and grown under controlled conditions at a temperature of 16 °C. The flowers emerged in late October or early November and were used for floral dip experiments. After floral dip, the pots were covered with polybags and kept back at the temperature mentioned above. The flowers were harvested 72 h after agroinfiltration, frozen in liquid nitrogen and stored at − 80 °C till further use.
Transcriptome wide identification and domain analysis of Crocus sativus MYB genes
The Crocus transcripts annotated as MYB genes based on the Blast homology results were retrieved from different Crocus transcriptome databases available in public domain (Baba et al. 2015; Jain et al. 2016; Ahrazem et al. 2018, 2019; Tan et al. 2019). The retrieved nucleotide sequences were scanned for duplications and were subjected to iTAK- Plant Transcription factor and Protein Kinase Identifier and Classifier (http://itak.feilab.net/cgi-bin/itak/online_itak.cgi) filtering (Zheng et al. 2016). Pfam and interPro scan was done to confirm presence of MYB domain. PROSITE was used to identify nuclear localization signals.
Phylogeny and motif analysis
CstMYB protein sequences were aligned using MUSCLE alignment tool. Neighbour-Joining (NJ) phylogenetic tree was constructed using MEGA6 (Molecular Evolutionary Genetic Analysis) Version 6.0 software (Tamura et al. 2013) with bootstrap value of 1000 replications. Two separate combined NJ phylogenetic trees for 1R and 2R genes from Crocus and Arabidopsis were also built using the same procedure. The Arabidopsis MYB proteins were downloaded from TAIR (The Arabidopsis Information Resource) (https://www.arabidopsis.org). Further, the CstMYB protein sequences were subjected to conserved motif analysis using MEME Suite (http://meme-suite.org). MEME search was performed for a total of 10 motifs.
RNA isolation, cDNA synthesis and quantitative RT-PCR Analysis
RNA isolation was done by RNeasy isolation kit (Qiagen) following manufacturer’s instructions. The quality of RNA samples was determined by gel electrophoresis and A260/A280 ratio using spectrophotometer. DNase I (Fermentas) treatment was given to remove genomic DNA contamination. cDNA was synthesized using Revert Aid cDNA synthesis kit (Fermentas) and following manufacturer’s instructions. For qPCR, the cDNA was diluted 10 times and reaction was performed in triplicates in a total volume of 10 μl which included 5 μL of 2X SYBR Green Master Mix, 0.1 μM gene specific primers and 100 ng of template cDNA using QuantStudio 3.0 Real time PCR (Applied biosystems). For each sample tested, three biological replicates were used for quantification and mean of three was calculated and presented in the results. The list of primers used is given in supplementary table S2. The cycling conditions were 95 °C for 20 s, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. For normalization, the expression of C. sativus 18S gene was used as an endogenous control (Baba et al. 2017). The relative quantification method (ΔΔ−CT) was used for further analysis (Livak and Schmittgen 2001).
Transient over-expression of CstMYB1 and CstMYB1R2 in C. sativus
For transient over-expression, full length ORFs of CstMYB1 and CstMYB1R2 were amplified and cloned in pBI121 vector at XbaI and SmaI using CstMYB1-F/CstMYB1-R1 and CstMYB1R2-F/CstMYB1R2-R1 primers (Supplementary table S2). For amplification of CstMYB1, the cycling parameters were 94 °C for 3 min, 34 cycles (94 °C for 30 s, 60 °C for 40 s, 72 °C for 1 min) and 72 °C for 7 min. For CstMYB1R2, the parameters were 94 °C for 3 min, 34 cycles (94 °C for 30 s, 53 °C for 40 s, 72 °C for 1 min) and 72 °C for 7 min. The amplification products were gel purified using gel extraction kit (Qiagen) and cloned into pBI121 plant binary vector containing CaMV 35S promoter.
Transient over-expression was studied in flowering Crocus plants grown under optimal conditions. A. tumefaciens strain GV3101 containing empty pBI121, pBI121-CstMYB1 and pBI121-CstMYB1R2 were cultured at 28 °C in YEM medium (Sigma) supplemented with 100 mg/L rifampicin (Sigma) and 100 mg/L kanamycin (Sigma). The overnight culture was harvested at OD600 of 1.5 to 2.0 by centrifugation at 5000 rpm for 10 min and re-suspended in 50 ml of infiltration liquid. The centrifugation and re-suspension procedure was repeated three to five times. The A. tumefaciens cells were diluted to OD600 of 0.10 in agro-infiltration liquid which composed of 41.65 mM D-glucose, 100 mM CaCl2, 100 mM MES-KOH (pH 5.6), 0.011 mM BAP, 0.01% Silwet L-77, 0.05 mM MgCl2 and 12.5 mM Acetosyringone (made with DMF, dimethylformamide) stock solution, and suitable amount of double distilled water to make final volume to 20 ml. All the chemicals used were procured from Sigma. Transformation was performed as per Fang et al. 2018 with some modifications. The Crocus flower was dipped into Agrobacterium suspension after slightly injuring the tip of stigma with a needle. The flower was kept dipped for five minutes with gentle agitation followed by covering with plastic bag and transferring to normal growth conditions. The stigma tissue was harvested 72 h after transformation for further analysis. The above mentioned method of transient overexpression in Crocus by floral dip was developed for the first time in our laboratory.
Extraction and quantification of crocin and carotenoids by HPLC quantification
Quantification of crocin was done following Moraga et al. 2009 with some modifications as described below. Stigma tissue (100 mg) was ground using liquid nitrogen and 2 ml of 50 mM Tris–HCl (pH 7.5) with 1 M NaCl were added for homogenization of powdered stigma mass. This was followed by the addition of one volume of chloroform. The samples were then incubated for 10 min on ice. Centrifugation at 3500 g for 5 min was performed at 4 °C for the separation of phases. The upper aqueous layer was used for the HPLC analysis of crocin. The dried residues obtained after evaporation of lower chloroform phase were stored at − 80 °C. Prior to HPLC, the residues were dissolved in ethyl acetate and used for HPLC quantification of carotenoids namely lycopene, β-carotene and zeaxanthin. The HPLC for the quantitative assessment of crocin and other carotenoids was performed on HPLC system (Shimadzu, Japan). The HPLC was run for 45 min with column temperature of 40 °C. Crocin was quantified using 250–4 mm 18e LiChrospher column with 5 µm particle size (Merck) with a flow of 0.8 ml/min. The mobile phase used was 90% H2O with 0.1% formic acid (FA) plus 10% acetonitile (ACN) at 0.01 min, 50% H2O with 0.1% FA plus 50% ACN at 20 min, 30% H2O with 0.1% FA plus 70% ACN at 25 min, 10% H2O with 0.1% FA plus 90% ACN at 35 min, 90% H2O with 0.1% FA plus 10% ACN at 40 min and 90% H2O with 0.1% FA plus 10% ACN at 45 min. Lycopene was quantified using same HPLC column with flow rate of 0.7 ml/min. The mobile phase consisted of 70% methanol and 30% ACN. The HPLC analysis of β-carotene and zeaxanthin was performed using 18e 100–4.6 mm Chromolith column of 5 µm particle size (Merck, USA). The mobile phase used consisted of 60% methanol, 30% ACN and 10% CHCl3. The standards were used at 1 mg/ml stock concentrations. All the standards were procured from Sigma and the catalogue numbers are β-carotene: C4582; lycopene: 75,051; zeaxanthin: 14,641; and crocin: 17,304.
HPLC quantification of quercetin
Quantification of quercetin was done following Trapero et al. 2012 with some modifications as described below. Crocus stigmas were crushed to fine powder in liquid nitrogen. The serial extraction of powdered samples was performed with MeOH: di-chloromethane (DCM): H2O (80: 1: 20) thrice at 4 °C using fresh solvent mix every time. The filtrates obtained were pooled together and then passed through Whatman No. 1 paper filter. Following, the solvents were evaporated at 45 °C under reduced pressure with the help of a rotary evaporator (Sigma-Aldrich). The stock solution (1 mg/ml) of quercetin along with those of the dried extracts was freshly prepared in methanol. The methanol dissolved samples were filtered with membrane filters (0.25 μm, Millipore) and filtrates were used for the quantification of quercetin by HPLC.
The HPLC for the quantitative analysis of Quercetin was performed on HPLC system (Shimadzu, Japan). The HPLC procedure was carried out for 45 min with column temperature of 40 °C with a flow of 0.8 ml/min. 250–4 nm 18e LiChrospher column with 5 µm particle size (Merck, USA) was used for performing HPLC. The mobile phase consisted of 90% H2O with 0.1% formic acid (FA) plus 10% acetonitile (ACN) at 0.01 min, 50% H2O with 0.1% FA plus 50% ACN at 20 min, 30% H2O with 0.1% FA plus 70% CAN at 25 min, 10% H2O with 0.1% FA plus 90% ACN at 35 min, 90% H2O with 0.1% FA plus 10% ACN at 40 min and 90% H2O with 0.1% FA plus 10% ACN at 45 min. The standard used was procured from Sigma Aldrich (USA) with CAS number Q4951.
Sub-cellular localization of CstMYB1 and CstMYB1R2
The full length ORFs of CstMYB1 and CstMYB1R2 were amplified and cloned in pBI121 vector at XbaI and SmaI sites in translational frame with β-GUS using CstMYB1-F/CstMYB1-R2 and CstMYB1R2-F/CstMYB1R2-R2 primers (Supplementary table S2). Genes were amplified as mentioned above. Agrobacterium tumefaciens GV3101 strain harbouring pBI121-CstMYB1–GUS, pBI121-CstMYB1R2–GUS (constructs without stop codon in MYB ORFs) and pBI121-GUS (empty vector) were transiently transformed into onion epidermal cells by agroinfiltration and sub-cellular localization of CstMYB1 and CstMYB1R1 was determined by GUS staining method as previously described by Xu et al. 2014. GUS assay was performed using X-Gluc assay buffer containing 1 mM X-Gluc (5-bromo-4-chloro-3-indolyl glucuronide), 50 mM sodium phosphate buffer pH 7.0 and 0.1% Triton X100. The peels from onion epidermis were dipped in different wells of a 24 well plate each containing 1 ml of X-Gluc assay buffer. The assay was carried out in triplicates and the plate was incubated at 37 °C till the blue color developed. The onion peels were then rinsed with 70% ethanol for 5 min before visualizing under microscope. For counterstaining of the nuclei, the onion epidermal cells were treated with 1 μg/ml (in PBS) of 4′-6-diamidino-2-phenylindole (DAPI; Sigma) for 20 min in dark at room temperature and the nuclear fluorescence was detected using fluorescent microscope.
Transactivation assay
For estimating the transactivation potential of CstMYB1 and CstMYB1R2, the full length ORFs of two Crocus MYB genes were cloned into yeast pGBKT7 (BD) vector at NdeI and XmaI restriction sites. The BD construct of each MYB gene was transformed into yeast Y187 cells by Yeast Transformation System 2.0 (Takara, Japan) and transformations were confirmed on synthetic dropout media plates lacking L-tryptophan (SD-Trp). The transformed Y187 were further grown on SD-Trp plates supplemented with bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal, 80 µg/ml), 7 mg/ml Na2HPO4.7H2O and 3 mg/ml NaH2PO4. Instead of D-glucose, D-galactose and D-raffinose were added to the media at final concentrations of 2% and 1%, respectively. A track was kept on the development of blue color on media plates. Y187 cells transformed with pGBKT7 vector were used as negative control and those co-transformed with pGBKT7-p53 and pGADT7-T plasmids were used as positive control in the transactivation assay.
To determine the β-galactosidase (β- gal) activity, the Y187 transformed cells were grown in liquid dropout media and ortho-Nitrophenyl-β-galactoside (ONPG) was used as substrate. Following the guidelines of Yeast protocols Handbook (Clontech), β-gal units were calculated by following equation:
where OD420 and OD600 are absorbance values of the culture at 420 and 600 nm, respectively; t is the elapsed time (min) till yellow color develops and V is equal to 0.5 ml (including concentration factor).
Yeast one hybrid assay
In vivo interaction analysis for CstMYB1 and CstMYB1R2 transcription factors with the promoter stretches of PSY and CCD2 was carried out in Y1H Gold yeast strain using Matchmaker® Gold Yeast One-Hybrid Library Screening System (Takara, Clontech). The promoter core region of CCD2 gene was amplified using primers CCD2-P-1, CCD2-P-2 by genome walking using Universal Genome Walker™ 2.0 Kit (Takara, Clontech) according to manufacturer’s protocol. The PSY gene promoter was amplified using PSY-F, PSY-R primers. Both the CCD2 and PSY promoters were scanned through PLACE online software tool in order to search for the transcription factor binding sites. The 100 bp and 170 bp promoter stretches with MYB binding region (s) were selected from PSY and CCD2 core promoters, respectively. These promoter stretches were cloned into MCS of pAbAi vector using primers (CCD2-P-F, CCD2-P-R; PSY-P-F, PSY-P-R). The MYB binding regions of PSY and CCD2 promoter stretches were mutated through insertion of transversions in primer sequences. Single MYB binding site of PSY was mutated to generate a mutant PSY promoter stretch using mutated reverse primer, PSY-P-R mut while as two mutant constructs were generated for CCD2 promoter fragment, one with single MYB binding site mutation and other with mutation of two MYB binding sites using forward primers CCD2-P-F mut1 and CCD2-P-F mut2, respectively. The native (wt) and mutant (mut) PSY and CCD2 promoter stretches were cloned in pAbAi yeast vector.
The pAbAi-promoter constructs of PSY (pPSYwt and pPSYmut) and CCD2 (pCCD2wt, pCCD2mut1 and pCCD2mut2) along with p53-pAbAi control were first linearized by digesting with BstBI restriction enzyme (NEB) in Cut Smart Buffer at 65 0C. After confirming the digestion, the purified linearized restriction products were introduced into ura3-52 locus of Y1H gold yeast strain genome to generate the PSY and CCD2 Y1H promoter specific bait reporter strains using Yeastmaker™ Yeast Transformation System 2 (Takara, Clontech). The transformed Y1H colonies were selected by growing on Synthetic Defined Medium without L-Uracil (SD-Ura). Further, the confirmation of bait strain generation was made by colony PCR using promoter specific primers.
On the other side, the full length ORFs of the two MYB genes were cloned in pGADT7 (AD) vector in frame with GAL4 region. The primers used for the cloning were pGADCstMYB1-F, pGADCstMYB1-R, pGADCstMYB1R2-F, pGADCstMYB1R2-R and the sequences are given in supplementary table S2. For interactional studies, each of the pGADT7-CstMYB1, pGADT7-CstMYB1R2 constructs and pGADT7 vector (as negative control) were transformed into CCD2 and PSY Y1H bait strains. Transformed colonies were selected on synthetic defined medium without L-Uracil and L-Leucine (SD-Ura/-Leu) and finally confirmed by colony PCR using CstMYB1 and CstMYB1R2 gene specific primers. DNA–protein interaction was assessed by growing the transformed bait strains on SD-Ura/-Leu media plates with AbA concentration slightly higher than minimal concentration for bait strain inhibition. Also, p53-control insert was transformed into p53 bait strain and confirmed by growing on SD-Ura/-Leu media plates with 200 ng/ml AbA concentration. The p53 control insert transformed bait strain was used as positive control.
Accession numbers
Sequences from Baba et al. 2015 bear accession numbers MK050815-MK050823.
Results
Identification of MYB genes from Crocus transcriptome and their phylogenetic analysis
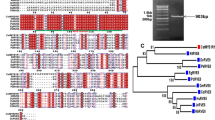
A total of 150 genes encoding MYB transcription factors were identified from Crocus transcriptome databases generated in our laboratory (Baba et al. 2015) and from other publicly available databases (Jain et al. 2016; Ahrazem et al. 2018, 2019; Tan et al. 2019) based on homology search using blast tool. Presence of MYB domain was confirmed by Pfam and interPro analysis which divided Crocus MYB genes into four categories depending upon the number of “R” repeats present. MYB DNA binding domains are highly conserved and consist of up to four imperfect amino acid sequence repeats (Rs). Each such “R” repeat consists of about 52 amino acids. R2R3 was the largest group with 78 members followed by MYB-related genes which were 65 in number. Further, we have three members from R1R2R3 (3R) and four from atypical group which had more than three R repeats (Supplementary table S1).
In order to identify sequence similarity and potential evolutionary relationship between Crocus MYB genes, we did their phylogenetic analysis. A neighbour-joining tree was constructed which grouped the Crocus MYB genes into 27 clusters (C1-27) (Fig. 1). C1-C9 consisted of mainly R2R3 MYB genes. Out of the three 3R MYBs, two clustered in C13 while the third one grouped with R2R3 MYB genes in cluster C7. All the four atypical MYBs clustered together in C20. C11-15, C17-19, C21-27 contained mostly MYB-related genes with a few members from R2R3 class. Further, a comprehensive motif analysis was performed which revealed that the members of same clad showed similar motif compositions (Supplementary fig S1).
Since R2R3 (2R) and MYB-related genes (1R) were the major groups, we wanted to learn about their relationship with their Arabidopsis homologs. For this we constructed another neighbour-joining phylogenetic tree using 78 R2R3 MYB sequences from Crocus and 126 from Arabidopsis. The gene sequences from both the plants separated into 25 different groups designated as G1-25 (Supplementary fig. S2). The MYB proteins from both the plants were distributed more or less uniformly, however, in a few groups there was no or less representation from Crocus. For example, groups like G6, 7, 9, 15, 20, 25 composed of only Arabidopsis homologs. It is important to note that these groups consisted of only a few genes. On the other hand, group 14 and 24 predominantly consisted of Crocus genes. In some cases, within a group, separate subgroups consisted of either Crocus or Arabidopsis genes. Another tree was constructed using 65 MYB-related gene sequences from Crocus and 66 from Arabidopsis (Supplementary fig. S3). The MYB-related proteins from both the plants were distributed into 21 different groups. Most of the groups had members from both the plant species. However, groups G2, 8, 13 and 21 consisted of Crocus genes only while as groups like G3, 11 and 12 had only Arabidopsis members.
Expression profile of CstMYB genes in various tissues of Crocus
In order to gain insights into the role of MYB genes in Crocus, we investigated their expression in different tissues like corm, tepal, anther and stigma. Transcriptome based expression profile was available for 71 MYB genes retrieved from two databases, one generated in house (Baba et al. 2015) and another from Jain et al. 2016. The in silico differential expression showed that out of the 71 MYB genes, 12 had higher expression (more than two fold) in stigma (Supplementary fig. S4). The expression of these 12 genes was further checked by qPCR which confirmed their stigma upregulated expression (Fig. 2a). Out of the 12 MYB genes, 8 belonged to R2R3 MYBs and 4 were MYB-related genes. Since stigma is the site of apocarotenoid biosynthesis, we selected these 12 stigma upregulated genes for further analysis.
Transcript levels of Crocus MYB genes in different tissue types and at different stages. a Heat map showing expression of 12 MYB genes in different tissues b at different developmental stages i.e., orange, red (before anthesis) and after anthesis. Expression was determined using quantitative real time PCR. Each data point represents mean of three replicates ± S.D. 18-S gene was used as endogenous control
Positive correlation between crocin accumulation and four CstMYB genes
The three most important apocarotenoids of Crocus are crocin, picrocrocin and saffranal. Crocin and picrocrocin are formed by cleavage of zeaxanthin by CCD2 and picrocrocin is subsequently converted into safranal. Previous reports show that these apocarotenoids accumulate in stigma in developmental stage specific manner and their concentration increases from yellow through orange to red stages and starts to decrease post anthesis (Moraga et al. 2009). In order to confirm previous reports, we estimated crocin by HPLC in stigmas collected at orange and red stages (prior to anthesis) and three days post anthesis. We observed that crocin accumulation increased from orange to red stages and then started to decrease after anthesis (Fig. 3).
With an aim to identify MYB genes which show a positive correlation between their expression profile and accumulation pattern of crocin, we investigated relative abundance of the 12 stigma upregulated genes at three stages i.e., orange and red stage three days before anthesis and three days after anthesis. We observed that three R2R3 MYBs (CstMYB1, CstMYB14, CstMYB16) and one MYB-related gene (CstMYB1R2) showed increase in expression from orange to red stages and this subsequently decreased after anthesis (Fig. 2b). This expression pattern correlated with the accumulation pattern of apocarotenoids in Crocus stigma, therefore, indicating possible involvement of these genes in regulating apocarotenoid metabolism.
Identification of CstMYB1 and CstMYB1R2 as potential regulators of apocarotenoid metabolism
Out of the four genes identified above, we selected two genes (CstMYB1 and CstMYB1R2) to confirm their role in apocarotenoid metabolism. The genes were cloned in pBI121 vector and transformed into stigma of Crocus flower. This is the first report on Agrobacterium mediated transformation of Crocus stigma. The method was standardized using pBI121 vector and checking GUS expression (Supplementary fig. S5). To confirm role of MYB genes on apocarotenoid regulation, we did quantification of carotenoids like lycopene, beta carotene, zeaxanthin and apocarotenoids like crocin by HPLC in vector transformed and 35S::CstMYB1 and 35S::CstMYB1R2 plants. We observed a significant increase in all the investigated carotenoids and apocarotenoids in 35S::CstMYB1 and 35S::CstMYB1R2 plants, respectively (Fig. 4, supplementary fig. S7, S8). This confirmed that CstMYB1 and CstMYB1R2 act as positive regulators of apocarotenoid metabolism.
For confirming the HPLC data, we investigated expression of carotenoid/apocarotenoid pathway genes in 35S::CstMYB1 and 35S::CstMYB1R2 plants. We observed that as compared to plants transformed with vector alone, 35S::CstMYB1 and 35S::CstMYB1R2 plants showed upregulation of important apocarotenoid pathway genes (Fig. 5). Phytoene synthase which is the rate limiting enzyme of the carotenoid pathway showed about 4.5 and fivefold induction in 35S::CstMYB1 and 35S::CstMYB1R2 plants respectively. Beta carotene hydroxylase (BCH) which converts beta carotene into zeaxanthin (precursor of Crocus apocarotenoids) showed around 7 and fourfold induction in CstMYB1 and CstMYB1R2 overexpressing plants. Further, CCD2 which cleaves zeaxanthin to form crocin and picrocrocin showed 4.4 and 3.6 fold induction in CstMYB1 and CstMYB1R2 overexpressing plants, respectively (Fig. 5).
Effect of overexpression of CstMYB1 and CstMYB1R2 on pathway genes of carotenoid/apocarotenoid metabolism in Crocus sativus. The expression levels of pathway genes in a CstMYB1 and b CstMYB1R2 overexpressing plants and those overexpressing empty vector were quantified by qPCR. All the values represent means of three independent biological replicates ± S.D. 18-S gene was used as endogenous control
There are many reports where MYB genes have been shown to regulate flavonoid metabolism. This prompted us to investigate if CstMYB1 and CstMYB1R2 have any role in regulating flavonoid metabolic pathways. For this we quantified quercetin which is one of the major flavonoids in Crocus in vector control and CstMYB1 and CstMYB1R2 overexpressing plants. There was no significant change in quercetin content in MYB overexpressing plants when compared to vector control ones (Supplementary fig. S6a). We also checked expression of flavonoid pathway genes in vector control and CstMYB1 and CstMYB1R2 overexpressing plants and observed that there was no significant change (Supplementary fig. S6b). This confirmed that CstMYB1 and CstMYB1R2 do not have any role in regulating flavonoid metabolism.
CstMYB1 and CstMYB1R2 are localized in nucleus and are transcriptionally active
To infer about the subcellular localization of CstMYB1 and CstMYB1R2, their sequences were analyzed for the presence of nuclear localization signals (NLS) using PROSITE which confirmed the presence of NLS. In order to validate these results, the genes were cloned in pBI121 fused with GUS. Each construct was agroinfiltered into onion epidermal cells which were then visualized under microscope. While control vector was spread in the entire cell, CstMYB1 and CstMYB1R2 were localized into nucleus (Fig. 6a).
Sub-cellular localization and transactivation of CstMYB1 and CstMYB1R2 genes a Nuclear localization of MYB genes: (i) control vector: pBI121-GUS (ii) CsMYB1-GUS fusion protein (iii) CsMYB1R2-GUS fusion protein b transactivation assay with the cells harbouring BD-CstMYB1 and BD-CstMYB1R2 and positive control (Y187 cells co-transformed with pGBKT7-p53 and pGADT7-T plasmids) developed the blue colour due to activation of β-galactosidase reporter gene c quantification of transactivation using β-galactosidase assay
To investigate the transcriptional activity of the two genes, they were cloned in pGBKT (BD) vector having GAL4 DNA binding domain and transformed into yeast strain Y187. After confirming the yeast transformations, the constructs were streaked on SD/-Trp plates containing D-raffinose, D-galactose and X-gal. Only the cells harbouring BD-CstMYB1 and BD-CstMYB1R2 and positive control (Y187 cells co-transformed with pGBKT7-p53 and pGADT7-T plasmids) could develop the blue colour which is attributed to activation of β-galactosidase reporter gene in host yeast strain. The Y187 cells transformed with only the pGBKT7 vector, treated as negative control, did not exhibit any blue coloration in galactosidase assay (Fig. 6b). Further, the measurement of β-galactosidase activity using ONPG revealed the presence of significantly higher β-galactosidase units in Y187 cells transformed with BD-CstMYB1 or BD-CstMYB1R2 plasmid constructs in comparison to negative control cells (Fig. 6c). The positive control cells possessed highest β-galactosidase activity. Taken together, these findings suggest that both CstMYB1 and CstMYB1R2 genes are transcriptionally active.
CstMYB1 and CstMYB1R2 directly bind promoters of carotenoid pathway genes
To gain an understanding on the mechanism of action of CstMYB1 and CstMYB1R2, we checked their interaction with promoters of two carotenoid pathway genes viz, PSY and CCD2 through yeast one hybrid (Y1H) assay. For this we amplified and cloned promoters of PSY and CCD2. We further amplified the native 100 bp and 170 bp fragments of PSY and CCD2 promoters, respectively. These stretches possessed one and two MYB binding regions, respectively. Mutant constructs were generated accordingly (one for PSY and two for CCD2). The bait strains were prepared with the native as well as mutant promoter stretches and their interactions were inferred with CstMYB1and CstMYB1R2 after examining the minimum aureobasidin A (AbA) concentrations for each bait strain. In case of PSY promoter, only CstMYB1R2 showed the interaction which was abolished when the MYB binding site was mutated in the PSY promoter stretch of mutant PSY bait strain (pPSYmut) indicating the specificity of CstMYB1R2-PSY interaction (Fig. 7a). We also observed that the CCD2 native bait (pCCD2wt) yeast cells co-transformed with CstMYB1and CstMYB1R2 could grow on selective medium supplemented with aureobasidin A which confirmed the interaction of CstMYB1 and CstMYB1R2 with CCD2 promoter. However, the mutant CCD2 strain with mutation in only one MYB binding site (pCCD2mut1) could grow on selection plates with 200 ng mL−1 AbA but the one with mutations in both MYB binding sites (pCCD2mut2) was unable to grow on the selection medium (Fig. 7b). Summing up, the results showed that while CstMYB1 directly binds to CCD2 promoter only, CstMYB1R2 binds to the promoters of both PSY and CCD2.
Discussion
Identification of transcription factors and their underlying mechanism of regulation of carotenoid/apocarotenoid metabolism has remained elusive in all plants particularly in Crocus. Crocus represents the only plant species which synthesizes apocarotenoids like crocin, picrocrocin and safranal in significant amounts (Ashraf et al. 2015). The biosynthetic pathway of apocarotenoids has been studied to a considerable extent (Frusciante et al. 2014; Bouvier et al. 2003; Rubio-Moraga et al. 2014). However, the mechanism that regulates expression of pathway genes in stigma and in developmentally regulated manner is still not known. There is only meagre information available on this aspect (Ahrazem et al. 2018; Ahrazem et al. 2019; Malik and Ashraf 2017; Ashraf et al. 2015).
MYB proteins represent functionally diverse and richest family, most of which act as transcription factors. They play key roles in regulatory networks which control many aspects of plant development and metabolism as well as stress response (Dubos et al. 2010; Ambawat et al. 2013; Liu et al. 2015). Recently MYB genes were demonstrated to play role in carotenoid metabolism in fruits and flowers of many plants (Ampomah-Dwamena et al. 2019; Sagawa et al. 2016; Meng et al. 2019; Fu et al. 2020; Wu et al. 2020). The Crocus transcriptome data developed in our laboratory (Baba et al. 2015) showed that MYB gene family was the largest transcription factor family in Crocus which indicated its diverse roles in this crop. Abundance of MYB genes in Crocus together with reports of involvement of MYBs in regulating carotenoid metabolism in a number of plants prompted us to investigate role of these genes in Crocus carotenoid/apocarotenoid metabolism. In this context we identified 150 MYB genes from Crocus transcriptome database generated in house (Baba et al., 2015) and other publicly available databases (Jain et al. 2016; Ahrazem et al. 2018; Ahrazem et al. 2019; Tan et al. 2019). These 150 genes may not represent the actual number of MYBs in Crocus because most of the transcriptome databases have been generated from stigma tissue only. Here, although the genes were identified using five databases, there were lots of duplications since most of the sequences were generated from stigma. The presence of MYB domain was confirmed by Pfam and interPro analysis. Out of the total 150 MYBs, 78 belonged to R2R3; 65 to MYB-related genes; three were R1R2R3 (3R) while four members were from atypical (5R) group (Supplementary table S1). This distribution was similar to that of other plant species. In most of the plants R2R3 forms the largest subfamily of MYB genes (Katiyar et al. 2012; Wang et al. 2015; Salih et al. 2016). There are around 126 R2R3 MYB genes in Arabidopsis, 121 in tomato (Zhao et al. 2014), 109 in Oryza sativa (Dubos et al. 2010), 192 in Populus (Wilkins et al. 2009) and 222 in apple (Cao et al. 2013). Further, 64 and 70 MYB-related genes have been identified from Arabidopsis and Oryza sativa respectively. R1R2R3 (3R) represents a small subfamily and in most plants is encoded by five genes. In Arabidopsis and Oryza sativa, there are 5 and 4 members respectively (Katiyar et al. 2012). We identified six members of this subfamily from Crocus. Further, the smallest subfamily is atypical group which is encoded by a single gene, however, we found four members in Crocus.
Phylogenetic analysis divided Crocus MYB genes into 27 clusters (C1-27) (Fig. 1). Phylogeny vis a vis analysis of conserved motifs (Supplementary fig. S1) showed that in most of the cases, MYBs with similar domain structure clustered together. This might provide a clue about conservation of functions of such genes. Since R2R3 MYBs and MYB-related genes were the dominant classes, we performed their phylogeny with their Arabidopsis counterparts. Two separate neighbour-joining trees were constructed for R2R3 MYBs (Supplementary fig. S2) and MYB-related genes (Supplementary fig. S3). In both the cases, we observed that most of the genes from Crocus were distributed uniformly among their Arabidopsis homologs. However, there were a few exceptions where whole group consisted of genes from Arabidopsis or Crocus only. For example, for R2R3 MYBS, groups like G6, 7, 9, 15, 20 and 25 consists of only Arabidopsis homologs. However, these groups had only few members. On the other hand, G14 and G24 clades had mostly Crocus homologs. Such clades might represent genes with specialized functions. In contrast to the 2R MYBS, the MYB-related (1R-MYB) subfamily has been studied to a lesser extent. The phylogenetic tree between Crocus and Arabidopsis MYB- related genes showed that three clades (G2, 8, 13 and 21) had only Crocus homologs while clades (G3, 11 and 12) were specific to Arabidopsis. More research would provide leads on species specific role of these genes. Further, the genes which fall in the same clade seem to have similar functions. Many MYB genes in Arabidopsis have been characterized and their functions revealed. This information would provide a platform to understand the functions of Crocus orthologs.
To gain more information about functional roles of MYB genes in Crocus, we checked their expression profile in various tissue types. Out of the 150 MYB genes identified, in silico tissue specific expression data was available for only 71 genes. The data showed that 12 out of 71 genes had higher expression in stigma suggesting their stigma specific role (Fig. 2a). Crocus apocarotenoids are accumulated in stigma only and in developmentally regulated manner (Moraga et al. 2009). We further confirmed this by quantification of crocin at orange, red stage before anthesis and at post anthesis stage and observed increase in crocin content from orange to red stages before anthesis which then starts to decrease (Fig. 3). We also checked the expression of the 12 stigma upregulated genes at the above mentioned three stages using qPCR. We observed that three R2R3 MYBs (CstMYB1, CstMYB14 and CstMYB16) and one MYB- related gene (CstMYB1R2) showed enhanced expression from orange to red stage before anthesis and decreased after anthesis (Fig. 2b). The expression profile of the above mentioned MYB genes corroborated with accumulation pattern of crocin, therefore, indicating possible involvement of these four genes in apocarotenoid regulation. Therefore, this study provides an important platform for working out mechanism of regulation of apocarotenoids.
In order to confirm whether the identified MYB genes actually have any role in apocarotenoid regulation, we chose one gene (CstMYB1) among the R2R3MYBs and one (CstMYB1R2) among MYB-related genes. These two genes were selected based on their higher expression. The phylogenetic analysis of CstMYB1 with its Arabidopsis homologs placed it in G18 clad (Supplementary fig. S2) which had six members from Crocus and three from Arabidopsis. Among Arabidopsis homologs AtMYB21 and AtMYB24 have been characterized and are known to show flower specific expression and control terpene synthesis (Yang et al. 2020) and anther development respectively (Yang et al. 2007). Inspite of falling in the same clad, CstMYB1 showed only around 57% homology with Arabidopsis counterparts. Also the close Arabidopsis homologs of CstMYB1R2 are not characterized yet. This further prompted us to unravel the function of these two MYB genes. Moreover, the phylogenetic analysis of other two genes (CstMYB14 and CstMYB16) grouped them with members from Arabidopsis which are known to play roles in flavonoid regulation. For example, CstMYB16 is present in group 11 (Supplementary figure S2) and the members of this group are known to repress phenyl-propanoid pathway. Here our aim was to choose the genes which could have novel roles. That is why CstMYB14 and CstMYB16 were not analyzed further. CstMYB1 and CstMYB1R2 were transiently overexpressed in stigma of Crocus flower by Agrobacterium mediated transformation (Supplementary fig. S5). It is important to mention here that we are reporting Agrobacterium mediated transformation first time in Crocus. The method was standardized using GUS gene in pBI121 vector. Further, the CstMYB1 and CstMYB1R2 showed around 6 and fourfold induction in the overexpressing plants. To ascertain role of these MYB genes in carotenoid/apocarotenoid metabolism, we quantified lycopene, beta carotene, zeaxanthin and crocin using HPLC and found significant increase in these compounds in MYB overexpressing plants compared to those transformed with vector control (Fig. 4). We also checked expression of key carotenoid pathway genes in CstMYB1 and CstMYB1R2 overexpressing plants and observed many fold increase in their expression (Fig. 5). These results confirmed role of CstMYB1 and CstMYB1R2 in regulating Crocus apocarotenoid metabolism.
Many MYB transcription factors have been implicated in flavonoid and anthocyanin pathway regulation (Allan et al. 2008; Zhai et al. 2016). To investigate if CstMYB1 and CstMYB1R2 have any role in regulating above said pathways, we checked quercetin content in vector control and MYB overexpressing plants which showed that there was no significant change (Supplementary fig. S6a). This was confirmed by expression of flavonoid pathway genes which too displayed no significant change (Supplementary fig. S6b). This again confirmed specific role played by CstMYB1 and CstMYB1R2 in regulating carotenoid/apocarotenoid metabolism.
Next, we wanted to investigate if there is any direct interaction between these MYB genes and promoters of pathway genes. We performed yeast one hybrid of PSY and CCD2 promoter stretches with the two selected MYBs. We chose PSY because this gene is considered as key determinant of carotenoid pathway. Further, CCD2 is the actual enzyme involved in production of Crocus apocarotenoids (Frusciante et al. 2014). The yeast one hybrid results indicated a positive interaction of CstMYB1 with CCD2 promoter and of CstMYB1R2 with both PSY and CCD2 promoters (Fig. 7). Even though overexpression of CstMYB1 enhanced expression of both, PSY and CCD2 genes, it showed interaction with promoter of CCD2 only. In other words there might be indirect interaction between CstMYB1 and PSY promoter. For further validation of these results, the MYB binding motifs of both the promoters were mutated and then the interaction of mutated promoter stretches was checked with the MYB genes. The results indicated that there was no interaction. This confirmed that these MYB genes directly bind to promoters of carotenoid pathway genes and influence their expression.
Taken together, we propose that CstMYB1 and CstMYB1R2 are novel regulators of Crocus apocarotenoid metabolism and exert their influence by binding to promoters of key pathway genes.
Change history
30 September 2021
A Correction to this paper has been published: https://doi.org/10.1007/s11103-021-01192-2
References
Abdullaev FI, Espinosa-Aguirre JJ (2004) Biomedical properties of saffron and its potential use in cancer therapy and chemoprevention trials. Cancer Detect Prev 28(6):426–432. https://doi.org/10.1016/j.cdp.2004.09.002
Ahrazem O, Argandoña J, Fiore A, Aguado C, Lujan R, Rubio-Moraga A, Marro M, Araujo-Andrade C, Loza-Alvarez P, Diretto G, Gómez-Gómez L (2018) Transcriptome analysis in tissue sectors with contrasting crocins accumulation provides novel insights into apocarotenoid biosynthesis and regulation during chromoplast biogenesis. Sci Rep 8(1):1–17. https://doi.org/10.1038/s41598-018-21225-z
Ahrazem O, Argandona J, Fiore A, Rujas A, Rubio-Moraga A, Castillo R, Gómez-Gómez L (2019) Multi-species transcriptome analyses for the regulation of crocins biosynthesis in Crocus. BMC Genomics 20(1):320. https://doi.org/10.1186/s12864-019-5666-5
Allan AC, Hellens RP, Laing WA (2008) MYB transcription factors that colour our fruit. Trends Plant Sci 13(3):99–102. https://doi.org/10.1016/j.tplants.2007.11.012
Ambawat S, Sharma P, Yadav NR, Yadav RC (2013) MYB transcription factor genes as regulators for plant responses: an overview. Physiol Mol Biol Plants 19(3):307–321. https://doi.org/10.1007/s12298-013-0179-1
Ampomah-Dwamena C, Thrimawithana AH, Dejnoprat S, Lewis D, Espley RV, Allan ACA, kiwifruit (Actinidia deliciosa), (2019) R2R3-MYB transcription factor modulates chlorophyll and carotenoid accumulation. New Phytol 221(1):309–325. https://doi.org/10.1111/nph.15362
Ashraf N, Jain D, Vishwakarma RA (2015) Identification, cloning and characterization of an ultrapetala transcription factor CsULT1 from Crocus: a novel regulator of apocarotenoid biosynthesis. BMC Plant Biol 15(1):25. https://doi.org/10.1186/s12870-015-0423-7
Auldridge ME, McCarty DR, Klee HJ (2006) Plant carotenoid cleavage oxygenases and their apocarotenoid products. Curr Opin Plant Biol 9(3):315–321. https://doi.org/10.1016/j.pbi.2006.03.005
Baba SA, Mohiuddin T, Basu S, Swarnkar MK, Malik AH, Wani ZA, Abbas N, Singh AK, Ashraf N (2015) Comprehensive transcriptome analysis of Crocus sativus for discovery and expression of genes involved in apocarotenoid biosynthesis. BMC Genomics 16(1):698. https://doi.org/10.1186/s12864-015-1894-5
Baba SA, Vishwakarma RA, Ashraf N (2017) Functional characterization of CsBGlu12, a β-glucosidase from Crocus sativus, provides insights into its role in abiotic stress through accumulation of antioxidant flavonols. J Biol Chem 292(11):4700–4713. https://doi.org/10.1074/jbc.M116.762161
Bouvier F, Suire C, Mutterer J, Camara B (2003) Oxidative remodeling of chromoplast carotenoids: identification of the carotenoid dioxygenase CsCCD and CsZCD genes involved in Crocus secondary metabolite biogenesis. Plant Cell 15(1):47–62. https://doi.org/10.1105/tpc.006536
Cao ZH, Zhang SZ, Wang RK, Zhang RF, Hao YJ (2013) Genome wide analysis of the apple MYB transcription factor family allows the identification of MdoMYB121 gene confering abiotic stress tolerance in plants. PLoS ONE 8(7):e69955. https://doi.org/10.1371/journal.pone.0069955
D’Agostino N, Pizzichini D, Chiusano ML, Giuliano G (2007) An EST database from saffron stigmas. BMC Plant Biol 7(1):53. https://doi.org/10.1186/1471-2229-7-53
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15(10):573–581. https://doi.org/10.1016/j.tplants.2010.06.005
Fang F, Oliva M, Ehi-Eromosele S, Zaccai M, Arazi T, Michal Oren-Shamir M (2018) Successful floral-dipping transformation of post-anthesis lisianthus (Eustoma grandiflorum) flowers. Plant J 96(4):869–879. https://doi.org/10.1111/tpj.14076
Frusciante S, Diretto G, Bruno M, Ferrante P, Pietrella M, Prado-Cabrero A, Rubio-Moraga A, Beyer P, Gomez-Gomez L, Al-Babili S, Giuliano G (2014) Novel carotenoid cleavage dioxygenase catalyzes the first dedicated step in saffron crocin biosynthesis. Proc Natl Acad Sci 111(33):12246–12251. https://doi.org/10.1073/pnas.1404629111
Fu C, Chen H, Gao H, Lu Y, Han C, Han Y (2020) Two papaya MYB proteins function in fruit ripening by regulating some genes involved in cell-wall degradation and carotenoid biosynthesis. J Sci Food Agric 100(12):4442–4448. https://doi.org/10.1002/jsfa.10484
Giuliano G, Tavazza R, Diretto G, Beyer P, Taylor MA (2008) Metabolic engineering of carotenoid biosynthesis in plants. Trends Biotechnol 26(3):139–145. https://doi.org/10.1016/j.tibtech.2007.12.003
Gonzalez A, Zhao M, Leavitt JM, Lloyd AM (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53(5):814–827. https://doi.org/10.1111/j.1365-313X.2007.03373.x
Jain M, Srivastava PL, Verma M, Ghangal R, Garg R (2016) De novo transcriptome assembly and comprehensive expression profiling in Crocus sativus to gain insights into apocarotenoid biosynthesis. Sci Rep 6(1):22456. https://doi.org/10.1038/srep22456
Katiyar A, Smita S, Lenka SK, Rajwanshi R, Chinnusamy V, Bansal KC (2012) Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics 13(1):544. https://doi.org/10.1186/1471-2164-13-544
Liu J, Osbourn A, Ma P (2015) MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol Plant 8(5):689–708. https://doi.org/10.1016/j.molp.2015.03.012
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Lu S, Li L (2008) Carotenoid metabolism: biosynthesis, regulation, and beyond. J Integr Plant Biol 50(7):778–785. https://doi.org/10.1111/j.1744-7909.2008.00708.x
Malik AH, Ashraf N (2017) Transcriptome wide identification, phylogenetic analysis, and expression profiling of zinc-finger transcription factors from Crocus sativus L. Mol Genet Genom 292(3):619–633. https://doi.org/10.1007/s00438-017-1295-3
Meng Y, Wang Z, Wang Y, Wang C, Zhu B, Liu H, Ji W, Wen J, Chu C, Tadege M, Niu L (2019) The MYB activator WHITE PETAL1 associates with MtTT8 and MtWD40-1 to regulate carotenoid-derived flower pigmentation in Medicago truncatula. Plant Cell 31(11):2751–2767. https://doi.org/10.1105/tpc.19.00480
Moraga ÁR, Rambla JL, Ahrazem O, Granell A, Gómez-Gómez L (2009) Metabolite and target transcript analyses during Crocus sativus stigma development. Phytochemistry 70(8):1009–1016. https://doi.org/10.1016/j.phytochem.2009.04.022
Roy S (2016) Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant Signal Behav 11(1):e1117723. https://doi.org/10.1080/15592324.2015.1117723
Rubio-Moraga A, Rambla JL, Fernández-de-Carmen A, Trapero-Mozos A, Ahrazem O, Orzáez D, Granell A, Gómez-Gómez L (2014) New target carotenoids for CCD4 enzymes are revealed with the characterization of a novel stress-induced carotenoid cleavage dioxygenase gene from Crocus sativus. Plant Mol Biol 86(4–5):555–569. https://doi.org/10.1007/s11103-014-0250-5
Sagawa JM, Stanley LE, LaFountain AM, Frank HA, Liu C, Yuan YW (2016) An R2R3-MYB transcription factor regulates carotenoid pigmentation in Mimulus lewisii flowers. New Phytol 209(3):1049–1057. https://doi.org/10.1111/nph.13647
Salih H, Gong W, He S, Sun G, Sun J, Du X (2016) Genome-wide characterization and expression analysis of MYB transcription factors in Gossypium hirsutum. BMC Genet 17(1):129. https://doi.org/10.1186/s12863-016-0436-8
Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B (2007) Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J 50(4):660–677. https://doi.org/10.1111/j.1365-313X.2007.03078.x
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 60. Mol Biol Evol 30(12):2725–2729. https://doi.org/10.1093/molbev/mst197
Tan H, Chen X, Liang N, Chen R, Chen J, Hu C, Li Q, Li Q, Pei W, Xiao W, Yuan Y (2019) Transcriptome analysis reveals novel enzymes for apo-carotenoid biosynthesis in saffron and allows construction of a pathway for crocetin synthesis in yeast. J Exp Bot 70(18):4819–4834. https://doi.org/10.1093/jxb/erz211
Trapero A, Ahrazem O, Rubio-Moraga A, Jimeno ML, Gómez MD, Gómez-Gómez L (2012) Characterization of a glucosyltransferase enzyme involved in the formation of kaempferol and quercetin sophorosides in Crocus sativus. Plant Physiol 159(4):1335–1354. https://doi.org/10.1104/pp.112.198069
Wang Z, Tang J, Hu R, Wu P, Hou XL, Song XM, Xiong AS (2015) Genome-wide analysis of the R2R3-MYB transcription factor genes in Chinese cabbage (Brassica rapa ssp. pekinensis) reveals their stress and hormone responsive patterns. BMC Genomics 16(1):17. https://doi.org/10.1186/s12864-015-1216-y
Wilkins O, Nahal H, Foong J, Provart NJ, Campbell MM (2009) Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol 149(2):981–993. https://doi.org/10.1104/pp.108.132795
Wu M, Xu X, Hu X, Liu Y, Cao H, Chan H, Gong Z, Yuan Y, Luo Y, Feng B, Li Z (2020) SlMYB72 regulates the metabolism of chlorophylls, carotenoids, and flavonoids in tomato fruit. Plant Physiol 183(3):854–868. https://doi.org/10.1104/pp.20.00156
Xu K, Huang X, Wu M, Wang Y, Chang Y, Liu K, Zhang J, Zhang Y, Zhang F, Yi L, Li T (2014) A rapid, highly efficient and economical method of Agrobacterium-mediated in planta transient transformation in living onion epidermis. PLoS ONE 9(1):e83556. https://doi.org/10.1371/journal.pone.0083556
Yang XY, Li JG, Pei M, Gu H, Chen ZL, Qu LJ (2007) Over-expression of a flower-specific transcription factor gene AtMYB24 causes aberrant anther development. Plant Cell Rep 26(2):219–228. https://doi.org/10.1007/s00299-006-0229-z
Yang Z, Li Y, Gao F, Jin W, Li S, Kimani S, Yang S, Bao T, Gao X, Wang L (2020) MYB21 interacts with MYC2 to control the expression of terpene synthase genes in flowers of Freesia hybrida and Arabidopsis thaliana. J Exp Bot 71(14):4140–4158. https://doi.org/10.1093/jxb/eraa184
Zhai R, Wang Z, Zhang S, Meng G, Song L, Wang Z, Li P, Ma F, Xu L (2016) Two MYB transcription factors regulate flavonoid biosynthesis in pear fruit (Pyrus bretschneideri Rehd.). J Exp Bot 67(5):1275–1284. https://doi.org/10.1093/jxb/erv524
Zhang Z, Wang CZ, Wen XD, Shoyama Y, Yuan CS (2013) Role of saffron and its constituents on cancer chemoprevention. Pharm Biol 51(7):920–924. https://doi.org/10.3109/13880209.2013.771190
Zhao PP, Li Q, Li J, Wang LN, Ren ZH (2014) Genome-wide identification and characterization of R2R3MYB family in Solanum lycopersicum. Mol Genet Genomics 289:1183–1207
Zheng Y, Jiao C, Sun H, Rosli HG, Pombo MA, Zhang P, Banf M, Dai X, Martin GB, Giovannoni JJ, Zhao PX (2016) iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol Plant 9(12):1667–1670. https://doi.org/10.1016/j.molp.2016.09.014
Acknowledgements
ZYB is thankful to UGC, New Delhi for Senior Research Fellowship. TM is thankful to CSIR, New Delhi for her fellowship. We are also thankful to Oussama Ahrazem and Lourdes Gómez-Gómez for providing MYB gene sequence data generated in their laboratory. The article bears institutional manuscript number CSIR-IIIM/IPR/00200.
Author information
Authors and Affiliations
Contributions
NA conceived and supervised the experiments and wrote the manuscript. ZYB and TM performed and analysed the experiments. AK did HPLC analysis and AJL performed assembly and annotation of MYB gene sequence databases.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original version of this article was revised: the second affiliation was incorrect.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bhat, Z.Y., Mohiuddin, T., Kumar, A. et al. Crocus transcription factors CstMYB1 and CstMYB1R2 modulate apocarotenoid metabolism by regulating carotenogenic genes. Plant Mol Biol 107, 49–62 (2021). https://doi.org/10.1007/s11103-021-01180-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-021-01180-6