Abstract
The R2R3MYB proteins comprise one of the largest families of transcription factors and play regulatory roles in developmental processes and defense responses in plants. However, there has been relatively little effort to systematically carry out comprehensive genomic and functional analyses of these genes in tomato (Solanum lycopersicum L.), a reference species for Solanaceae plants, and the model plant for fruit development. In this study, a total of 121 R2R3MYB genes were identified in the tomato genome released recently and further classified into 29 subgroups based on the phylogenetic analysis of the complete protein sequences. Phylogenetic comparison of the members of this superfamily among tomato, Arabidopsis, grape, rice, poplar, soybean, cucumber and apple revealed that the putative functions of some tomato R2R3MYB proteins were clustered into the Arabidopsis functional clades. The chromosome distribution pattern revealed that tomato R2R3MYB genes were enriched on several chromosomes and 52 % of the family members were tandemly duplicated genes. Tissue specificity or different expression levels of SlR2R3MYBs in different tissues suggested differential regulation of tissue development as well as metabolic regulation. The transcript abundance level analysis during abiotic conditions identified a group of R2R3MYB genes that responded to one or more treatments suggesting that the SlR2R3MYBs played major roles in the plant response to abiotic conditions and involved in signal transduction pathways. This study not only provides a solid foundation for further functional dissection of tomato R2R3MYB family genes, but may also be profitable for, in the future, the improvement of tomato stress tolerance and fruit quality.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The MYB family is large, functionally diverse and represented in all eukaryotes, and most of them function as transcription factors with MYB binding domain conferring the ability to bind DNA (Ogata et al. 1994). The MYB domain generally consists of up to four imperfect amino acid sequence repeats (R) of about 52 amino acids, and each repeat encodes three α-helices, with the second and third helices forming a helix–turn–helix (HTH) structure which intercalates in the major groove of DNA (Ogata et al. 1994; Dubos et al. 2010). MYB proteins can be divided into different types according to the number of repeat(s) in the MYB domain: 4R-MYB contains four R1/R2-like repeats, 3R-MYB (R1R2R3MYB) three consecutive repeats, R2R3MYB two repeats, and the MYB-related type usually, but not always, has a single repeat (Dubos et al. 2010; Li et al. 2012a). Among the four classes identified, the R2R3 subfamily is the most abundant type in plants and play important roles in many plant-specific processes (Romero et al. 1998; Jin and Martin 1999; Dubos et al. 2010). Although many R2R3-MYB genes have been characterized in many species (Dubos et al. 2010), little information is available about the R2R3MYB genes and their functions in fruit development and abiotic stress tolerance of tomato (Solanum lycopersicum L.), a word-wide vegetable.
With the genomes of an ever increasing number of plant species being fully sequenced, the genome-wide analysis of R2R3MYB genes have soared in recent time. Based on their well conserved DNA-binding domains, R2R3MYB family has been annotated genome-wide in both monocotyledonous and dicotyledonous genomes, such as Zea mays (more than 200 members) (Dias et al. 2003), Oryza sativa (102 members) (Chen et al. 2006; Li et al. 2012a), Arabidopsis thaliana (126 members) (Stracke et al. 2001), Vitis vinifera (117 members) (Matus et al. 2008), Populus trichocarpa (192 members) (Wilkins et al. 2009), Glycine max (244 members) (Du et al. 2012), Cucumis sativus (47 members) (Li et al. 2012a; Table S2 in this study) and Malus domestica (222 members) (Cao et al. 2013). The members of the R2R3MYB family from Arabidopsis have been divided into 25 subgroups (Dubos et al. 2010). Comparative phylogenetic studies have identified new R2R3MYB subgroups in other plant species for which there are no representatives in Arabidopsis (e.g. in grape, poplar, rice, soybean, cucumber and apple), which suggested that these proteins might have specialized roles which have been either lost in Arabidopsis or were acquired after divergence from the last common ancestor (Matus et al. 2008; Wilkins et al. 2009; Du et al. 2012; Li et al. 2012a; Cao et al. 2013). Functional diversification in gene families encoding transcription factors is emerging as a major source of morphological and physiological diversity underlying evolution (Carretero-Paulet et al. 2010; Doebley and Lukens 1998; Riechmann et al. 2000; Tsiantis and Hay 2003; Kellogg 2004). From the plant R2R3MYB genes that have been characterized to date, the majority have been implicated in diverse pathways and appear to play crucial roles in plant-specific processes, including secondary metabolism such as flavonoids (Mehrtens et al. 2005; Stracke et al. 2007) and anthocyanin (Teng et al. 2005; Deluc et al. 2006; Li et al. 2012b), cell fate and identity such as trichomes (Oppenheimer et al. 1991; Higginson et al. 2003), root hairs (Wada et al. 1997; Tominaga et al. 2007; Tominaga-Wada et al. 2012) and petal epidermis (Perez-Rodriguez et al. 2005; Baumann et al. 2007), developmental processes such as anther and pollen development (Brownfield et al. 2009; Millar and Gubler 2005) and axillary meristem formation (Müller et al. 2006; Keller et al. 2006), plant defense and response to biotic and abiotic stresses (Urao et al. 1993; Daniel et al. 1999; Sugimoto et al. 2000; Hemm et al. 2001; Stockinger et al. 2001; Vailleau et al. 2002; Abe et al. 2003; Denekamp and Smeekens 2003; Nagaoka and Takano 2003), and so on.
Tomato is not only one of the most important vegetables all over the world, but is also a model system for fruit development (The Tomato Genome Consortium 2012). Tomato fleshy fruits are unique because they typically undergo a ripening process after seed maturation that involves irreversible changes in color, texture, sugar content, aroma and flavor to become succulent and appealing to seed-dispersal vectors (including humans). Regulation of this process ensures accurate and tissue-specific control of a developmental transition that would be highly detrimental if it were to occur in the wrong tissue or at the wrong stage of fruit maturity (Zhong et al. 2013). To date, in tomato, only few R2R3MYB genes were cloned and identified (Lin et al. 1996; Schmitz et al. 2002; Mathews et al. 2003; Gong and Bewley 2008; Abuqamar et al. 2009; Ballester et al. 2010; Wang et al. 2011; Naz et al. 2013) (Table S1). Since numerous R2R3MYB proteins have been found to be involved in the control of many plant-specific processes (Dubos et al. 2010), it is of interest to determine that how many R2R3MYB genes exist in tomato. Moreover, it is important to uncover that which R2R3MYBs take part in different biological processes in tomato, such as abiotic stress tolerance and fruit development. In addition, the completion of tomato genome sequence provides an opportunity for genome-wide analysis of R2R3MYB genes encoding transcription factors (The Tomato Genome Consortium 2012).
In this study, we have systematically examined the putative R2R3MYB gene subfamily and revealed that the tomato genome contained a total of 121 gene members encoding R2R3MYB transcription factors. The detailed information on the genomic structures, chromosomal locations and phylogenetic analysis among R2R3MYB genes in tomato, Arabidopsis, grape, rice, poplar, soybean, cucumber and apple were presented. In addition, the expression profiles during fruit development and in response to abiotic conditions were compared for 51 selected R2R3MYB genes by quantitative real-time PCR. Our results may provide the first insight, to our knowledge, into the possible mechanisms of R2R3MYB proteins in the diversification of plant form and function through analyzing the entire R2R3MYB family encoded in the tomato genome.
Materials and methods
Database search and sequence conservation analysis of tomato R2R3MYB genes
126 Arabidopsis R2R3MYB protein sequences were downloaded from TAIR (Dubos et al. 2010; Matus et al. 2008; Stracke et al. 2001). 117 Vitis vinifera and 197 Populus trichocarpa R2R3MYB genes come from Wilkins et al. (2009), and the corresponding protein sequences were downloaded from the International Grape Genome Program’s (IGGP) Web site (http://www.genoscope.cns.fr/externe/English/Projets/Projet_ML/projet.html) and Joint Genome Institute P. trichocarpa version 1.1 Web site (http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html), respectively. 102 Oryza sativa R2R3MYB genes come from Chen et al. (2006) and Li et al. (2012a, b) and the corresponding protein sequences were downloaded from the Rice Genome Annotation Project’s Web site (http://rice.plantbiology.msu.edu). 244 Glycine max R2R3MYB genes come from Du et al. (2012), and the corresponding protein sequences were downloaded from the Joint Genome Institute (JGI) Glycine max version 7.0 website (http://www.phytozome.net/cgi-bin/gbrowse/soybean/).
It is important to note that, here, the number of R2R3MYB proteins from cucumber is different from the previous study (Li et al. 2012a). We identified 46 CsR2R3MYB genes from the updated Version 2.0 cucumber genome database (http://www.icugi.org/cgi-bin/ICuGI/genome/home.cgi?ver=2&organism=cucumer), while (Li et al. 2012a) found 55 R2R3MYB genes from the Version 1.0 out of date. Of all the 55 CsR2R3MYB genes, 14 genes were deleted (Csa004708, Csa005219, Csa007739, Csa009054, Csa014650, Csa015156, Csa017164, Csa017539, Csa018350, Csa022057, Csa024079, Csa008474, Csa000156, Csa020800), while 5 genes were newly added (Csa3M535090.1, Csa1M575180.1, Csa3M182040.1, Csa6M046460.1, Csa6M046240.1) and named CsMYB55–CsMYB59. In addition, a new cucumber R2R3MYB was discovered by (Li et al. 2013) and named CsMYB60 (Table S2). The tomato annotated (predicted) genes and proteins were downloaded from Tomato Genome Sequencing Project. This annotated data can be downloaded from Tomato Genome DataBase (http://mips.helmholtz-muenchen.de/plant/tomato/searchjsp/blast.jsp).
126 Arabidopsis R2R3MYB proteins were used as query sequences and Blastp searches against the predicted tomato proteins. In addition, the Hidden Markov Model (HMM) profile for the MYB binding domain (PF00249) from the Pfam database (http://pfam.janelia.org) was also applied as a query to search the tomato genome database using BlastP program for identifying all MYB containing sequences in tomato. To further verify the reliability of these candidate sequences, the Pfam database (http://pfam.sanger.ac.uk/search) and SMART (http://smart.embl-heidelberg.de/) (Letunic et al. 2009) were used to confirm each candidate SlR2R3MYB protein as a member of R2R3MYB family.
To analyze the features of the MYB domain of tomato R2R3MYB proteins, the sequences of R2 and R3 MYB repeats of 121 SlR2R3MYB proteins were aligned with the ClustalX 1.81 and adjusted manually. The sequence logos for R2 and R3 MYB repeats were obtained by submitting the multiple alignment sequences to the website (http://weblogo.berkeley.edu/logo.cgi) (Crooks et al. 2004).
Phylogenetic analysis
Multiple sequence alignments were performed using ClustalX 1.81 with default parameters, and the alignments were then adjusted manually before phylogenetic tree was constructed. A phylogenetic tree was constructed with the aligned R2R3MYB binding domain and full predicted protein sequences of 121 SlR2R3MYB genes using MEGA 4 (Tamura et al. 2007), respectively. The neighbor-joining (NJ) method was used with the following parameters: Poisson correction, pairwise deletion, and bootstrap (1,000 replicates; random seed). The complete amino acid sequences of 1175 R2R3MYB proteins, including 126 AtR2R3MYB, 117 VvR2R3MYB, 102 OsR2R3MYB, 197 PtR2R3MYB, 244 GmR2R3MYB, 47 CsR2R3MYB and 222 MdR2R3MYB, were used to construct NJ tree using MEGA 4 (Tamura et al. 2007). Classification of the SlR2R3MYB genes was then performed according to their phylogenetic relationships with their corresponding Arabidopsis, grape, rice, poplar and soybean R2R3MYB genes.
Intron–exon structure analysis
The DNA and cDNA sequences corresponding to each predicted gene from the tomato genome were downloaded, and then the intron distribution pattern and splicing phase were analyzed using the web-based bioinformatics tool GSDS (http://gsds.cbi.pku.edu.cn/) (Guo et al. 2007).
An intron was designated as occurring in one of three phases. In phase 1, splicing occurred after the first nucleotide of the codon; in phase 2, splicing occurred after the second nucleotide; and in phase 0, splicing occurred after the third nucleotide of the codon (Li et al. 2006; Sharp 1981). The numbers above…to phase 0, 1, 2 were corrected.
Genome distribution and gene duplication analysis
Genes were mapped on chromosomes by identifying their chromosomal position provided in the Tomato Genome Database. The distribution of SlR2R3MYB family members throughout the tomato genome was drawn by MapInspect. To detect the tandemly duplicated genes, the method of Huang et al. (2012) was used. Software DNAMAN 5.2.2 was used to analyze the SlR2R3MYB homologs in the phylogenetic tree for similarity.
Expression analysis
Tomato (Solanum lycopersicum L. cv. Micro-Tom) seeds were germinated on moist filter paper in an incubator at 28 °C for 1 day. The germinated seeds were sown into soil mixture in the greenhouse at Shandong Agricultural University. After 10 days, batches of ten seedlings were transferred to a plastic tank filled with an aerated nutrient solution (pH 6.0–6.5) containing: Ca(NO3)2: 3.5 mM, KNO3: 7 mM, KH2PO4: 0.78 mM, MgSO4: 2 mM, H3BO3: 29.6 μM, MnSO4: 10 μM, Fe-EDTA: 50 μM, ZnSO4: 1.0 μM, H2MoO4: 0.05 μM and CuSO4: 0.95 μM (Li et al. 2012a). The experiment was carried out in an illuminated incubator and the air temperature (28 °C during the day and 18 °C during the night) and light intensity (400 μmol m−2 s−1) regimes were maintained throughout each treatment. When the tomato seedlings were at the three-true-leaf stage, three treatments were conducted, respectively: 100 mM NaCl, 100 μM ABA, 4 °C. Leaves for RNA extraction were harvested at 0, 1, 3, 6 and 12 h after the three treatments, respectively. The roots, stems, leaves, flowers and different developmental stages of fruits were collected separately used for tissue specific expression analysis.
Total RNA was prepared from different tissues with an RNAprep pure Plant Kit (TIANGEN, China), according to the manufacturers’ instructions. First strand cDNA was synthesized using 1 μg total RNA and PrimeScript 1st Strand cDNA Synthesis Kit (TaKaRa, Japan).
Gene structures of the differently spliced transcripts were analyzed using GSDS (http://gsds.cbi.pku.edu.cn/) (Guo et al. 2007). The ORFs were predicted for the transcripts that were cloned using ORF Finder software (http://www.ncbi.nlm.nih.gov/gorf/gorf.html).
To analyze expression patterns of SlR2R3MYB genes, quantitative real-time PCR was carried out using the RealMasterMix (SYBR Green) kit (TIANGEN, China) and quantified the PCR amplification according to the manufacturers’ protocol. β-actin gene (GenBank 101262163) was used as an internal control. Outside of the conserved region, the PCR primers were designed to avoid the conserved region and to amplify products of ranging from 100 to 450 bp long in length. Primer sequences are shown in detail in Table S3. Amplification was performed on an iCycler iQ™ multicolor real-time PCR detection system (Bio-Rad, hercules, USA) and the analysis of each type of sample was repeated three times. The analysis of relative mRNA expression data was performed using the \(2^{{ - \varDelta \varDelta C_{\text{t}} }}\) method (Livak and Schmittgen 2001). Each expression profile was independently verified in 3 replicate experiments performed under identical conditions. Heat map representation was performed using centring and normalized ΔC t value, with Cluster 3.0 software and JavaTreeview to visualize the dendrogram.
Results
Identification and sequence conservation of tomato R2R3MYB genes
To identify putative R2R3MYB proteins in tomato, we performed a BLASTP search against tomato genome database (http://mips.helmholtzmuenchen.de/plant/tomato/searchjsp/blast.jsp) using one hundred and twenty-six R2R3MYB protein sequences in Arabidopsis and Hidden Markov Model (HMM) profile of the MYB-binding domain as queries. By removing the redundant ones, a total of 135 genes in the tomato genome were identified as putative members of the SlR2R3MYB family. Finally, 121 typical R2R3MYB genes were confirmed by Pfam and SMART program. To check the reliability of the 121 indentified R2R3MYB genes, the coding sequences of the 121 R2R3MYB genes were searched against tomato genome (http://blast.ncbi.nlm.nih.gov) using TBLASTX. Of all the 121 members of the R2R3MYB family, 119 were found to have the corresponding full-length cDNA clones, two R2R3MYB genes, Solyc06g009410.1.1 and Solyc05g009230.1.1 were found to have truncated coding sequences. To better reflect the proper orthologous relationship between SlR2R3MYBs and AtR2R3MYBs, we named each SlR2R3MYBs based on their phylogenies and sequence similarity corresponding to individual AtR2R3MYB. Finally, the 121 typical R2R3MYB genes (named SlMYB0 to SlMYB120) were subjected to further analysis (Table 1).
To investigate the homologous domain sequence features, and the frequency of the most prevalent amino acids at each position within each repeat of the tomato R2R3MYB domain, sequence logos were produced through multiple alignment analysis using the 121 homologous domain amino acid sequences of R2 and R3 repeats, respectively. As shown in Fig. 1, ten and seven conserved amino acid residues were identical among the members detected in the R2 and R3 MYB repeat regions, respectively. Within the 121 tomato R2R3MYB proteins, all the R2 repeat sequences contained three tryptophan residues. However, in the R3 repeats, the first tryptophan residue was replaced by phenylalanine. The second tryptophan residue was conserved in all the members and the third was mainly conserved. These results were consistent with those from Arabidopsis (Stracke et al. 2001), poplar (Wilkins et al. 2009), Triticum (Zhang et al. 2012) and cucumber (Li et al. 2012a).
The R2 and R3 MYB repeats are highly conserved across all SlR2R3MYB proteins. The sequence logos of the R2 (top) and R3 (bottom) MYB repeats are based on conserved alignments of all SlR2R3MYB proteins. The overall height of each stack indicates the conservation of the sequence at that position, whereas the height of letters within each stack represents the relative frequency of the corresponding amino acid. The asterisks indicates the position of the conserved amino acid that is identical among all the 121 tomato R2R3MYB proteins
Phylogenetic analysis of the tomato R2R3MYB family
An un-rooted neighbor-joining (NJ) phylogenetic tree was generated based on the alignment of the corresponding tomato R2R3MYB complete protein sequences (Fig. 2). For statistical reliability, we conducted bootstrap analysis with 1,000 replicates. The 121 members of the SlR2R3MYB family were subdivided into 29 subgroups, designated S1–S29, according to clades with at least 50 % bootstrap support. In this phylogenetic tree, forty-three gene pairs were formed with strong bootstrap (≥70 %) support. Sequence alignment and phylogenetic tree analysis on the basis of tomato R2R3MYB conserved domain were also produced, similar subgroups were obtained, though the classifications of only a few members varied (Fig. 2, Fig. S1). This indicated that the conserved R2R3MYB domain was an important unit in SlR2R3MYB protein and the dramatic divergence of the C-terminal regions did not appear to have a large influence on the regulatory function of the corresponding proteins (Dias et al. 2003).
Neighbor-joining (NJ) phylogenetic tree and intron–exon structures of SlR2R3MYB family genes. The unrooted phylogenetic tree (the part of left side) from the complete protein sequence was depicted by the MEGA 4.0 program with the NJ method. The tree shows the 29 phylogenetic subgroups (S1–S29) with high bootstrap value. The bootstrap values lower than 50 are not shown in the phylogenetic tree. All of 121 gene’s intron–exon structures are described in the middle part. Exons and introns are indicated by green boxes and single lines, respectively. Intron phases 0, 1 and 2 are indicated by numbers 0, 1 and 2, respectively. The length of each SlR2R3MYB gene can be estimated using the scale at the bottom. The exon number and length of each gene are listed in the table at right (color figure online)
To uncover the evolutionary relationship of the R2R3MYB genes, an un-rooted NJ phylogenetic tree was built from alignments of the R2R3MYB complete protein sequences from 8 species: tomato (121), Arabidopsis (126) (Dubos et al. 2010), grape (117) (Matus et al. 2008), rice (102) (Chen et al. 2006; Li et al. 2012a), poplar (197) (Wilkins et al. 2009), soybean (244) (Du et al. 2012), cucumber (47) (Li et al. 2012a; this study) and apple (222) (Cao et al. 2013) (Fig. 3, Fig. S2). The phylogeny was very similar to a previously published phylogeny that included all known Arabidopsis, grape, poplar, soybean and cucumber R2R3MYB proteins (Dubos et al. 2010; Matus et al. 2008; Wilkins et al. 2009; Du et al. 2012; Li et al. 2012a).
Phylogenetic relationships and subgroup designations in R2R3MYB proteins from tomato (Sl), cucumber (Cs), Arabidopsis (At), grape (Vv), rice (Os), poplar (Pt) soybean (Gm) and apple (Md). The neighbor-joining tree includes 121 R2R3MYB proteins from tomato, 47 R2R3MYB proteins from cucumber, 126 from Arabidopsis, 117 from Vitis, 102 from rice, 197 from poplar, 244 from soybean and 222 from apple. The bootstrap values lower than 50 are not shown in the phylogenetic tree. The proteins are clustered into 130 subgroups (triangles), designated with a subgroup number (e.g. C1). Eighty-six proteins did not fit well into subgroups (lines). The membership of each subgroup is described in the table at right. Several subgroups are highlighted. 9 subgroups (yellow) are shared in all the 8 species. 10 subgroups (red) are shared only in tomato. The uncompressed tree with full taxa names is available as Figure S2 (color figure online)
The resulting tree generated 130 groupings (triangles), which were designated with a subgroup number (C1–C130). However, 86 proteins did not fit well into any subgroup (lines) (Fig. 3, Fig. S2). The 86 proteins were considered orphans, most likely representing highly diverged lineage-specific R2R3MYB protein sequences. Phylogenetic analysis of the predicted R2R3MYB protein sequences revealed that there was not equal representation of these 8 species within the given subgroups (Fig. 3, Fig. S2). Nine (C1, 2, 7, 28, 34, 39, 63, 122 and 126) were shared in all the 8 species. Eighty-four subgroups (C4, 5, 9, 10, 12–18, 22–24, 26, 27, 29–33, 35, 36, 38, 41, 44–47, 49–53, 55–60, 64–66, 68, 69, 71, 73–78, 80–85, 87–91, 95, 96, 99, 100, 103–106, 108–110, 113, 115–118, 120, 121, 127, 128 and 130) were absent in the tomato genome. Meanwhile, some species-specific subgroups were also found, For example, 13 subgroups (C29, 35, 45, 51, 60, 65, 74, 76, 103, 110, 113, 118 and 120) only contained Arabidopsis members, 4 subgroups (C5, 27, 68 and 116) only grape members, 14 subgroups (C4, 9, 14, 38, 44, 49, 52, 59, 71, 87, 91, 99, 100, 105) only rice members, 3 subgroup (C17, 66 and 75) only poplar members, 8 subgroups (C30, 36, 50, 55, 69, 73, 83 and 57) only soybean members, 5 subgroups (C31, 47, 77, 90 and 96) only apple members and 10 subgroups (C19, 21, 54, 61, 67, 79, 93, 98, 102 and 119) only tomato members. Interestingly, in 4 subgroups (C94, 97, 107 and 124) only cucumber member was absent, and in 2 subgroups (C40 and C111), only rice member was absent.
A phylogenetic tree combining tomato, Arabidopsis, grape, rice, poplar, soybean, cucumber and apple R2R3MYB proteins would not only help to understand the phylogenetic relationships among the R2R3MYB proteins from these 8 species, but would also allow speculation on the putative functions of the tomato R2R3MYB proteins based on the functional clades that are identified in Arabidopsis. So, we summarized SlR2R3MYB proteins of known function and predicted the possible biological roles of the SlR2R3MYB proteins of unknown function based on the experimentally characterized AtR2R3MYBs (Table S1).
In conclusion, although evolutionary relationship could not be clearly deciphered for all families, the analysis did yield some interesting results.
Intron–exon structure of the tomato R2R3MYB family
According to the results of intron–exon structure analysis (Table 2), within the 121 SlR2R3MYB genes, the number of exons ranged from one to twelve and about 95.9 % (116 out of 121) had more than one exon. As shown in Fig. 4a, exon 1 and exon 2 coded for almost the entire R2R3 DNA binding domain, although this pattern differ in complex multiexonic genes (e.g. SlMYB88 and 110), appeared to be more restricted in length, while exon 3 was more variable (101–1,407 bp). Exon 3 coded for the last region of the R3 repeat and for the C-terminal of the protein. Changes both in length and sequence of this exon could have generated functional divergence between MYB homologues within and between species, leading to different functional motifs and domains (Dias et al. 2003; Matus et al. 2008). The presence of fifth to twelfth exon was unique to some specific genes. Despite this variability, the lengths of the first two exons were very similar (exon 1,133 bp; exon 2,130 bp) and highly conserved (exon 1, 40.5 % occurrence; exon 2, 68.6 % occurrence). Although exon 3 was the most diverse in size, R2R3MYB families from tomato, Arabidopsis (Matus et al. 2008), grape (Matus et al. 2008), soybean (Du et al. 2012) and cucumber (Li et al. 2012a) species were similarly distributed when the first three exon lengths were considered (Fig. 4b).
Exon length distribution analysis of the tomato R2R3MYB genes. a Exon length (Exon 1–Exon 5) values were analyzed using Box plot depicted by SigmaPlot 10.0. Each box represents the exon size range in which 50 % of the values for a particular exon are grouped. The mean value is shown as a dotted line (red) and the median as a continuous line. b First, second and third exon lengths distribution of tomato R2R3MYB genes using 3D Scatter Plot depicted by SigmaPlot 10.0 (color figure online)
To gain further insights into the SlR2R3MYB gene structures, the number of introns contained in their R2 and R3 domains was determined. All 121 genes, according to 20 relative positions and 3 phases, could be arranged into 14 (A–N) different splicing patterns (Fig. 5). Patterns A–C, composed of one or two intron(s) distributed at two highly conserved specific positions (indicated by white inverted triangles), accounted for approximately 72.7 % of SlR2R3MYB genes. Approximately 5 % of these 121 genes (patterns N) had no introns at the MYB binding domain. The other patterns had introns at varying positions in the R2 or R3 domain. It was interesting that the two genes (SlR2R3MYB118 and 119) from pattern L–M had three introns at R2 and two introns at R3 domain.
Intron distribution patterns of 121 tomato R2R3MYB genes. Red, blue and green rectangles represented the R2 domain, R3 domain and five amino acids between the R2 and R3 domain, respectively. The intron splicing patterns were designated A–N, according to relative positions and phases within the MYB binding domains of the SlR2R3MYB proteins. The white triangles are used when the position of the intron coincides with the intron 1 and intron 2 in example. The black triangles indicate that the location of the intron within the MYB binding domain corresponding to the example (line) is different from the intron 1 and intron 2. The numbers above the triangles indicate the splicing phases, 0, 1, 2 refers to phase 0, 1, 2. The markers 1–20 beside the triangles show different positions of the introns. The number of tomato R2R3MYB proteins with each pattern is given at right. Here the position of introns in the variable region has been adjusted manually to make them more contracted (color figure online)
Figure 5 also show that the introns 1 and 2 in the two conserved positions (indicated by white inverted triangles) have phases 1 and 2, respectively. The other introns, with less conserved positions (black inverted triangles), are in phases 0, 1 or 2. Among the 210 introns analyzed here, approximately 43.8 % (92 out of 210) had phase 1 and 49.5 % (104 out 210) had phase 2, whereas only 6.7 % had phases 0.
Genome distribution and gene duplication of tomato R2R3MYB genes
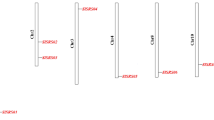
To determine the genomic distribution of the SlR2R3MYB genes, the DNA sequence of each SlR2R3MYB gene was used to search the tomato genome database using BLASTN. All of 121 R2R3MYB genes could be mapped on chromosomes 1–12 (Table 1; Fig. 6). Although each of the twelve tomato chromosomes contained some SlR2R3MYB genes, the distribution seemed to be uneven and the majorities are clustered at the chromosomal ends (Fig. 6). The largest number of R2R3MYB genes was found on chromosome 6 (sixteen genes), followed by chromosome 10 (fifteen genes), chromosome 5 (thirteen genes), chromosome 4 (eleven genes), chromosomes 1 and 2 (ten genes). Nine genes were distributed on each of chromosomes 3 and 7, and eight genes on each of chromosomes 8 and 12. Only five genes were located on chromosome 11. Relatively high densities of SlR2R3MYB genes were found in some chromosomal regions, such as the top arm of chromosome 5, 6, 9 and 10, and the bottom arm of chromosome 1, 2, 3, 4. In contrast, several large chromosomal regions lacked SlR2R3MYB genes, such as the top of chromosome 2 and the central sections of chromosome 1, 4, 5, 7, 8 and 11.
Chromosomal locations and predicted cluster for SlR2R3MYB genes. Chromosomal positions of the SlR2R3MYB genes are indicated by SlR2R3MYB number. The scale is in megabases (Mb). The numbers below the name of the chromosome show the number of SlR2R3MYB genes in this chromosome. Tandemly duplicated gene clusters are indicated in different colors
We further determined the tandem duplications of SlR2R3MYB genes along the 12 tomato chromosomes. According to the description reported by (Huang et al. 2012), the tandemly duplicated genes were defined as an array of two or more homologous genes within a range of 100-kb distance. As shown in Fig. 6, 24 SlR2R3MYB gene clusters (genes labeled by different color background) containing 63 tandemly duplicated genes were identified on each chromosome without chromosome 1. Totally, more than 50 % SlR2R3MYB genes on chromosome two (7 out of 10), five (8 out of 13), six (10 out of 16), seven (7 out of 9), nine (4 out of 7) and ten (10 out of 15) are tandemly duplicated, indicating that the high density of SlR2R3MYB genes on these chromosomes are partially due to the tandem gene duplication events. Similar results were also found in the tomato WRKY (Huang et al. 2012), SUN, OFP and YABBY (Huang et al. 2013) families.
Expression profiles for tomato R2R3MYB genes in different tissues and under different abiotic conditions
According to the Arabidopsis functional clades where SlR2R3MYB genes were in the phylogenetic tree (Fig. 3, Fig. S2, Table S1), we selected 51 putative tomato R2R3MYB genes responding to abiotic stress to gain insights into their role in tomato growth and development, and in abiotic stress tolerance. Their expression patterns in different tissues and three abiotic treatments were detected using real-time quantitative RT-PCR. The expression profiles of the 51 tomato R2R3MYB genes showed different patterns of tissue-specific expression (Fig. 7, Fig. S3). 44 out of 51 SlR2R3MYBs were expressed in all tissues tested, including both vegetative and floral tissues, although the transcript abundance of some genes in specific tissues was very low. However, some SlR2R3MYBs showed organ/tissue-specific expression patterns in tomato (Fig. 7; Fig. S3). For example, SlMYB16 showed high levels of transcript abundance in stems, leaves, flowers and immature green fruits but low levels in the roots. The transcript abundances of SlMYB2, SlMYB20, SlMYB29, SlMYB43, SlMYB45, SlMYB53, SlMYB58, SlMYB73, SlMYB74, SlMYB79, SlMYB92, SlMYB93 and SlMYB102 were higher in the roots than any other tissues. The expression of SlMYB3, 7, 21, 77, 112, 113, 115 and 116 were higher in the flowers than any other tissues. For the expression of other genes, two were restricted to vegetative tissues: SlMYB23 and SlMYB117, two were limited to reproductive tissues: SlMYB3 and SlMYB62.
Heatmap showing the expression of SlR2R3MYB genes in different tissues. Quantitative RT-PCR was used to assess SlR2R3MYB transcript accumulation in total RNA samples extracted from seedling, roots (R), stem (S), leaf (L), flowers (Fl), different development fruits: immature green (IM), mature green (MG), breaker (Br), red-ripe (7 days after breaker stage) (RR). Values represent the best experiment among three independent biological replicates. Genes highly or weakly expressed in the tissues are colored red and green, respectively. Genes without expression are colored gray. The heat map was generated using cluster 3.0 software (color figure online)
The expression of most SlR2R3MYB genes could also be detected in different developmental stages of the tomato fruits (Fig. 7, Fig. S3). During tomato fruit development, four genes, including SlMYB1, 32, 69 and 70 were down-regulated, and three genes, including SlMYB49, 71 and 78 were up-regulated along with the fruit development. In addition, three genes (SlMYB16, 30, 44) showed a similar expression pattern, with a lower expression at breaker stage.
In this study, the transcript abundances of the 51 SlR2R3MYB genes were investigated under NaCl (100 mM), low temperature (4 °C) and ABA (100 μM) treatments at the three-true-leaves stage, respectively. The results indicated that 19 (~37.3 %) genes responded to at least one treatment, which included 10 genes responding to NaCl treatment, 9 genes to ABA and 14 genes to low temperature (Fig. 8, Fig. S4, Fig. S5, Fig. S6). Among these genes, 6 genes (SlMYB11, 23, 28, 41, 49 and 116) were able to respond to two treatments and 3 (SlMYB62, 74 and 102) genes to all three treatments. The rest 10 genes only responded to a single treatment. The expression of 8, 7 and 10 genes were induced by NaCl, ABA and low temperature treatment, respectively, whereas 2, 2 and 4 genes were repressed, respectively (Fig. 8, Fig. S4, Fig. S5, Fig. S6). Interestingly, some genes behaved in an opposite manner to their expression profile when subjected to different treatments. For example, SlMYB64 was induced by high salinity but was repressed by low temperature, and SlMYB28 was induced by high salinity and ABA, respectively, but was repressed by low temperature.
Heatmap showing the expression patterns of tomato R2R3MYB genes under three abiotic stresses. Quantitative RT-PCR was used to assess SlR2R3MYB transcript accumulation in total RNA samples extracted from three-true-leave seedlings with three abiotic stresses. Of 51 selected R2R3MYB genes, 10 genes responded to NaCl (100 mM) treatment, 9 genes to ABA (100 μM) and 14 genes to low temperature (4 °C). Values represent the best experiment among three independent biological replicates. Genes highly or weakly expressed in the tissues are colored red and green, respectively. Genes without expression are colored gray. The heat map was generated using cluster 3.0 software (color figure online)
Discussion
Characterization of the tomato R2R3MYB family
R2R3MYBs constitute the most abundant group of transcription factors described in plants (Dias et al. 2003; Wilkins et al. 2009). The MYB transcription factors have been implicated in multiple biological processes in plants, especially in regulating defense against biotic and abiotic stresses (Dubos et al. 2010). However, very little information is available about the R2R3MYBs in tomato. Complete and accurate annotation of genes is an essential starting point for further evolution and function study in gene family. This study identified and characterized 121 tomato R2R3MYB genes based on the published tomato annotated genes in tomato genome through genome-wide analysis. Moreover, the genome of the inbred tomato cultivar ‘Heinz 1706’ was sequenced and assembled using a combination of Sanger and ‘next generation’ technologies. Base accuracy is approximately one substitution error per 29.4 kilobases (kb) and one indel error per 6.4 kb. The scaffolds were linked with two bacterial artificial chromosome (BAC)-based physical maps and anchored/oriented using a high-density genetic map, introgression line mapping and BAC fluorescence in situ hybridization (FISH) (The Tomato Genome Consortium 2012). Therefore, the number of SlR2R3MYB family was not the result of inadequate depth of genome coverage and may be complete.
Gene duplication played an important role in evolution (Ohno 1970). The plant genomes have experienced comprehensive sequence diversification (Bowers et al. 2005), such as small fragment insertions, deletions, inversions, translocations, duplication (Navajas-Pérez and Paterson 2009), chromosomal rearrangement and fusion (Simillion et al. 2004) from an ancient-whole genome duplication event. These processes eventually led to species diversification (Ohno 1970). The recent gene duplication events were the most important for the rapid expansion and evolution of gene families (Cannon et al. 2004; Ling et al. 2011). Arabidopsis (as well as rice and poplar) underwent the recent duplication events, which led to the large-scale expansion of the R2R3MYB family in their genome (Cannon et al. 2004; Taylor and Raes 2004). Song et al. (2012) used tomato whole genome to detect the WGD (Whole Genome Duplication) event during the long evolutionary history of Solanaceae and two WGD events had been detected after the divergence of rice–Arabidopsis (monocot–dicot). The method utilized by (Huang et al. 2012) was used to detect whether recent small duplication blocks occurred in the SlR2R3MYB family. About 52 % (63/121) SlR2R3MYB genes were found to evolve from tandem gene duplication (Fig. 6), indicating that tandem gene duplication probably played a pivotal role in R2R3MYB gene expansion in tomato genome. Moreover, eight of the R2R3MYB gene pairs in tomato were genetically linked to each other on their corresponding chromosomal locations, which indicated the existence of recent tandem duplication event in SlR2R3MYB genes. In addition, the tomato genome contained the relatively large gene family size compared to grape, rice and cucumber. This may explain, in part, to the large number of genes found in tomato.
Phylogenetic analysis and evolution of tomato R2R3MYB genes
The evolutionary relationship of this gene family within and among the different species has been systematically studied, which divided the 1175 R2R3MYBs into 130 clades (Fig. 3) and the 121 SlR2R3MYB members into 29 clades (Fig. 2). There are quantitative differences in some clades among these 8 species, such as the subgroup C126 which included 4 CsR2R3MYB, 6 AtR2R3MYB, 4 VvR2R3MYB, 7 OsR2R3MYB, 8 PtR2R3MYB, 15 GmR2R3MYB and 6 MdR2R3MYB proteins. This indicated that C126 is an expanded subgroup in tomato compared with the cucumber, Arabidopsis, grape, rice, poplar and apple R2R3MYB families except soybean. In addition, the gene loss and lineage-specific expansions were likely to be accounted for by genomic drift (Nozawa et al. 2007), so it is possible that some clades could have expanded differently in the R2R3MYB families of these 8 species.
The phylogenetic analysis and expression studies can serve as predictors of possible gene function in tomato. Similarities or differences in the expression patterns between tomato and Arabidopsis orthologs can point to conservation or diversification of gene function. Eighty-four clades did not include any tomato R2R3MYB, which suggested that these clades were either lost in tomato or were acquired after divergence from the last common ancestor (Fig. 3). For example, three genes in subclade C110, AtMYB0 (GL1), AtMYB23, AtMYB66 (WER), are known to be involved in epidermis cell-fate determination in Arabidopsis (Bloomer et al. 2012; Kang et al. 2009; Tominaga-Wada et al. 2012). In tomato, no subgroup C110 genes were observed, which indicated that the possible gene loss and/or lineage-specific expansions may reflect species-specific adaptations (Nozawa et al. 2007). The possible reason could be that multi-cellular trichomes in tomato (as well as Cucumis sativus) develop through a transcriptional regulatory network that differs from those regulating unicellular trichome formation in Arabidopsis (and perhaps cotton) (Serna and Martin 2006; Yang et al. 2011). The AtMYB75, 90, 113 and 114 genes in subgroup C65 play a role in the regulation of anthocyanin biosynthesis (Gonzalez et al. 2007; Borevitz et al. 2000), but none of the tomato R2R3MYB genes was grouped into C65. So, it would be interesting to characterize the anthocyanin-related R2R3MYB genes in tomato.
Some tomato R2R3MYB proteins were clustered into Arabidopsis functional clades (Fig. S2). This provided an excellent reference to explore the functions of the tomato R2R3MYB genes. For instance, SlMYB104 and SlMYB120 shared a high level of sequence similarity with male gamete cell formation protein AtMYB125 (DUO1). This implied that the possible functions of SlMYB104 and 120 were related to male gamete cell division and differentiation (Brownfield et al. 2009). SlMYB16, 23, 25, 64 and 106 were grouped into clade 34 with two Arabidopsis proteins, AtMYB16 (MIXTA), proposed to control the shape of petal epidermal cells (Baumann et al. 2007) and AtMYB106 (NOK), a negative regulator of trichome branching (Jakoby et al. 2008). This represented a functional clade containing proteins responsible for cell development or morphogenesis.
Four subgroups (C94, 97, 107 and 124) did not include any cucumber R2R3MYB proteins but only members from tomato, Arabidopsis, grape, rice, poplar, soybean and apple (Fig. 3, Fig. S2), suggesting that the genes in these clades may have been lost in cucumber during the evolutionary process. This could also explain why two subgroups (C40, and 111) were absent in the rice genome but not in tomato, cucumber, Arabidopsis, grape, poplar, soybean and apple (Fig. 3, Fig. S2). Furthermore, the species-specific subgroups were also observed (Fig. 3, Fig. S2), suggesting that R2R3MYB genes may have evolved in one species, or been lost in other species, following divergence of these 8 species from the last common ancestor.
In addition, 9 R2R3MYB genes (SlMYB10, 13, 34, 66, 76, 82, 84, 90 and 101) did not fit well into any of the clades, implying that these MYB genes may have specialized roles that were acquired in tomato during the process of tomato genome evolution (Fig. 3; Fig. S2). Our expression analysis revealed that tomato R2R3MYBs had a variety of expression patterns in different tissues (Fig. 7; Fig. S3). Therefore, we believe that these genes may regulate essential biological processes during tomato development.
Usually, the pattern of intron positioning provided an important evidence for evolutionary relationships. Previous studies demonstrated that the intron–exon structure was conserved within the same subgroup, but differed between subgroups in the MYB gene family in Arabidopsis, rice (Jiang et al. 2004) and soybean (Du et al. 2012). Unexpectedly, among the 29 subgroups, SlR2R3MYB genes in twelve subgroups (S1, 2, 3, 4, 10, 15, 16, 18, 21, 22, 23, 26 and 27) did not always show similar intron–exon structures in the same subgroup. In addition, intron–exon structure was not conserved, even in the same sister pair (SlMYB22 and 27; SlMYB5 and 82; SlMYB94 and 96) (Fig. 2). As previously observed in cucumber (Li et al. 2012a), Arabidopsis, grape (Matus et al. 2008) and soybean (Du et al. 2012) R2R3MYB genes, the modal lengths of the first two exons were very similar (exon 1, 133 bp; exon 2, 130 bp) and highly conserved. The exon length of the SlR2R3MYB family was also investigated and the results showed that the first two exon lengths were very similar to Arabidopsis, grape and soybean, which suggested that MYB binding domains could be partially conserved because exons coding for this domain have all evolved with restricted lengths (Fig. 4).
Spatio-temporal expression of tomato R2R3MYB genes and their responses to abiotic conditions
The SlR2R3MYB genes in various tissues and fruit developmental stages with organ/tissue-specific or same expression patterns suggested that the encoded proteins may perform specific or redundant functions. The high expression divergence in different tissues reflected the complexity of gene family functions (Audran-Delalande et al. 2012). For example, the eight genes are expressed in flowers with high expression levels than the other tissues, indicating that these tomato R2R3MYB genes are probably involved in the development of reproductive organs. Actually, many SlR2R3MYB genes were found to be differentially expressed during fruit development, indicating that SlR2R3MYB gene family might play an important role in the fruit development and/or ripening of tomato. Overall, the tissue-preferential expression displayed by some SlR2R3MYB genes could be indicative of their involvement in specific plant tissues and various aspects of physiological and developmental processes.
Amounts of R2R3MYB proteins have been characterized by genetic analysis and have been found to occur in responses to various abiotic stresses (Dubos et al. 2010; Zhang et al. 2012). However, relatively few R2R3MYB family genes have been shown to respond to abiotic conditions in tomato. To date, only one tomato R2R3MYB gene, AIM1, has been functionally characterized to respond to biotic and abiotic stresses (Abuqamar et al. 2009) (Table S1). For this reason, the expression patterns of tomato R2R3MYB genes were investigated under NaCl (100 mM), low temperature (4 °C) and ABA (100 μM) treatment, respectively. Out of 51 candidate genes, we identified 19 (~37.3 %) genes responded to at least one treatment of NaCl, ABA and low temperature (Fig. 8, Fig. S4, Fig. S5, Fig. S6). Interestingly, some genes, such as SlMYB28 and SlMYB64, showed opposing expression patterns under different stress conditions (Fig. 8, Fig. S4, Fig. S5, Fig. S6), this also occurred in cucumber R2R3MYB genes. These results indicated that they played a major role in the plant responses to abiotic conditions and involved in communication between different signal transduction pathways.
In conclusion, we identified a total of 121 R2R3MYB genes grouped into 29 clades in the tomato genome and found a number of tandem duplications which contributed to the expansion of this superfamily in tomato. The structural characteristics and the comparison of the phylogenetic relationships among SlR2R3MYBs will provide insight into the identification and comprehensive functional characterization of the R2R3MYB gene family from tomato and other species. Expression analysis revealed that SlR2R3MYB genes in tomato had different expression patterns in various tissues, multiple developmental stages of fruits and abiotic conditions. A number of SlR2R3MYB genes were discovered to be involved in the development of flowers and fruits in tomato. To further explore the functions of these SlR2R3MYBs might provide a subset of candidate target genes for gene engineering to improve agronomic traits and/or stress tolerance in the future.
References
Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15:63–78
Abuqamar S, Luo H, Laluk K, Mickelbart MV, Mengiste T (2009) Crosstalk between biotic and abiotic stress responses in tomato is mediated by the AIM1 transcription factor. Plant J 58:347–360
Audran-Delalande C, Bassa C, Mila I, Regad F, Zouine M, Bouzayen M (2012) Genome-wide identification, functional analysis and expression profiling of the Aux/IAA gene family in tomato. Plant Cell Physiol 53:659–672
Ballester AR, Molthoff J, de Vos R, Hekkert Bt, Orzaez D, Fernández-Moreno JP, Tripodi P, Grandillo S, Martin C, Heldens J, Ykema M, Granell A, Bovy A (2010) Biochemical and molecular analysis of pink tomatoes: deregulated expression of the gene encoding transcription factor SlMYB12 leads to pink tomato fruit color. Plant Physiol 152:71–84
Baumann K, Perez-Rodriguez M, Bradley D, Venail J, Bailey P, Jin H, Koes R, Roberts K, Martin C (2007) Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development 134:1691–1701
Bloomer RH, Juenger TE, Symonds VV (2012) Natural variation in GL1 and its effects on trichome density in Arabidopsis thaliana. Mol Ecol 21:3501–3515
Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12:2383–2394
Bowers JE, Arias MA, Asher R, Avise JA, Ball RT, Brewer GA, Buss RW, Chen AH, Edwards TM, Estill JC, Exum HE, Goff VH, Herrick KL, Steele CL, Karunakaran S, Lafayette GK, Lemke C, Marler BS, Masters SL, McMillan JM, Nelson LK, Newsome GA, Nwakanma CC, Odeh RN, Phelps CA, Rarick EA, Rogers CJ, Ryan SP, Slaughter KA, Soderlund CA, Tang H, Wing RA, Paterson AH (2005) Comparative physical mapping links conservation of microsynteny to chromosome structure and recombination in grasses. Proc Natl Acad Sci USA 102:13206–13211
Brownfield L, Hafidh S, Borg M, Sidorova A, Mori T, Twell D (2009) A plant germline-specific integrator of sperm specification and cell cycle progression. PLoS Genet 5:e1000430
Cannon SB, Mitra A, Baumgarten A, Young ND, May G (2004) The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 1:4–10
Cao ZH, Zhang SZ, Wang RK, Zhang RF, Hao YJ (2013) Genome wide analysis of the apple MYB transcription factor family allows the identification of MdoMYB121 gene conferring abiotic stress tolerance in plants. PLoS One 8:e69955
Carretero-Paulet L, Galstyan A, Roig-Villanova I, Martínez-García JF, Bilbao-Castro JR, Robertson DL (2010) Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol 153:1398–1412
Chen YH, Yang XY, He K, Liu MH, Li JG, Gao ZF, Lin ZQ, Zhang YF, Wang XX, Qiu XM, Shen YP, Zhang L, Deng XH, Luo JC, Deng XW, Chen ZL, Guo HY, Qu LJ (2006) The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol 60:107–124
Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14:1188–1190
Daniel X, Lacomme C, Morel JB, Roby D (1999) A novel myb oncogene homologue in Arabidopsis thaliana related to hypersensitive cell death. Plant J 20:57–66
Deluc L, Barrieu F, Marchive C, Lauvergeat V, Decendit A, Richard T, Carde JP, Merillon JM, Hamdi S (2006) Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol 140:499–511
Denekamp M, Smeekens SC (2003) Integration of wounding and osmotic stress signals determines the expression of the AtMYB102 transcription factor gene. Plant Physiol 132:1415–1423
Dias AP, Braun EL, McMullen MD, Grotewold E (2003) Recently duplicated maize R2R3 Myb genes provide evidence for distinct mechanisms of evolutionary divergence after duplication. Plant Physiol 131:610–620
Doebley J, Lukens L (1998) Transcriptional regulators and the evolution of plant form. Plant Cell 10:1075–1082
Du H, Yang SS, Feng BR, Liu L, Tang YX, Huang YB, Liang Z (2012) Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol 12:106
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15:573–581
Gong X, Bewley DJ (2008) A GAMYB-like gene in tomato and its expression during seed germination. Planta 228:563–572
Gonzalez A, Zhao M, Leavitt JM, Lloyd AM (2007) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53:814–827
Guo AY, Zhu QH, Chen X, Luo JC (2007) GSDS: a gene structure display server. Yi Chuan 29:1023–1026
Hemm MR, Herrmann KM, Chapple C (2001) AtMYB4: a transcription factor general in the battle against UV. Trends Plant Sci 6:135–136
Higginson T, Li SF, Parish RW (2003) AtMYB103 regulates tapetum and trichome development in Arabidopsis thaliana. Plant J 35:177–192
Huang S, Gao Y, Liu J, Peng X, Niu X, Fei Z, Cao S, Liu Y (2012) Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol Genet Genomics 287:495–513
Huang Z, Van Houten J, Gonzalez G, Xiao H, van der Knaap E (2013) Genome-wide identification, phylogeny and expression analysis of SUN, OFP and YABBY gene family in tomato. Mol Genet Genomics 288:111–129
Jakoby MJ, Falkenhan D, Mader MT, Brininstool G, Wischnitzki E, Platz N, Hudson A, Hülskamp M, Larkin J, Schnittger A (2008) Transcriptional profiling of mature Arabidopsis trichomes reveals that NOECK encodes the MIXTA like transcriptional regulator MYB106. Plant Physiol 148:1583–1602
Jiang CZ, Gu X, Peterson T (2004) Identification of conserved gene structures and carboxy-terminal motifs in the Myb gene family of Arabidopsis and Oryza sativa L. ssp. Genome Biol 5:R46
Jin H, Martin H (1999) Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol 41:577–585
Kang YH, Kirik V, Hulskamp M, Nam KH, Hagely K, Lee MM, Schiefelbein J (2009) The MYB23 gene provides a positive feedback loop for cell fate specification in the Arabidopsis root epidermis. Plant Cell 21:1080–1094
Keller T, Abbott J, Moritz T, Doerner P (2006) Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell 18:598–611
Kellogg EA (2004) Evolution of developmental traits. Curr Opin Plant Biol 7:92–98
Letunic I, Doerks T, Bork P (2009) SMART 6: recent updates and new developments. Nucleic Acids Res 37:D229–D232
Li X, Duan X, Jiang H, Sun Y, Tang Y, Yuan Z, Guo J, Liang W, Chen L, Yin J, Ma H, Wang J, Zhang D (2006) Genome-wide analysis of basic/helix–loop–helix transcription factor family in rice and Arabidopsis. Plant Physiol 141:1167–1184
Li Q, Zhang C, Li J, Wang L, Ren Z (2012a) Genome-wide identification and characterization of R2R3MYB family in Cucumis sativus. PLoS One 7:e47576
Li YY, Mao K, Zhao C, Zhao XY, Zhang HL, Shu HR, Hao YJ (2012b) MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol 160:1011–1022
Li Y, Wen C, Weng Y (2013) Fine mapping of the pleiotropic locus B for black spine and orange mature fruit color in cucumber identifies a 50 kb region containing a R2R3-MYB transcription factor. Theor Appl Genet 126:2187–2196
Lin Q, Hamilton WD, Merryweather A (1996) Cloning and initial characterization of 14 myb2 related cDNAs from tomato (Lycopersicon esculentum cv. Ailsa Craig). Plant Mol Biol 30:1009–1020
Ling J, Jiang W, Zhang Y, Yu H, Mao Z, Gu X, Huang S, Xie B (2011) Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genom 12:471
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔ Ct method. Methods 25:402–408
Mathews H, Clendennen SK, Caldwell CG, Liu XL, Connors K, Matheis N, Schuster DK, Menasco DJ, Wagoner W, Lightner J, Wagner DR (2003) Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 15:1689–1703
Matus JT, Aquea F, Arce-Johnson P (2008) Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biol 8:83
Mehrtens F, Kranz H, Bedn arek P, Weisshaar B (2005) The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenyl-propanoid biosynthesis. Plant Physiol 138:1083–1096
Millar AA, Gubler F (2005) The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17:705–721
Müller D, Schmitz G, Theres K (2006) Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 18:586–597
Nagaoka S, Takano T (2003) Salt tolerance-related protein STO binds to a Myb transcription factor homologue and confers salt tolerance in Arabidopsis. J Exp Bot 54:2231–2237
Navajas-Pérez R, Paterson AH (2009) Patterns of tandem repetition in plant whole genome assemblies. Mol Genet Genomics 281:579–590
Naz AA, Raman S, Martinez CC, Sinha NR, Schmitz G, Theres K (2013) Trifoliate encodes an MYB transcription factor that modulates leaf and shoot architecture in tomato. Proc Natl Acad Sci USA 110:2401–2406
Nozawa M, Kawahara Y, Masatoshi Nei M (2007) Genomic drift and copy number variation of sensory receptor genes in humans. Proc Natl Acad Sci USA 104:20421–20426
Ogata K, Morikawa S, Nakamura H, Sekikawa A, Inoue T, Kanai H, Sarai A, Ishii S, Nishimura Y (1994) Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell 79:639–648
Ohno S (1970) Evolution by gene duplication. Springer, New York
Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD (1991) A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67:483–493
Perez-Rodriguez M, Jaffe FW, Butelli E, Glover BJ, Martin C (2005) Development of three different cell types is associated with the activity of a specific MYB transcription factor in the ventral petal of Antirrhinum majus flowers. Development 132:359–370
Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, Pilgrim M, Broun P, Zhang JZ, Ghandehari D, Sherman BK, Yu G (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 2:2105–2110
Romero I, Fuertes A, Benito MJ, Malpica JM, Leyva A, Paz-Ares J (1998) More than 80 R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J 14:273–284
Schmitz G, Tillmann E, Carriero F, Fiore C, Cellini F, Theres K (2002) The tomato Blind gene encodes a MYB transcription factor that controls the formation of lateral meristems. Proc Natl Acad Sci USA 99:1064–1069
Serna L, Martin C (2006) Trichomes: different regulatory networks lead to convergent structures. Trends Plant Sci 11:274–280
Sharp PA (1981) Speculations on RNA splicing. Cell 23:643–646
Simillion C, Vandepoele K, Saeys Y, Van de Peer Y (2004) Building genomic profiles for uncovering segmental homology in the twilight zone. Genome Res 14:1095–1106
Song C, Guo J, Sun W, Wang Y (2012) Whole genome duplication of intra- and inter-chromosomes in the tomato genome. J Genet Genomics 39:361–368
Stockinger EJ, Mao Y, Regier MK, Triezenberg SJ, Thomashow MF (2001) Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res 29:1524–1533
Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4:447–456
Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B (2007) Differential regulation of closely related R2R3-MY B transcription factors controls flavonol accumulation indifferent parts of the Arabidopsis thaliana seedling. Plant J 50:660–677
Sugimoto K, Takeda S, Hirochika H (2000) MYB-related transcription factor NtMYB2 induced by wounding and elicitors is a regulator of the tobacco retrotransposon Tto1 and defense-related genes. Plant Cell 12:2511–2528
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Taylor JS, Raes J (2004) Duplication and divergence: the evolution of new genes and old ideas. Annu Rev Genet 38:615–643
Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S (2005) Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol 139:1840–1852
The Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485:635–641
Tominaga R, Iwata M, Okada K, Wada T (2007) Functional analysis of the epidermal specific MYB genes CAPRICE and WEREWOLF in Arabidopsis. Plant Cell 19:2264–2277
Tominaga-Wada R, Nukumizu Y, Sato S, Kato T, Tabata S, Wada T (2012) Functional divergence of MYB-related genes, WEREWOLF and AtMYB23 in Arabidopsis. Biosci Biotechnol Biochem 76:883–887
Tsiantis M, Hay A (2003) Comparative plant development: the time of the leaf. Nat Rev Genet 4:169–180
Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K (1993) An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5:1529–1539
Vailleau F, Daniel X, Tronchet M, Montillet JL, Triantaphylides C, Roby D (2002) A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc Natl Acad Sci USA 99:10179–10184
Wada T, Tachibana T, Shimura Y, Okada K (1997) Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277:1113–1116
Wang TT, Jongsma MA, Yu CY, Li HX, Zhang JH, Yang CX, Ye ZB (2011) Cloning and expression analysis of ShMYB1 related to the trichome initiation in tomato (Solanum habrochaites). J Agric Biotechnol 19:407–416
Wilkins O, Nahal H, Foong J, Provart NJ, Campbell MM (2009) Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol 149:981–993
Yang C, Li H, Zhang J, Luo Z, Gong P, Zhang C, Li J, Wang T, Zhang Y, Lu Y, Ye Z (2011) A regulatory gene induces trichome formation and embryo lethality in tomato. Proc Natl Acad Sci USA 108:11836–11841
Zhang L, Zhao G, Jia J, Liu X, Kong X (2012) Molecular characterization of 60 isolated wheat MYB genes and analysis of their expression during abiotic stress. J Exp Bot 63:203–214
Zhong S, Fei Z, Chen YR, Zheng Y, Huang M, Vrebalov J, McQuinn R, Gapper N, Liu B, Xiang J, Shao Y, Giovannoni JJ (2013) Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat Biotechnol 31:154–159
Acknowledgments
This work was supported by fundings from ‘Taishan Scholar’ Foundation of the People’s Government of Shandong Province, the National Natural Science Foundation of China (NSFC; 31222048 and 31171977), and the Program for Changjiang Scholars and Innovative Research Team in University (IRT1155).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhao, P., Li, Q., Li, J. et al. Genome-wide identification and characterization of R2R3MYB family in Solanum lycopersicum . Mol Genet Genomics 289, 1183–1207 (2014). https://doi.org/10.1007/s00438-014-0879-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-014-0879-4