Abstract
Key message
Two new TDZ derivatives (HETDZ and 3FMTDZ) are very potent inhibitors of CKX and are promising candidates for in vivo studies.
Abstract
Cytokinin hormones regulate a wide range of essential processes in plants. Thidiazuron (N-phenyl-N′-1,2,3-thiadiazol-5-yl urea, TDZ), formerly registered as a cotton defoliant, is a well known inhibitor of cytokinin oxidase/dehydrogenase (CKX), an enzyme catalyzing the degradation of cytokinins. TDZ thus increases the lifetime of cytokinins and their effects in plants. We used in silico modeling to design, synthesize and characterize twenty new TDZ derivatives with improved inhibitory properties. Two compounds, namely 1-[1,2,3]thiadiazol-5-yl-3-(3-trifluoromethoxy-phenyl)urea (3FMTDZ) and 1-[2-(2-hydroxyethyl)phenyl]-3-(1,2,3-thiadiazol-5-yl)urea (HETDZ), displayed up to 15-fold lower IC 50 values compared with TDZ for AtCKX2 from Arabidopsis thaliana and ZmCKX1 and ZmCKX4a from Zea mays. Binding modes of 3FMTDZ and HETDZ were analyzed by X-ray crystallography. Crystal structure complexes, solved at 2.0 Å resolution, revealed that HETDZ and 3FMTDZ bound differently in the active site of ZmCKX4a: the thiadiazolyl ring of 3FMTDZ was positioned over the isoalloxazine ring of FAD, whereas that of HETDZ had the opposite orientation, pointing toward the entrance of the active site. The compounds were further tested for cytokinin activity in several cytokinin bioassays. We suggest that the combination of simple synthesis, lowered cytokinin activity, and enhanced inhibitory effects on CKX isoforms, makes 3FMTDZ and HETDZ suitable candidates for in vivo studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
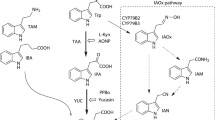
Cytokinins are hormones that influence a wide range of essential processes in plants, including cell division, shoot and root development, seed and fruit development, germination, senescence and response to environmental stresses (Mok and Mok 2001). They are adenine derivatives with a distinct N 6 side-chain. Nowadays, use of plant growth regulators with cytokinin-like activity is a common practice in agriculture and horticulture, and thus much effort has been devoted to the development of new compounds with improved cytokinin-like properties. Thidiazuron (TDZ) is one of the most widely used synthetic compounds in agriculture, showing a strong cytokinin effect based on a combination of two activities: TDZ possesses strong cytokinin activity (Mok et al. 1982; Yamada et al. 2001; Spíchal et al. 2004) and also inhibits the enzyme cytokinin oxidase/dehydrogenase (abbreviated as CKO or CKX, EC 1.5.99.12, Chatfield and Armstrong 1986; Hare and Van Staden 1994). CKX catalyzes the irreversible oxidative breakdown of cytokinins to form adenine/adenosine and the corresponding aldehyde (Whitty and Hall 1974; Brownlee et al. 1975; Chatfield and Armstrong 1986). Therefore, in addition to a direct effect at the receptor level, TDZ increases endogenous cytokinin concentrations in plants.
CKXs are encoded by small multigene families. The model plant Arabidopsis thaliana contains seven CKX isoforms, which are involved in the regulation of endogenous cytokinin levels (Werner et al. 2003), of which AtCKX2 is the most active and well-studied isoform (Galuszka et al. 2007). In contrast, the model plant Zea mays contains 13 CKX isoforms, which differ in substrate specificity, spatial and temporal expression patterns and subcellular localization (Massonneau et al. 2004; Vyroubalová et al. 2009; Šmehilová et al. 2009; Zalabák et al. 2014). Among these, ZmCKX1 is the best studied isoform, which has been shown to play a crucial role in cytokinin degradation in maize (Houba-Hérin et al. 1999; Morris et al. 1999). ZmCKX1 preferentially oxidizes cytokinin bases and ribosides (R), with N 6-(∆2-isopentenyl)adenine (iP) being the best substrate (Bilyeu et al. 2001; Kopečný et al. 2005). In contrast, other cytokinins, including cytokinin glucosides (G) and monophosphates (MP), have been reported to be poor substrates (Zalabák et al. 2014; Kopečný et al. 2015). ZmCKX2, 3, 4a, 4b and 5, expressed mainly in reproductive organs, have been shown to preferentially oxidize iPRMP and iP9G as substrates, followed by iP. Such a change in substrate specificity is caused by substitution of the Glu381 residue in ZmCKX1 by a serine, alanine or glycine residue in the other isoforms. Moreover, the crystal structures of ZmCKX2 and ZmCKX4a display a mobile domain, which is well defined in the ZmCKX1 structure (Kopečný et al. 2015). The domain includes two helices and a loop belonging to the substrate binding domain and likely contributes to binding of the N 9 substituted cytokinins. ZmCKX1, ZmCKX4a and AtCKX2 have been expressed in Yarrowia lipolytica, Escherichia coli and Saccharomyces cerevisiae (Kopečný et al. 2005; Zalabák et al. 2014; Frébortová et al. 2007), allowing production of large amounts of recombinant CKX.
To date, several groups of compounds are known to inhibit CKX. One such group includes urea compounds, such as diphenyl urea, TDZ and 1-phenyl-3-(2-chloro-pyridin-4-yl)urea (CPPU) (Chatfield and Armstrong 1986; Laloue and Fox 1989; Burch and Horgan 1989; Hare and van; Staden 1994). TDZ and CPPU are important promoters of growth of various horticultural crops, such as kiwi, melons, watermelons and grapes (Arima et al. 1995). TDZ has also been extensively used as a cotton defoliant in the USA (Arndt et al. 1976). A new series of CPPU derivatives were described recently and tested on ZmCKX1 (Kopečný et al. 2010). The two best compounds were found to display IC 50 values 25 times lower than that for TDZ. However, the synthesis of these compounds was complicated. Another group of compounds known to inhibit CKX is the suicide substrates N 6-(buta-2,3-dienyl)adenine and N 6-(penta-2,3-dienyl)adenine, which are strong CKX inhibitors with significant cytokinin activity (Suttle and Mornet 2005; Kopečný et al. 2008). Yet another group includes anilinopurine derivatives (Zatloukal et al. 2008). However, their synthesis includes the use of hydrogen fluoride, which is highly corrosive and toxic. The strongest inhibitor from anilinopurine group was successfully used to support in vitro organogenesis (Aremu et al. 2015) and to enhance plant stress tolerance toward salinity (Aremu et al. 2014) and cadmium (Gemrotová et al. 2013). Therefore, the development of new urea derivatives that are easy to synthesize and inhibit various CKX isoforms is desired for potential applications in horticulture, agriculture and biotechnology.
In the present study, we employed in silico drug design to identify new TDZ derivatives with improved CKX inhibitory properties. The designed inhibitors were synthesized in a two-step reaction by fusing 1,2,3-thiadiazol-5-isocyanate with phenylamine or benzylamine derivatives and the products were characterized. The binding properties of the TDZ derivatives obtained in silico were compared to IC 50 values (concentration at which the enzyme activity is inhibited by 50 %) determined in vitro using AtCKX2. A subset of these derivatives was further screened using ZmCKX1 and ZmCKX4a and binding modes were analyzed by X-ray crystallography using ZmCKX4a. TDZ is also known to strongly activate all three cytokinin receptors in A. thaliana: ARABIDOPSIS HISTIDINE KINASE2 (AHK2), AHK3, and CYTOKININ RESPONSE1/AHK4 as well as primary cytokinin-sensitive genes including the Arabidopsis Response Regulator 5 (ARR5; Spíchal et al. 2004; Stolz et al. 2011; Lomin et al. 2015). AHK genes are expressed in almost all cells of different organs although with different strength (Nishimura et al. 2004). While AHK2 shows a lower sensitivity in planta, apparent affinity constant (K D) values for major natural cytokinins and synthetic TDZ for all three receptors are in the low nanomolar range (Yamada et al. 2001; Romanov et al. 2006; Stolz et al. 2011). We were able to screen TDZ derivatives for binding to AHK3 and CRE1/AHK4 receptors and for their cytokinin activity in Arabidopsis using transgenic plants harboring the ARR5::GUS reporter gene (GUS = β-glucuronidase; D’Agostino et al. 2000). Finally, the cytokinin activity of the compounds was also assessed in three classical cytokinin bioassays, i.e., wheat leaf senescence, tobacco callus and Amaranthus bioassays.
Results and discussion
Active sites in ZmCKX1, ZmCKX4a and AtCKX2
The active site of ZmCKX1 is composed of two pockets connected by a narrow tunnel bordered by Asp169. The crystal structure in complex with CPPU (PDB ID: 2QKN, Kopečný et al. 2010) shows that Asp169 forms hydrogen bonds to both urea nitrogen atoms of CPPU. The smaller of the two pockets is occupied by the isoalloxazine ring of FAD, whereas the larger pocket is exposed to solvent and is guarded by Glu381. CPPU can adopt two possible orientations, i.e., with either the phenyl ring or 2-chloropyridin-4-yl ring in direct π-stacking interaction with the isoalloxazine ring of FAD. The crystal structure of ZmCKX4a shows that CPPU is bound only in the orientation with the 2-chloropyridin-4-yl ring pointing towards the entrance of the active site and phenyl ring in direct π-stacking interaction with the isoalloxazine ring (PDB ID: 4O95 and 4OAL, Kopečný et al. 2015). ZmCKX4a has an alanine in place of Glu381 found in ZmCKX1 and this residue apparently affects the binding of urea-derived cytokinins.
A homology model of AtCKX2, prepared based on the ZmCKX1 structure, revealed that it shares a similar shape of active site. Amino acids in the active site of AtCKX2 and ZmCKX1 are almost identical and include Asp150/Asp169, Val345/Val378, Trp358/Trp391, Asn366/Asn399, Tyr460/Tyr491, Leu461/Leu492 and especially Glu348/Glu381. A small difference is replacement of Leu397 by Ile429. To confirm the ligand binding, CPPU and TDZ were docked into the active site of ZmCKX1 and AtCKX2 using AutoDock Vina. The results were nearly identical and agreed with positions observed for CPPU in known crystal structures. CPPU was found in two orientations with binding energies of −6.6 and −8.2 kcal mol−1. The latter corresponded to preferred binding with the phenyl ring in π-stacking interaction with the isoalloxazine ring. In contrast, when docking TDZ, the thiadiazolyl ring was located over the isoalloxazine ring.
Design and chemistry of TDZ derivatives
Molecular docking to the active site of AtCKX2 was performed in parallel to preparation of new N,N′-bis substituted urea derivatives possessing the 1,2,3-thiadiazolyl ring of TDZ. Twenty of the most promising compounds, based on the obtained ligand conformations, binding energies (Table 1) and availability of phenyl- and benzyl-amines for the synthesis, were selected and synthesized (Table 1; Fig. 1). Commercially available 1,2,3-thiadiazol-5-ylamine was a starting compound for the synthesis of the intermediate 5-isocyanato-1,2,3-thiadiazole, which was prepared using diphosgene (Kurita et al. 1976). Although, many TDZ derivatives have been synthesized before (Mok et al. 1982; Abad et al. 2004), use of 5-isocyanato-1,2,3-thiadiazole has not yet been reported. Final compounds were prepared by mild heating of this isocyanate and the corresponding amine in the presence of a catalytic amount of triethylamine in tetrahydrofuran (THF) (Goldschmidt and Bardach 1892). The compounds were characterized by LC-PDA-MS and 1H NMR (Supplementary material and Figure S3 and S4). Two compounds HETDZ and 3FMTDZ were also characterized in MS/MS experiments (Figure S5).
Reaction scheme of the synthesis of new TZD derivatives. In the first step, a 1,2,3-thiadiazol-5-ylamine was dissolved in THF and added dropwise into a solution of diphosgene to form an intermediate 5-isocyanato-1,2,3-thiadiazole. The isocyanate was further reacted with a phenyl- or benzyl-amine derivative in the presence of a catalytic amount of triethylamine to form the final product
Inhibition of AtCKX2 and correlation with molecular docking
TDZ derivatives all docked into the active site of the AtCKX2 model with apparent preferred orientation of the 1,2,3-thiadiazolyl ring towards the isoalloxazine ring of FAD. Both urea nitrogen atoms bound to Asp150. The polar functional groups attached to the phenyl ring interacted with Glu348 at the entrance, whereas those attached to the benzyl ring were too large to be accommodated easily. This finding is in line with the calculated binding energies, which were more favorable for phenyl derivatives (∆G from −8.1 to −9.3 kcal mol−1) than those for benzyl derivatives (∆G from −6.3 to −8.2 kcal mol−1). However, ∆G differences among phenyl derivative series are too small to predict their real in vivo binding and thus data correlate rather roughly with IC 50 values measured with AtCKX2 (Table 1). No correlation was found for compound 12 despite it displayed together with FMTDZ the best binding energy (∆G value of −9.3 kcal mol−1) found by docking. As the compound 12 is more hydrophobic due to the presence of chlorine atoms, it is likely that lower solubility or non-specific binding may have negatively affected its properties.
The ability of the prepared derivatives to inhibit the activity of AtCKX2 was compared with that of TDZ (Fig. 2). IC 50 values were determined using a saturating concentration of the substrate, i.e., 45 µM iP (Table 1). An IC 50 value of 62 μM was determined for TDZ. Similar IC 50 values were determined for compounds 2 and 4, which were ortho- and meta-hydroxylated TDZ derivatives, respectively. No significant change was observed for compounds 1 (ortho-methoxy TDZ), 7 (ortho-methyl, meta-hydroxy TDZ) and 11 (meta-trifluoromethyl TDZ). Lower IC 50 values were observed for compounds 8 (IC 50 = 18 μM) and 3 (IC 50 = 22 μM) bearing ortho-methylhydroxy and meta-methoxy substituents, respectively. Compounds with IC 50 values below 10 μM were compounds 5 (IC 50 = 7.6 μM), 10 (3FMTDZ, IC 50 = 5.5 μM) and 9 (HETDZ, IC 50 = 3.9 μM) bearing 2,5-dimethoxy-, meta-trifluoro-methoxy- and ortho-ethylhydroxy groups. Compounds bearing a dichloro-phenyl ring or substituted benzyl ring were less active or even inactive. 3FMTDZ was one of the top two compounds with the best binding energy found by docking (∆G = −9.3 kcal mol−1), whereas the other compound, 12, was not effective at all. As the latter compound is more hydrophobic due to the presence of chlorine atoms, it is likely that lower solubility or non-specific binding may have negatively affected its properties. Zatloukal et al. (2008) reported an IC 50 value for TDZ of 29 μM toward AtCKX2. This is about half that of our result (62 μM). This difference can be explained by the use of lower substrate concentration in the previous study (30 μM iP). Zatloukal’s best inhibitors reached an IC 50 of about 1–2 μM, which is very close to the inhibitory strength of HETDZ (IC 50 = 3.9 μM). In contrast, our IC 50 was obtained with 45 μM iP as a substrate.
Inhibitor strength of selected TDZ derivatives against AtCKX2, ZmCKX1 and ZmCKX4a. CKX activity was measured by PMS/MTT kinetic assay (Frébort et al. 2002). The standard deviation of three replicates did not exceed 10 %. The whole assays were performed twice and presented graphs are representative examples
Inhibition of ZmCKX1 and ZmCKX4a
Based on the results obtained with AtCKX2, selected TDZ derivatives were further tested using ZmCKX1 and ZmCKX4a. Compounds 1–12 displayed similar properties toward ZmCKX1, probably because it has a similar active site composition to that of AtCKX2. HETDZ and 3FMTDZ were the strongest inhibitors of ZmCKX1, with IC 50 values of 2.8 and 4.8 μM, respectively. Only compounds 7, 8, HETDZ and 3FMTDZ inhibited ZmCKX4a better than TDZ itself: the IC 50 value for TDZ was slightly higher than 200 μM, whereas it was 117, 91 and 120 μM for compounds 7, 8 and HETDZ, respectively. The best inhibitor was 3FMTDZ, with IC 50 of 35 μM (Table 1; Fig. 2). Previously, the two strongest CPPU derivatives toward the ZmCKX1 isoform were reported to have IC 50 values of 1.8 and 2 µM (Kopečný et al. 2010). However, as mentioned above, the synthesis of these compounds is much more complicated than that of HETDZ and 3FMTDZ.
Although earlier work suggested that TDZ was a non-competitive inhibitor (Hare and Van Staden 1994), more recent studies have shown that it is a competitive inhibitor of CKX when 2,6-dichlorophenol indophenol (DCPIP) is used as an electron acceptor (Bilyeu et al. 2001; Kopečný et al. 2010). Using a method based on the coupled redox reaction of PMS and MTT (Frébort et al. 2002), we showed that TDZ and the two strongest inhibitors HETDZ and 3FMTDZ inhibit AtCKX2 as well as ZmCKX4a in a competitive manner (Fig. 3). With AtCKX2, K i values for HETDZ and 3FMTDZ were 1.25 and 1.8 μM, respectively, whereas that for TDZ was one order of magnitude higher (K i = 21 μM). With ZmCKX4a, K i values for TDZ and 3FMTDZ were 7.2 and 3.6 μM, respectively.
Inhibition of ZmCKX4a and AtCKX2 with selected TDZ derivatives. a Structures of the two best inhibitors HETDZ and 3FMTDZ. b, c Double-reciprocal plots of competitive inhibition of ZmCKX4a by TDZ and 3FMTDZ. Lineweaver–Burk plots were constructed using iP as a substrate with concentration range 5–15 μM. The respective inhibitor concentrations were 0 μM (open diamond), 15 μM (filled square) and 30 μM (filled triangle) for TDZ and 0 μM (open diamond), 4 μM (filled square) and 8 μM (filled triangle) for 3FMTDZ. d–f Double-reciprocal plots showing competitive inhibition of AtCKX2 by TDZ, HETDZ and 3FMTDZ. The respective inhibitor concentrations were 0 μM (open diamond), 7.5 μM (filled square) and 15 μM (filled triangle) for TDZ and 0 μM (open diamond), 1.5 μM (filled square) and 3 μM (filled triangle) for HETDZ and 3FMTDZ. The insets show secondary plots of slope against inhibitor concentration, which were used for K i determination
Crystal structures of ZmCKX4a in complex with 3FMTDZ and HETDZ
Two crystal structures of ZmCKX4a in complex with two urea inhibitors 3FMTDZ and HETDZ were solved at 2.0 Å resolution (PDB IDs 5HMR and 5HQX). A summary of the refinement results together with model statistics is given in Table 2. The crystal structure complexes revealed that HETDZ and 3FMTDZ occupy the same place in the active site as cytokinin substrates. Surprisingly, HETDZ and 3FMTDZ bind in opposite orientations (Fig. 4). Their urea nitrogen atoms are at a distance of 2.7–3.1 Å from the side chain oxygen atom of catalytic aspartate D170, allowing hydrogen bond interactions. The urea’s carbonyl oxygen is 3.3 Å distant from the N5 atom of the isoalloxazine plane of FAD. The thiadiazolyl ring of 3FMTDZ is positioned over the flavin ring and one of its two nitrogen atoms can establish a hydrogen bond to Asn391 directly (3.3 Å distance) or via a water molecule. One of the urea nitrogens forms an H-bond to a water molecule, which is further bound to the side-chain oxygen atom of D170, O4 atom of isoalloxazine, hydroxyl group of Tyr425 and the main-chain oxygen of Ile185. The 3-trifluoromethoxy group is oriented toward the hydrophobic pocket composed of side chains of Trp389, Trp383, Phe57, Leu377 and His387. In contrast, in the case of HETDZ, the 2-(2-hydroxyethyl)phenyl ring is positioned over the flavin ring, whereas the thiadiazolyl ring points toward the entrance of the active site. The oxygen atom of the hydroxyethyl side chain replaces a water molecule present in the ZmCKX4a-3FMTDZ structure and makes a H-bond to the side-chain oxygen atom of Asp170 and hydroxyl group of Tyr425. This water molecule is present in most of the CKX structures, including substrate and inhibitor complexes, and is H-bonded to a catalytic aspartate residue. Orientation and position of 3FMTDZ found in the crystal structure was very similar to that predicted by molecular docking for AtCKX2. Interestingly, orientation of the docked HETDZ was similar to that of 3FMTDZ with thiadiazolyl ring positioned over the flavin ring and hydroxyethyl side chain pointing towards and interacting with Arg344 (Arg369 in ZmCKX4a). However, this interaction does not appear in the crystal structure of ZmCKX4a and further crystallization experiments are needed to verify whether HETDZ binding may differ among CKX isoforms.
Binding of the two urea inhibitors 3FMTDZ and HETDZ in the active site of ZmCKX4a. a Binding of 3FMTDZ (PDB ID: 5HMR). The inhibitor is colored green and shown in a Fo–Fc omit map contoured at 4.0 σ. b Binding of HETDZ (PDB ID: 5HQX). The inhibitor is colored blue and shown in a Fo–Fc omit map contoured at 3.0 σ. c Superposition of bound 3FMTDZ and HETDZ. Both ligands form a hydrogen bond to the oxygen atom of D170 via two nitrogen atoms of the urea backbone. The oxygen atom of the hydroxyethyl side chain of HETDZ replaces a water molecule observed in the ZmCKX4a-3FMTDZ structure. The FAD cofactor is colored yellow and neighboring residues are labeled
Activity in classical cytokinin bioassays
Three classical cytokinin bioassays, i.e., wheat leaf senescence, Amaranth and tobacco callus assay, were used to investigate the cytokinin activity of the selected compounds using N 6-benzyladenine (BA) and TDZ as controls (Table 3). TDZ is a very potent cytokinin and none of the tested derivatives possessed higher activity than TDZ itself in any bioassay. In the senescence assay, where the degradation of chlorophyll is assessed, eight compounds were completely inactive and only three had an IC 50 (concentration at which the chlorophyll degradation is inhibited by 50 %) lower than 100 μM, although the IC 50 of TDZ in this assay was about 10 μM. The most active compounds were those bearing benzyl substituted in the meta-position by fluorine or hydroxyl group (compounds 17 and 15). In the amaranth assay, which is based on the dark induction of betacyanin synthesis in amaranth cotyledons by cytokinins, EC 50 values (concentration at which the activation response reaches 50 %) were determined. Compound 8 was the most active in this assay, slightly exceeding the activity of BA. This compound had an ortho-methylhydroxy phenyl ring. Moderately less active were compounds 1, 2 and 3, which had an ortho-methoxy, ortho-hydroxy and meta-methoxy substituted phenyl ring, respectively, and compound 17, which had a meta-fluorobenzyl group. In the tobacco callus assay, where the growth of callus is cytokinin dependent, none of the compounds reached the TDZ activity. The most active compounds (8, 15 and 17) showed similar activity to that of BA. In contrast, substances 3, 11, 14, 16 and 3FMTDZ showed maximum activity for a concentration one order of magnitude higher.
Activation of cytokinin primary response gene ARR5
We employed transgenic Arabidopsis plants (A. thaliana) harboring the ARR5::GUS reporter gene (D’Agostino et al. 2000) to monitor complex in vivo cytokinin activity of each compound. ARR5 is the primary response gene with a cytokinin-dependent promoter, and its activation integrates the responses of several putative cytokinin signaling pathways as well as the contribution of individual cytokinin receptors. Compounds were tested at 0.5, 1.0 and 10 μM concentrations (Table 3) and their ability to activate the ARR5::GUS expression is presented for their optimal concentration. TDZ had highest activity in the nanomolar range, whereas BA reached its maximum activity at 1 µM. The most active TDZ derivatives, namely compounds 1, 8 and HETDZ (9), possessed a similar activity to that of BA. For these compounds, there was a clear structure–activity relationship, with the activity decreasing in the order ortho-methoxy group (1) > ortho-methylhydroxy group (8) > ortho-ethylhydroxy group (HETDZ). Slightly lower activities were exhibited by compounds 3, 4 and 10 (3FMTDZ) bearing methoxy, hydroxy and trifluoromethoxy groups, respectively, in the meta position of the phenyl ring. Compounds 2, 5, 11, 12 and 15 caused maximal activation only at 10 μM concentration. Other compounds, which did not activate the ARR5::GUS expression even at 10 μM concentration, were categorized as very poor cytokinins. These included TDZ derivatives with a substituted benzyl ring rather than phenyl ring.
Interaction with Arabidopsis cytokinin receptors AHK3 and CRE1/AHK4
Transformed E. coli cells expressing the genes of cytokinin receptor AHK3 or CRE1/AHK4 and the cytokinin-activated reporter gene cps::lacZ (Suzuki et al. 2001; Yamada et al. 2001) were employed to examine the ability of the synthesized compounds to activate the receptor (in case of CRE1/AHK4 receptor only) or bind to the cytokinin binding site. In the CRE1/AHK4 receptor activation assay, EC 50 values (half maximal effective concentration for receptor activation) of the compounds were determined and compared with those for trans-zeatin (tZ) and TDZ (see Figure S1). The EC 50 values for tZ and TDZ were 0.25 and 0.72 μM, respectively. This is in line with previous results reporting an EC 50 value for tZ of 0.2 μM (Nisler et al. 2010). The receptor was activated by several compounds at the two highest concentrations tested (10 and 50 μM). All of the derivatives showed decreased cytokinin activity when compared to TDZ. The most active was compound 2 bearing an ortho-hydroxyphenyl ring (EC 50 = 5.6 μM), followed by compounds 15 (EC 50 = 17 μM) > 4 (EC 50 = 22 μM) ≅ 17 (EC 50 = 25 μM) > 8 (EC 50 = 32 μM) > 3 (EC 50 > 50 μM) bearing a meta-hydroxybenzyl, meta-hydroxyphenyl, meta-fluorobenzyl, ortho-hydroxymethylphenyl and meta-methoxyphenyl ring, respectively. Interestingly, except for the fluorinated derivative (17), only compounds possessing hydroxyl group or methoxy group were capable of the activation. None of the other compounds, including HETDZ and 3FMTDZ, activated the CRE1/AHK4 receptor (Figure S1).
The same E. coli cells were also used to determine whether 3FMTDZ or HETDZ possessed affinity toward the CRE1/AHK4 receptor. Their affinities were compared with those of tZ, TDZ and adenine (Figure S1). While tZ at 10 nM concentration (used as a positive control) displaced 66 % of [2-3H]tZ from the receptor, TDZ was inactive at this concentration. TDZ displaced 85 % of tritiated tZ from the receptor at much higher 1 μM concentration whereas both 3FMTDZ and HETDZ were inactive at this concentration. They both displaced only about 15 % of tritiated tZ from the receptor at 10 μM concentration. The same result was achieved with the negative control, adenine. This confirms the results of the receptor activation assay and demonstrates limited interaction of 3FMTDZ and HETDZ with this receptor.
The AHK3 receptor is known to display better affinity (lower KD values) and lower selectivity towards natural and synthetic cytokinins than the AHK4 receptor (Spíchal et al. 2004; Romanov et al. 2006). The K D values for TDZ were reported as 13 nM (AHK3) and 40 nM (AHK4). In our measurements (Figure S1), 10 nM TDZ displaced about 80 % of tritiated tZ from the AHK3 receptor while all phenyl derivatives (series I) and compound 15 (series II) displaced tritiated tZ at 20 μM concentration. Remaining compounds of the series II did not bind at the concentration tested. Compounds 2, 3, 4, 6, 8, HETDZ and 15 displayed stronger binding as they displaced more than 70 % of tritiated tZ. Compounds 5, 7, 3FMTDZ, 11 and 12 displaced less than 50 % of tritiated tZ from the AHK3 receptor. In summary, both compounds HETDZ and 3FMTDZ bind to AHK3 receptor at much higher concentrations (and much lower affinity) compared with TDZ.
Conclusion—cytokinin activity versus CKX inhibitory activity
Previously described CKX inhibitors, including TDZ, CPPU and its derivatives, suicide substrates or anilinopurines, have been shown to activate Arabidopsis cytokinin receptors AHK3 and/or AHK4 (Spíchal et al. 2004; Kopečný et al. 2010, 2008; Zatloukal et al. 2008). These compounds therefore possess intrinsic cytokinin activity including TDZ, the urea-derived compound with the highest cytokinin activity. In contrast, the new TDZ inhibitors 3FMTDZ or HETDZ described in this work do not interact with the CRE1/AHK4 receptor. They both possess lower intrinsic cytokinin activity than the cytokinins TDZ and tZ and display wide and increased inhibitory properties to various CKX isoforms. Their lower affinity to cytokinin receptors AHK3 and AHK4 (AHK2 receptors were not tested in this work) makes both attractive candidates for manipulations with endogenous cytokinins in planta and for further development of new derivatives. It has been proposed that enhancing the content of endogenous cytokinins in plants via inhibiting CKX is more beneficial to plant growth and development than using the high intrinsic cytokinin activity of exogenously applied compounds (Gemrotová et al. 2013). This indicates that plants favor very gentle regulation of cytokinin biosynthesis, metabolism and signaling. Therefore, CKX inhibitors might affect different cytokinin functions in plants, thereby having possible positive effects on seed filling, delayed senescence and stress tolerance toward biotic and abiotic stresses, thus improving crop yields.
Materials and methods
Molecular modeling and docking
A model of AtCKX2 was built in Modeller 9.8 (Eswar et al. 2006) within UCSF Chimera 1.9 (Pettersen et al. 2004) by aligning the sequence (UNIPROT ID: Q9FUJ3) to a template structure of ZmCKX1 (PDB ID: 2QKN, Kopečný et al. 2010). Sequence alignment between both proteins can be found in the supplement (Figure S2). Modeller was instructed to build 5 models of AtCKX2 without explicit hydrogens with non-protein residues treated as rigid bodies. We have selected the best performing model in the zDOPE value with rotamer of Asp150 in the same interaction position as Asp169 in ZmCKX1. Model also did not suffer from sterical clashes and RMSD for all Cα atoms between final model and template was just 0.176 A. Ligands for molecular docking were constructed using Marvin 14.9.8 software (ChemAxon). Polar hydrogens were added to all ligands and proteins using AutoDock Tools (ADT) software (Morris et al. 2009) prior to docking using Autodock Vina 1.05 (Trott and Olson 2010). A grid box with size of 16 Å was centered on the active site around Asp150. The exhaustiveness parameter was set to 20 (default 8). After docking, the best docked ligand was selected and the best crystal-like poses of each ligand were analyzed with Pymol 1.7.4 (The PyMOL Molecular Graphics System, Version 1.7.4 Schrödinger, LLC.) and Maestro 2014-3 (Schrödinger Release 2014-3: Maestro, version 9.9, Schrödinger, LLC, New York, NY, 2014). RMSD cutoff for crystal-like pose is default defined below 2 Å.
Chemicals
1,2,3-Thiadiazol-5-ylamine was supplied by TCI Europe (Zwijndrecht, Belgium). TDZ, tZ and BA were supplied by Olchemim (Olomouc, Czech Republic). Other compounds, including 2-methoxy-phenylamine, 2-amino-phenol, 3-methoxy-phenylamine, 3-amino-phenol, 2,5-dimethoxy-phenylamine, 2-hydroxy-3-methyl-phenylamine, (2-amino-phenyl)-methanol, (2-amino-phenyl)-ethanol, 3-trifluoromethoxy-phenylamine, 3-trifluormethyl-phenylamine, 2,4-dichloro-phenylamine, 2-hydroxy-benzylamine, 2-methyl-benzylamine, 3-hydroxy-benzylamine, 3-methoxy-benzylamine, 3-fluoro-benzylamine, 3-chloro-benzylamine, 2,5-dimethoxy-benzylamine and tert-butyl-dimethyl-sillyl chloride, were purchased from Sigma Aldrich (Germany). All solvents and chemicals used were of standard p.a. quality.
Synthesis of 5-isocyanato-1,2,3-thiadiazole and final products
The synthesis of isocyanates from amines has been described elsewhere (Kurita et al. 1976). Briefly, 1,2,3-thiadiazol-5-ylamine (1.01 g, 10 mmol) was dissolved in THF (40 mL) and added dropwise into a solution of diphosgene (2.6 g, 13 mmol) in THF (100 mL). The reaction mixture was stirred for 40 min at 0 °C and then heated to 30 °C. After another 30 min, the solvent and excess of diphosgene were evaporated. The yellow solid residue of 5-isocyanato-1,2,3-thiadiazole was re-suspended in diethyl ether and filtered. Yield: 95, 1H NMR (δ, ppm, DMSO-d 6): 7.75(1H, s, CH).
All the prepared compounds were synthesized according to common protocols for the synthesis of disubstituted urea derivatives, using amine and isocyanate as starting materials in the presence of a catalytic amount of triethylamine (Goldschmidt and Bardach 1892). Firstly, 5-isocyanato-1,2,3-thiadiazole was mixed with THF (1:100) in a high pressure tube. Then, a catalytic amount of triethylamine was added and the suspension was allowed to dissolve. The starting amine was added in an equimolar amount into the solution and the reaction mixture was stirred for 5–12 h at 60 °C. The conversion was monitored by TLC (CHCl3:MeOH, 9:1). After reaction, the solvent was evaporated to a solid/semi-solid residue, which was purified by flash silica column chromatography (mobile phase CHCl3:MeOH, 9:1). Compounds were usually prepared in milligram quantities and yields varied between 30 and 80 %. If the starting amine contained a free hydroxyl group, it was protected by tert-butyl-dimethylsilyl chloride prior to the condensation with 5-isocyanato-1,2,3-thiadiazole and then de-protected in 2-propanolic HCl (Wuts and Greene 1991). All products (Table 1) were characterized by 1H NMR and LC-PDA-MS (Supplementary material). The MS/MS spectra of HETDZ and 3FMTDZ were also recorded using tandem mass analyser Quatro Micro API with MassLynx data system software (Waters, Manchester, UK; see Supplementary material for details).
General experimental procedures
The purity and mass spectra of the synthesized compounds were recorded using method LC-PDA-MS. Compounds (1 mg) were dissolved in 1 ml of 10 % methanol and injected (10 µL) onto a reverse-phased column (Symmetry C18, 5 μm, 150 mm × 2.1 mm; Waters, Milford, MA, USA) incubated at 25 °C. Solvent (A) consisted of 15 mM ammonium formate adjusted to pH 4.0. Solvent (B) consisted of methanol. At flow-rate of 200 µL/min, following binary gradient was used: 0 min, 10 % B; 0–24 min; linear gradient to 90 % B; 25–34 min; isocratic elution of 90 % B; 35–45 min; linear gradient to 10 % B. The effluent was introduced then to PDA detector (scanning range 210–700 nm with 1.2 nm resolution) and a tandem mass analyser Quatro Micro API (Waters, Manchester, UK) with an electrospray source (source temperature 120 °C, desolvation temperature 300 °C, capillary voltage 3 kV). Nitrogen was used as well as cone gas (50 L/h) and desolvation gas (500 L/h). Data acquisition was performed in the full scan mode (50-1000 Da), scan time of 0.5 s and cone voltage 20 V. Analyses were performed in positive mode (ESI+) therefore data were collected as protonated ions [M + H]+. Analytical thin-layer chromatography (TLC) was carried out using silica gel 60 WF254 plates (Merck). If necessary, compounds were separated on a flash chromatography column using 63 µm chromatographic silica (Davisil) and eluted with a mobile phase containing CHCl3/MeOH (9:1, v/v). 1H NMR spectra were measured on a Jeol 500 SS spectrometer operating at a temperature of 300 K and a frequency of 500.13 MHz (1H). Samples were prepared by dissolving the compounds in DMSO-d 6 or CDCl3. Tetramethylsilane (TMS) was used as an internal standard.
CKX inhibition measurements
The ability of the prepared compounds to inhibit AtCKX2, ZmCKX1 and ZmCKX4a was evaluated by determining their IC 50 values in the PMS/MTT (phenazine methosulfate/3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) activity assay previously adapted for screening in ELISA microtitre plates (Table 1, Frébort et al. 2002). Each well contained 100 μL of a reaction mixture consisting of 0.1 M KH2PO4 (pH adjusted to 7.4 with KOH), 1.5 mM PMS, 0.3 mM MTT, the tested compound (at various concentrations) and substrate iP at saturating concentration (45 μM for AtCKX2 and ZmCKX4a and 10 μM for ZmCKX1). Cell-free growth medium of S. cerevisiae strain 23344c ura- harbouring the plasmid pYES2-AtCKX2 (50 μL) was used directly as a source of AtCKX2 (Frébortová et al. 2007). Pure enzymes were used in the case of ZmCKX1 and ZmCKX4a. ZmCKX1 was expressed in yeast Y. lipolytica and purified according to a published protocol (Kopečný et al. 2005). E. coli BL21 STAR (DE3) cells carrying pTYB12-ZmCKX4a plasmid were grown for 16 h at 18 °C to produce ZmCKX4a, which was further purified by affinity chromatography on a column filled with a chitin resin, followed by ion-exchange chromatography (Zalabák et al. 2014). Plates were incubated in the dark for 20 min at 37 °C and the enzymatic reaction was monitored every 2 min by measuring the absorbance of the mixture in each well at 578 nm (spectrophotometer Synergy H4 Hybrid Reader). The absorbance of samples without substrate was subtracted as a baseline. Compounds were tested with two replicates and the entire test was repeated at least twice. Gene 5 software was used to calculate the residual CKX activity for each compound and concentration. IC 50 values for each compound were determined in Origin Pro software. Lineweaver–Burk plots for K i determination were constructed using iP as a substrate with concentration range from 5 to 15 μM.
Crystallization and structure determination of 3FMTDZ and HETDZ complexes with ZmCKX4a
Preliminary crystallization conditions were identified in sitting drops using Crystal Screen and Crystal Screen 2 (Hampton Research). ZmCKX4a was co-crystallized with 3FMTDZ in drops containing 100 mM HEPES pH 7.5, 80 % MPD, 0.16 mM ZmCKX4a and 10 mM 3FMTDZ in DMSO. For HETDZ, crystals of ZmCKX4a were grown in 22 % ethylene–glycol and then infiltrated by 5 mM HETDZ in DMSO for 1 h. Crystals were directly flash frozen in liquid nitrogen. Diffraction data were collected at 100 K on the Proxima 1 beamline at the SOLEIL synchrotron (Saint-Aubin, France) at 2.0 Å resolution. Intensities were integrated using the XDS program (Kabsch 2010) and data quality was assessed using the correlation coefficient CC 1/2 (Karplus and Diederichs 2012). Crystal structures were determined by performing molecular replacement with Phaser (Storoni et al. 2004) using a monomer of ZmCKX1 (PDB 2QKN) as search model. Model refinement was achieved with BUSTER-TNT (Bricogne et al. 2011). Electron density maps were evaluated using COOT (Emsley and Cowtan 2004). Refinement statistics are presented in Table 2. Molecular graphics images were generated using PYMOL (www.pymol.org).
Cytokinin bioassays and Arabidopsis ARR5::GUS reporter gene assay
Classical cytokinin bioassays, i.e., tobacco callus assay, Amaranthus assay and wheat senescence assay with excised wheat leaves in the dark, were performed as described in Holub et al. (1998). The tobacco callus bioassay was performed in 6-well microtiter plates (3 mL of MS medium in each well, into which 0.1 g of callus was placed). In the wheat leaf senescence and Amaranthus assays, IC 50 values (concentration at which chlorophyll degradation is inhibited by 50 %) and EC 50 values (half maximal effective concentration) for each compound were determined (Table 3). In the tobacco callus bioassay, the effect at optimal concentration was compared with BA activity (100 % corresponded to 10−6 M BA). Quantitative determination of GUS activity was performed by measuring fluorescence on a Synergy H4 hybrid reader (Biotek, USA) at excitation and emission wavelengths of 365 and 445 nm, respectively. The assay was carried out as described previously (Romanov et al. 2002). GUS specific activity was expressed in relative % and was calculated as RFU (relative fluorescence units) divided by protein content. Determination of protein content was done using the bicinchoninic acid assay (Smith et al. 1985).
Live-cell cytokinin-binding assays and CRE1/AHK4 receptor activation assay
Escherichia coli strain KMI001, harboring either the plasmid pIN-IIIAHK4 or pSTV28-AHK3, which express the Arabidopsis histidine kinases CRE1/AHK4 or AHK3 (Suzuki et al. 2001; Yamada et al. 2001) was used in the experiments. Bacterial strains were kindly provided by T. Mizuno (Nagoya, Japan). The CRE1/AHK4 receptor activation assay was performed as previously described in Spíchal et al. (2004). The live-cell cytokinin-binding assay was performed essentially as described in Romanov et al. (2005). The competition reaction was allowed to proceed with 3 nM [2-3H]tZ and various concentrations of the tested compounds for 30 min at 4 °C. When a binding equilibrium was reached, the suspension was centrifuged (6000g), the supernatant was removed and the bacterial pellet was re-suspended in scintillation cocktail (Beckman, Ramsey, MN, USA). Radioactivity was measured by scintillation counting on a Beckman LS 6500 scintillation counter (Beckman, Ramsey, MN, USA).
Accession numbers
The atomic coordinates and structure factors have been deposited in the Protein Data Bank (www.rcsb.org) under the accession codes 5HMR for ZmCKO4a in complex with 3FMTDZ and 5HQX for ZmCKO4a in complex with HETDZ.
References
Abad A, Agulló C, Cuñat AC, Jiménez R, Vilanova C (2004) Preparation and promotion of fruit growth in kiwifruit of fluorinated N-phenyl-N′-1,2,3-thiadiazol-5-yl ureas. J Agr Food Chem 52:4675–4683
Aremu AO, Masondo NA, Sunmonu TO, Kulkarni MG, Zatloukal M, Spichal L, Doležal K, Van Staden J (2014) A novel inhibitor of cytokinin degradation (INCYDE) influences the biochemical parameters and photosynthetic apparatus in NaCl-stressed tomato plants. Planta 240:877–889
Aremu AO, Stirk WA, Masondo NA, Plačková L, Novák O, Pěnčík A, Zatloukal M, Nisler J, Spíchal L, Doležal K, Finniea JF, Van Staden J (2015) Dissecting the role of two cytokinin analogues (INCYDE and PI-55) on in vitro organogenesis, phytohormone accumulation, phytochemical content and antioxidant activity. Plant Sci 238:81–94
Arima Y, Oshima K, Shudo K (1995) Evolution of a novel urea-type cytokinin: horticultural uses of forchlorofenuron. Acta Hortic 394:75–83
Arndt F, Rusch R, Stillfried HV (1976) SN49537, a new cotton defoliant. Plant Physiol 57:599
Bilyeu KD, Cole JL, Laskey JG, Riekhof WR, Esparza TJ, Kramer MD, Morris RO (2001) Molecular and biochemical characterization of a cytokinin oxidase from maize. Plant Physiol 125:378–386
Bricogne G, Blanc E, Brandl M, Flensburg C, Keller P, Paciorek W, Roversi P, Sharff A, Smart OS, Vonrhein C, Womack TO (2011) BUSTER version 2.1.0 Cambridge. Global Phasing Ltd, UK
Brownlee BG, Hall RH, Whitty CD (1975) 3-Methyl-2-butenal: an enzymatic degradation product of the cytokinin, N-6-(delta-2 isopentenyl) adenine. Can J Biochem 53:37–41
Burch LR, Horgan R (1989) The purification of cytokinin oxidase from Zea mays kernels. Phytochemistry 28:1313–1319
Chatfield JM, Armstrong DJ (1986) Regulation of cytokinin oxidase activity in callus tissues of Phaseolus vulgaris L. cv Great Northern. Plant Physiol 80:493–499
D’Agostino IB, Deruere J, Kieber JJ (2000) Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 124(:):1706–1717
Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132
Eswar N, Webb B, Marti-Renom M, Madhusudhan M, Eramian D, Shen M, Pieper U, Sali A (2006) Comparative protein structure modeling using Modeller. Curr Protoc Bioinform, Chap 5: Unit 5
Frébort I, Šebela M, Galuszka P, Werner T, Schmülling T, Peč P (2002) Cytokinin oxidase/cytokinin dehydrogenase assay: optimized procedures and applications. Anal Biochem 306:1–7
Frébortová J, Galuszka P, Werner T, Schmülling T, Frébort I (2007) Functional expression and purification of cytokinin dehydrogenase from Arabidopsis thaliana (AtCKX2) in Saccharomyces cerevisiae. Biol Plantarum 51:673–682
Galuszka P, Popelková H, Werner T, Frébortová J, Pospíšilová H, Mik V, Köllmer I, Schmülling T, Frébort I (2007) Biochemical characterization and histochemical localization of cytokinin oxidases/dehydrogenases from Arabidopsis thaliana expressed in Nicotiana tabaccum L. J Plant Growth Regul 26:255–267
Gemrotová M, Kulkarni MG, Stirk WA, Strnad M, Van Staden J, Spíchal L (2013) Seedlings of medicinal plants treated with either a cytokinin antagonist (PI-55) or an inhibitor of cytokinin degradation (INCYDE) are protected against the negative effects of cadmium. Plant Growth Regul 71:137–145
Goldschmidt H, Bardach B (1892) Zur Kenntniss der Diazoamidokörper. Chem Ber 25:1347–1378
Hare PD, Van Staden J (1994) Inhibitory effect of thidiazuron on the activity of cytokinin oxidase isolated from soybean callus. Plant Cell Physiol 35:1121–1125
Holub J, Hanuš J, Hanke DE, Strnad M (1998) Biological activity of cytokinins derived from ortho- and meta-hydroxybenzyladenine. Plant Growth Regul 26:109–115
Houba-Hérin N, Pethe C, d’Alayer J, Laloue M (1999) Cytokinin oxidase from Zea mays: purification, cDNA cloning and expression in moss protoplasts. Plant J 17:615–626
Kabsch W (2010) XDS. Acta Crystallogr D Biol Crystallogr 66:125–132
Karplus PA, Diederichs K (2012) Linking crystallographic model and data quality. Science 336:1030–1033
Kopečný D, Pethe C, Šebela M, Houba-Hérin N, Madzak C, Majira A, Laloue M (2005) High-level expression and characterization of Zea mays cytokinin oxidase/dehydrogenase in Yarrowia lipolytica. Biochimie 87:1011–1022
Kopečný D, Šebela M, Briozzo P, Spíchal L, Houba-Hérin N, Mašek V, Joly N, Madzak C, Anzenbacher P, Laloue M (2008) Mechanism-based inhibitors of cytokinin oxidase/dehydrogenase attack FAD cofactor. J Mol Biol 380:886–899
Kopečný D, Briozzo P, Popelková H, Šebela M, Končitíková R, Spíchal L, Nisler J, Madzak C, Frébort I, Laloue M, Houba-Hérin N (2010) Phenyl- and benzylurea cytokinins as competitive inhibitors of cytokinin oxidase/dehydrogenase: a structural study. Biochimie 92:1052–1062
Kopečný D, Končitíková R, Popelka H, Briozzo P, Vigouroux A, Kopečná M, Zalabák D, Šebela M, Skopalová J, Frébort I, Moréra S (2015) Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site. FEBS J 283:361–377
Kurita K, Matsumura T, Iwakura Y (1976) Trichloromethyl chloroformate. Reaction with amines, amino acids, and amino alcohols. J Org Chem 41:2070–2071
Laloue M, Fox JE (1989) Cytokinin oxidase from wheat: partial purification and general properties. Plant Physiol 90:899–906
Lomin SN, Krivosheev DM, Steklov MY, Arkhipov DV, Osolodkin DI, Schmülling T, Romanov GA (2015) Plant membrane assays with cytokinin receptors underpin the unique role of free cytokinin bases as biologically active ligands. J Exp Bot 66:1851–1863
Massonneau A, Houba-Hérin N, Pethe C, Madzak C, Falque M, Mercy M, Kopečný D, Majira A, Rogowsky P, Laloue M (2004) Maize cytokinin oxidase genes: differential expression and cloning of two new cDNAs. J Exp Bot 55:2549–2557
Mok DW, Mok MC (2001) Cytokinin metabolism and action. Annu Rev Plant Physiol Plant Mol Biol 52:89–118
Mok MC, Mok, DWS, Armstrong DJ, Shudo K, Isogai Y, Okamoto T (1982) Cytokinin activity of N-phenyl-N′-1,2,3-thiadiazol-5-ylurea (thidiazuron). Phytochem 21:1509–1511
Morris RO, Bilyeu KD, Laskey JG, Cheikh NN (1999) Isolation of a gene encoding a glycosylated cytokinin oxidase from maize. Biochem Biophys Res Commun 255:328–333
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) Autodock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 16:2785–2791
Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, Ueguchi C (2004) Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16:1365–1377
Nisler J, Zatloukal M, Popa I, Doležal K, Strnad M, Spíchal L (2010) Cytokinin receptor antagonists derived from 6-benzylaminopurine. Phytochem 71:823–830
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612
Romanov GA, Kieber JJ, Schmülling T (2002) A rapid cytokinin response assay in Arabidopsis indicates a role for phospholipase D in cytokinin signalling. FEBS Lett 515(:):39–43
Romanov GA, Spíchal L, Lomin SN, Strnad M, Schmülling T (2005) A live cell hormone-binding assay on transgenic bacteria expressing a eukaryotic receptor protein. Anal Biochem 347:129–134
Romanov GA, Lomin SN, Schmülling T (2006) Biochemical characteristics and ligand-binding properties of Arabidopsis cytokinin receptor AHK3 compared to CRE1/AHK4 as revealed by a direct binding assay. J Exp Bot 57:4051–4058
Schrödinger Release 2014-3 (2014) Maestro, version 9.9. Schrödinger, LLC, New York
Šmehilová M, Galuszka P, Bilyeu KD, Jaworek P, Kowalska M, Šebela M, Sedlářová M, English JT, Frébort I (2009) Subcellular localization and biochemical comparison of cytosolic and secreted cytokinin dehydrogenase enzymes from maize. J Exp Bot 60:2701–2712
Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC (1985) Measurement of protein using bicinchoninic acid. Anal Biochem 150:76–85
Spíchal L, Rakova NY, Riefler M, Mizuno T, Romanov GA, Strnad M, Schmülling T (2004) Two cytokinin receptors of Arabidopsis thaliana, CRE1/AHK4 and AHK3, differ in their ligand specificity in a bacterial assay. Plant Cell Physiol 45:1299–1305
Stolz A, Riefler M, Lomin SN, Achazi K, Romanov GA, Schmülling T (2011) The specificity of cytokinin signalling in Arabidopsis thaliana is mediated by differing ligand affinities and expression profiles of the receptors. Plant J 67:157–168
Storoni LC, McCoy AJ, Read RJ (2004) Likelihood-enhanced fast rotation functions. Acta Crystallogr D Biol Crystallogr 60:432–438
Suttle JC, Mornet R (2005) Mechanism-based irreversible inhibitors of cytokinin dehydrogenase. J Plant Physiol 162:1189–1196
Suzuki T, Miwa K, Ishikawa K, Yamada H, Aiba H, Mizuno T (2001) The Arabidopsis sensor His-kinase, AHK4, can respond to cytokinins. Plant Cell Physiol 42:107–113
The PyMOL Molecular Graphics System, Version 1.7.4 Schrödinger. LLC, New York
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem 31:455–461
Vyroubalová Š, Václavíková K, Turečková V, Novák O, Šmehilová M, Hluska, T, Ohnoutková L, Frébort I, Galuszka P (2009) Characterization of new maize genes putatively involved in cytokinin metabolism and their expression during osmotic stress in relation to cytokinin levels. Plant Physiol 151:433–447
Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15:2532–2550
Whitty CD, Hall RH (1974) A cytokinin oxidase in Zea mays. Can. J Biochem 52:789–799
Wuts PGM, Greene TW (1991) Greene’s protective groups in organic synthesis, 4th edition, Wiley, New York
Yamada H, Suzuki T, Terada K, Takei K, Ishikawa K, Miwa K, Yamashino T, Mizuno T (2001) The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol 42:1017–1023
Zalabák D, Galuszka P, Mrízová K, Podlešáková K, Gu R, Frébortová J (2014) Biochemical characterization of the maize cytokinin dehydrogenase family and cytokinin profiling in developing maize plantlets in relation to the expression of cytokinin dehydrogenase genes. Plant Physiol Biochem 74:283–293
Zatloukal M, Gemrotová M, Doležal K, Havlíček L, Spíchal L, Strnad M (2008) Novel potent inhibitors of A. thaliana cytokinin oxidase/dehydrogenase. Bioorg Med Chem 16:9268–9275
Acknowledgments
The authors gratefully acknowledge support through the projects LO1204 and LO1305 from the Ministry of Education, Youth and Sports of the Czech Republic and the grant 15-22322S and 15-19266S from the Czech Science Foundation. The work was also supported by student projects IGA_PrF_2016_028 and IGA_PrF_2016_018 of Palacký University, Olomouc.
Author contributions
J. Nisler, L. Spíchal and M. Strnad designed the research, V. Bazgier and K. Berka performed molecular modeling and docking, J. Nisler and M. Zatloukal synthesized the compounds, J. Nisler, R. Končitíková, D. Zalabák contributed to enzyme kinetics, R. Končitíková, D. Kopečný and P. Briozzo preformed X-ray crystallographic study, J. Nisler and D. Kopečný wrote the paper.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nisler, J., Kopečný, D., Končitíková, R. et al. Novel thidiazuron-derived inhibitors of cytokinin oxidase/dehydrogenase. Plant Mol Biol 92, 235–248 (2016). https://doi.org/10.1007/s11103-016-0509-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-016-0509-0