Abstract
Floral senescence involves an ordered set of events coordinated at the plant, flower, organ and cellular level. This review assesses our current understanding of the input signals, signal transduction and cellular processes that regulate petal senescence and cell death. In many species a visible sign of petal senescence is wilting. This is accompanied by remobilization of nutrients from the flower to the developing ovary or to other parts of the plant. In other species, petals abscise while still turgid. Coordinating signals for floral senescence also vary across species. In some species ethylene acts as a central regulator, in others floral senescence is ethylene insensitive and other growth regulators are implicated. Due to the variability in this coordination and sequence of events across species, identifying suitable models to study petal senescence has been challenging, and the best candidates are reviewed. Transcriptomic studies provide an overview of the MAP kinases and transcription factors that are activated during petal senescence in several species including Arabidopsis. Our understanding of downstream regulators such as autophagy genes and proteases is also improving. This gives us insights into possible signalling cascades that regulate initiation of senescence and coordination of cell death processes. It also identifies the gaps in our knowledge such as the role of microRNAs. Finally future prospects for using all this information from model to non-model species to extend vase life in ornamental species is reviewed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Higher plants are radically different to animals in that the germ line is not set down during embryogenesis, but rather develops from somatic cells. This process is regulated by a combination of environmental and developmental cues that trigger the formation of a transient reproductive structure: the flower. In some species, the individual plant will go on to recreate a new germ line and new reproductive apparatus season after season until its death. However each individual flower has a lifespan which is usually a small fraction of the lifespan of the parent plant (Rogers 2006). The life of the flower typically terminates in senescence and ultimately a form of programmed cell death (PCD).
There has been much debate on the use of the terms “senescence” and “programmed cell death” (Thomas et al. 2003; van Doorn and Woltering 2004). In the context of petal senescence it seems appropriate to use these words almost interchangeably (Rogers 2006), in agreement with others in the field (van Doorn and Woltering 2008). This is because cell death, at least in some cells, starts very early during petal senescence (Wagstaff et al. 2003). However at an organ level, senescence implies a degree of remobilization of nutrients (Thomas et al. 2003) and cell death can also be seen as the terminal event, following the remobilization process.
Flowers are not the only plant organ to undergo senescence, however compared to leaves, the lifespan of flowers attached to the plant is less influenced by environmental factors. In many cases floral senescence is activated or greatly accelerated by pollination, and is thus tightly regulated by developmental signals (van Doorn and Woltering 2008; Rogers 2006). Petals have often been the focus of attention in floral senescence. They provide a relatively simple organ that consists of many similar cell types and structures to those found in a leaf. Arguably petal senescence can therefore provide a useful model for studying the regulation of senescence in general, largely freed from environmental constraints.
This review starts by evaluating progress in identifying good models for studying petal senescence. It then focuses on those species in which most progress has been made to gauge our understanding of the key regulators of senescence, and prospects for regulating vase-life in ornamentals, particularly cut flowers.
The need for good models
Despite the many obvious advantages of studying petal senescence, our understanding of its regulation has been hampered by the lack of a good model species. This is due to several factors. Firstly two key features of petal senescence vary across species: sensitivity to ethylene and timing of abscission in relation to petal wilting. In many species floral senescence is coordinated by endogenous ethylene production often induced by pollination These include established models such as Arabidopsis thaliana, and many important ornamental species such as orchids (e.g. Phalaenopsis spp.), roses (Rosa spp.) and petunia (Petunia spp.) (Rogers 2006). In these species processes of remobilization, characteristic of senescence and leading to cell death, are coordinated by the ethylene signal. Furthermore, treatment with exogenous ethylene accelerates senescence, while treatment with inhibitors of ethylene synthesis or ethylene signalling, delay it (Stead and van Doorn 1994). In other species, however, little or no ethylene is produced in association with floral senescence. Furthermore floral senescence is insensitive to exogenous ethylene or ethylene inhibitors. This latter group also includes many important ornamental species such as lilies (Lilium spp.,) daffodils (Narcissus pseudonarcissus), tulips (Tulipa spp.) and Iris (Woltering and van Doorn 1988).
Independently of whether or not floral senescence is ethylene-regulated, species also differ in the progression of petal senescence in relation to abscission. Thus in some species, such as rose and magnolia (Magnolia grandiflora), petals abscise while still turgid. In contrast, in other species such as Iris and petunia, petals wilt and deteriorate while still attached to the plant (van Doorn and Woltering 2008). Petal deterioration is associated with variable levels of nutrient remobilization followed by PCD (Verlinden 2003; Rogers 2006). This is often associated with increasing vacuole size, followed by vacuole rupture in a process that resembles autophagic cell death (e.g. in Japanese morning glory, Ipomoea nil; Shibuya et al. 2009a). In some species that abscise turgid petals, cell death markers are not apparent prior to abscission (Yamada et al. 2007). PCD markers are however detected in other abscising species suggesting that the timing of PCD and abscission are independently regulated.
Thus identifying a single model that explains all the patterns of petal senescence is not attainable. However in other areas of plant development much has been achieved by focussing attention on Arabidopsis even though it cannot model all features of higher plant development. Unfortunately Arabidopsis has proven to be a somewhat limited model for studying detailed progression of petal senescence, although it has been used successfully to study petal abscission (Cho et al. 2008), and gross changes in petal senescence (Chen et al. 2011a). One of the key prerequisites of a good model for petal senescence is the ability to stage flowers based on key changes, usually associated to the development and maturation of the sexual organs. Unfortunately in Arabidopsis, although a developmental chart is available (Smyth et al. 1990) the detailed sequence of events is not as predictable as the chart would suggest. Furthermore Arabidopsis flowers abscise without substantial wilting, calling into question their role as a useful model for species whose petals wilt before abscission (Jones et al. 2009). Nevertheless, tools for Arabidopsis research outshine all other suitable models, and array data comparing young (stage 12) with older (stage 15) petals do provide the most complete dataset for the senescent petal transcriptome. The choice of these stages was validated by a clear up-regulation of the SAG12 gene expression in stage 15 petals (Wagstaff et al. 2009). SAG12 expression is an accepted marker for senescence in leaves and petals (Weaver et al. 1998). Its expression is closely linked to other petal senescence markers in wilting species where stages of senescence are better defined such as wallflowers (Erysimum linifolium; Price et al. 2008).
There are at least a couple of good alternative candidates as models for ethylene-sensitive petal senescence. Wallflowers are closely related to Arabidopsis and hence benefit from the use of cross-species microarrays. Approximately 80 % of genes tested cross-hybridised between wallflowers and Arabidopsis with very similar patterns of expression (Price et al. 2008). Wallflowers have the advantage over Arabidopsis of larger flowers with a range of petal colours, a predictable lifespan ending with wilting prior to abscission. Furthermore some varieties such as Bowles mauve are perennial (Price et al. 2008). However to date this species lacks many of the key tools required for an ideal model such as a transformation system, stocks of mutants and a genome sequence.
Petal senescence in petunia can be conveniently induced by pollination resulting in substantial wilting followed by abscission (Jones et al. 2009) thus allowing the study of both these processes. It boasts both transient and stable transformation systems (Conner et al. 2009; Chen et al. 2004), increasingly complete genome and transcriptome sequences (Zenoni et al. 2011) and an increasing bank of available mutant lines. As yet these resources have not been fully exploited, but the future seems rosy for the study of ethylene-sensitive petal senescence.
Brassica species also have potential as models for petal senescence. Cross species arrays are available and have been tested for leaf transcripts (Trick et al. 2009). Software has also been developed for facilitating analysis of cross species arrays (Hammond et al. 2005) and tested across Arabidopsis and Brassica oleracea. Interest in Brassica species has also provided resources including genome sequencing projects, genetic maps and markers, and proteomic and metabolomic datasets (http://www.brassica.info/resources.php). However although Brassica species are used as ornamentals for their colourful foliage, they are not directly related to flower ornamentals and thus information gained from them would need to be translated to ornamental crop species.
Progress in finding good models for ethylene-insensitive petal senescence however is lagging behind. Ethylene sensitivity in petunia can be suppressed by expression of the etr1-1 mutant allele, enabling the study of the processes that are essentially ethylene independent (Wilkinson et al. 1997). However, this is easily criticizable as a somewhat artificial system. Four o’clock (Mirabilis jalapa) is one of the few dicotyledonous species with apparently ethylene-independent petal senescence (Xu et al. 2007a). Although floral senescence is accelerated in this species by exogenous ethylene, little endogenous ethylene is produced during senescence and inhibitors of ethylene synthesis or action have little effect on its progression. However research on four o’clocks has not flourished even though a transformation system was established (Zaccai et al. 2007).
The lack of a good model has lead to the proliferation of papers studying floral senescence in a wide range of both monocotyledonous and dicotyledonous ornamental species (van Doorn and Woltering 2008; Rogers 2006), for which few tools and no complete genome sequences are available. However, this situation is rapidly improving with the advent of new much cheaper high throughput sequencing and the increasing use of virus induced gene silencing (VIGS; Edwards and Batley 2010; Senthil-Kumar and Mysore 2011).
Developmental and environmental signals
Although petal senescence is relatively unaffected by environmental signals there is an interaction between stress and senescence. Clearly the situation changes dramatically in cut flowers compared to those still attached to the plant. In petunias, growth at higher temperatures resulted in a shorter floral life span of unpollinated flowers attached to the plant (Gubrium et al. 2000). However, vase life of cut flowers was longer when plants had been grown at a higher compared to a lower temperature (Shvarts et al. 1997). In the cut flower Alstroemeria vase life was reduced by up to 20 % following stress treatments (Wagstaff et al. 2010) and was accompanied by substantial changes in gene expression.
Where comparisons have been made, longevity is generally reduced in cut flowers compared to those on the plant (e.g. in lilies: Arrom and Munné-Bosch (2010); carnation, Dianthus caryophyllus L.: van Staden and Dimalla 1980). In Lilium (Lilium longiflorum x Asiatic Hybrid var. Courier) antioxidants increased more sharply in the tepals of cut flowers although whether these might have a regulatory role is not known (Arrom and Munné-Bosch 2010). Furthermore changes in growth regulators differed between cut and uncut Lilium flowers (Arrom and Munné-Bosch 2012a). In particular levels of IAA and ABA were less in the outer petals of intact compared to cut flowers at anthesis. An interaction between environmental stress signals and plant growth regulators (PGRs) is also indicated by studies on transgenic petunias in which ethylene signalling has been down-regulated. In these flowers longevity is much more sensitive to growth at a higher temperature compared to wild type (Gubrium et al. 2000). More studies comparing global changes in transcription and metabolic status in stressed and unstressed and cut versus uncut flowers may reveal to what extent the regulatory signals and down-stream processes differ.
In species where multiple flowers are produced on the same stem, there is another important signal based on the interaction between the longevity of the less-developed and more-developed flowers. For example trimming younger flowers from Alstroemeria cymes increased longevity of the remaining flower (Chanasut et al. 2003). This is probably due to the important sink-source nutrient relationships within the inflorescence (Picchioni et al. 2007).
Nutrient status and energy depletion have been proposed as important regulators of floral senescence in cut flowers (Azad et al. 2008), and treatment with sucrose delayed floral senescence both in ethylene-sensitive (e.g. carnation: Hoeberichts et al. 2007) and ethylene-insensitive species (e.g. Alstroemeria: Chanasut et al. 2003; tulip: Azad et al. 2008; Lilium: Arrom and Munné-Bosch 2012a). In carnation (Hoeberichts et al. 2007) and wallflowers (Rogers, unpublished) sucrose treatment was very similar to treatment with the ethylene inhibitor STS indicating that it may be acting by repressing ethylene signal transduction. How the sucrose signal exerts its effects in ethylene-insensitive species remains to be determined. However, in Lilium at least, it affected the PGR balance (Arrom and Munné-Bosch 2012a) and thus may be acting via PGRs other than ethylene such as abscisic acid (ABA) or auxins.
Plant growth regulators (PGRs)
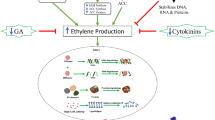
In species with ethylene-sensitive floral senescence, pollination induces a burst of ethylene production associated with up-regulation of ethylene biosynthetic enzymes: S-adenosylmethionine (SAM) synthase, 1-aminocyclopropane-1-carboxylic acid (ACC) synthase and ACC oxidase (Jones 2004; Hoeberichts et al. 2007). Ethylene production is associated with pollen tube growth in the style and later with fertilisation (Hoekstra and Weges 1986; ten Have and Woltering 1997). This results in autocatalytic ethylene production in the petals (Woodson and Lawton 1988; Graham et al. 2012). In rose (Müller et al. 2000) but not carnation (Shibuya et al. 2002), petal senescence is associated with changes in expression levels of the ethylene receptor genes. However expression of a mutated ethylene receptor gene (etr1-1) resulted in delayed floral senescence in several species including petunia. Components of the ethylene downstream signalling pathway (Yoo et al. 2009; Fig. 1) are expressed in petals and have also been targets for manipulation to delay flower senescence. Expression of CTR1-like genes in rose (Rosa hybrida) were up-regulated by exogenous ethylene (Ma et al. 2006), and expression of Rh-CTR1 also increased during floral senescence (Müller et al. 2002). Mutation of EIN2 delayed floral senescence in both petunia and Arabidopsis (Shibuya et al. 2004; Graham et al. 2012). EIN3-like gene expression increased in carnation during floral senescence and in response to ethylene (Iordachescu and Verlinden 2005), and its accumulation was delayed in sucrose-treated flowers. Down regulation (Tieman et al. 2001) or up-regulation (Yang et al. 2008) of tomato EIN3-like genes resulted in delayed or accelerated petal senescence respectively. This confirms a role for these genes too in ethylene-induced petal senescence.
Pollination and/or age related processes, resulting in a reduction of cytokinins and rise in ABA, trigger up-regulation of ethylene biosynthetic genes and an increase in ethylene sensitivity. Ethylene-responsive TFs are activated directly or via a MAP kinase signalling cascade. Other TFs are activated by auxin and other signals including probably ROS. Protease transcription is up-regulated presumably by a combination of transcription factors. KDEL proteases are localised to the ER and then ricinosomes, VPEs are targeted to the vacuole where they may be involved in processing of downstream proteases, metacaspases are activated and contribute to cell death. Autophagy genes are also up-regulated and presumably result in the formation of auotophagosomes which later fuse with the vacuole
In ethylene sensitive flowers this PGR clearly plays a decisive role in the initiation and coordination of senescence and ultimately cell death. However, even in these species, other PGRs also play important roles. In several species studied there was a senescence-associated fall in cytokinins and rise in ABA levels (Borohov et al. 1976; Mayak and Halevy 1970; Chang et al. 2003; van Staden et al. 1987). Both these changes seem to be connected to ethylene signalling. In carnations sensitivity to ethylene was reduced by treatment with cytokinin and inhibition of cytokinin metabolism delayed senescence (Taverner et al. 2000). Furthermore treatment of carnation petals with cytokinins blocked the conversion of exogenously supplied ACC to ethylene (Mor et al. 1983). In transgenic petunia, expression of the IPT gene, resulting in overproduction of cytokinin, led to a decrease in ethylene sensitivity and a reduction in ABA accumulation (Chang et al. 2003). Blocking ethylene signalling in petunia expressing etr1-1 abolished senescence-associated ABA accumulation in corollas (Jones et al. 2009) which is consistent with these results. Furthermore in carnation, exogenous ABA treatment accelerated senescence and increased both production of and sensitivity to ethylene (Ronen and Mayak 1981). However in hibiscus (Hibiscus rosa-sinensis L.;Trivellini et al. 2011) although exogenous ABA treatment accelerated flower senescence, it reduced ethylene biosynthesis and sensitivity. The complex relationship between ethylene, ABA and senescence in species with ethylene-sensitive floral senescence is therefore worthy of further investigation. There is a need for more physiological, transcriptomic and metabolomic studies in more species to make results more comparable and establish whether apparent species-specific differences are supported.
In ethylene insensitive cut flowers there is accumulating evidence that ABA may play an important role in petal senescence. Endogenous levels of ABA are found to rise in several species with ethylene insensitive floral senescence: e.g. daylily (Hemerocallis hybrid, cv. Stella d’Oro; Panavas et al. 1998), daffodil (Hunter et al. 2004a, b) and Lilium (Arrom and Munné-Bosch 2012a, b). Exogenous application of ABA to day lilies, resulted in an acceleration of natural senescence events including changes in transcriptional patterns (Panavas et al. 1998). Furthermore the effect could also be mimicked by treatment with sorbitol that induced an increase of endogenous ABA levels. Similar results were also obtained in daffodil (Hunter et al. 2004a) where it was further shown that the effects were ethylene-independent. However in daffodil, inhibitors of ABA biosynthesis did not delay senescence, although treatment with gibberellic acid (GA3) did mitigate the effects of exogenous ABA. This suggests that the ABA might be acting via GA. In all three species, however, the rise in endogenous ABA levels occurred quite late. This suggests that although this PGR may have a role in coordination of the later stages of petal senescence it is not likely to be involved in the initiation of the senescence programme.
The role of other PGRs such as auxin, jasmonates (JA) or salycilic acid (SA) in petal senescence remains unclear (van Doorn and Woltering 2008). Endogenous auxin levels fell in Lilium post-anthesis, while SA increased and JA remained stable (Arrom and Munné-Bosch 2012b). However in wallflowers free IAA increased during petal senescence (Mohd Salleh et al. unpublished results). Furthermore, in senescent Arabidopsis petals auxin biosynthetic and responsive gene expression was strongly up-regulated (Wagstaff et al. 2009). Application of exogenous auxin also accelerated senescence and ethylene production in carnation petals (Wulster et al. 1982; van Staden 1995).
Endogenous levels of reactive oxygen species (ROS) and ROS-related enzyme activities have been measured in numerous flowers with both ethylene-sensitive or insensitive senescence programmes (Rogers 2012). In general ROS levels rise and antioxidant levels fall during both forms of petal senescence. However, changes appear to start too late for ROS to be a key initiator of petal senescence. Nevertheless, this does not exclude a signalling role for redox status and or the mitochondrion in the coordination of later events. More information on the intracellular localisation of ROS production, levels of different ROS moieties, and on ROS signal transduction is needed in order to be able to resolve their role in petal senescence with confidence.
Signal transduction and transcription factors (TFs)
MAP kinases are central to signal transduction in many environmental and developmentally regulated cellular processes (Fiil et al. 2009), and senescence is no exception. The Arabidopsis genome contains 20 MAPKs, 10 MAPKKs, and more than 60 MAPKKKs (MAPK Group 2002). In leaf senescence MEKK1 bypasses the cascade by acting directly on the promoter of WRKY53, a senescence-associated TF (Miao et al. 2007). However MKK9 and MPK6 are also involved (Zhou et al. 2009). Less is known about MAPKs that might be involved in petal senescence. Expression of relatively few MAPK genes were up-or down regulated in Arabidopsis silique, leaf or petal senescence microarray analyses (Wagstaff et al. 2009) and none were specifically up or down-regulated in petal senescence. However expression of MPK11 was up-regulated in both petal and silique senescence and may be an interesting target for further study. This MAPK is also involved in both biotic and abiotic stresses and responds to both ROS and ABA, providing a potential link between these processes. In wallflowers, expression of a homologue of MKK9 was strongly up-regulated in both senescent leaves and petals (Price et al. 2008) and is also up-regulated in these tissues in Arabidopsis (Schmid et al. 2005). It is not surprising that at least some of the signal transduction mechanisms in leaf and petal are likely to be shared given the high levels of shared senescence-induced transcriptional changes in these tissues (Price et al. 2008; Wagstaff et al. 2009).
In ethylene-insensitive species MAPK-like transcripts have also been identified: in Alstroemeria a MAP3Ka-like transcript was up-regulated with tepal senescence (Wagstaff et al. 2010), although further work is needed to verify its expression pattern and function. Further work on MAP kinases involved in petal senescence is urgently needed.
Downstream of the MAP kinase cascade, TFs expressed during floral senescence have been identified in numerous EST, microarray, and SSH experiments including both ethylene-regulated species (e.g. wallflowers: Price et al. 2008; petunia: Liu et al. 2011; Arabidopsis: Wagstaff et al. 2009; carnation Hoeberichts et al. 2007) and ethylene-insensitive species (e.g. Alstroemeria: Breeze et al. 2004; Iris: van Doorn et al. 2003). A detailed analysis of the petal data from Wagstaff et al. (2009) revealed changes in the expression of 316 putative TFs of which 130 changed expression only in petal senescence and not in leaf or silique senescence. Forty-seven TF families were represented (as classified by Guo et al. 2005). The best represented was the AP2-EREB (ERF) family representing over 10 % of up-regulated TFs in petals (Fig. 2). Interesting differences are revealed by comparing TFs in three expression classes: (1) all those whose expression changed in petals (2) those whose expression was only up-regulated in petals but not in the other senescing tissues, and (3) those whose expression was up-regulated in all three senescing tissues (Fig. 2). Whereas the three most represented TF families up-regulated in class (1) were AP2-EREBP (ERF), NAC and WRKY accounting for almost 30 % of all the TFs expressed in this class, in class (2) the most represented families amongst TFs specifically up-regulated in petals were AP2-EREBP, homeobox (HB) and AUX-IAA (again accounting for about 30 % of the total). In contrast, in class (3) i.e. TFs up-regulated in all senescent tissues studied, the most represented families of up-regulated TFs were NAC (32 %), WRKY (18 %) and bZIP (11 %). Thus AP2-EREBP, HB and AUX-IAA TFs stand out as up-regulated TF classes that are most highly represented specifically in ethylene-regulated petal senescence and may be of significance in regulating specific senescence-associated events in this tissue. Although WRKY TFs were up-regulated in all three tissues, more family members were up-regulated in petals compared to leaves. Interestingly, WRKY53 which has been identified as an important hub regulator in leaf senescence, integrating MeJA and SA signals (Balbi and Devoto 2008; Miao and Zentgraf 2007; Miao et al. 2007), was not up-regulated in petal senescence.
Top 15 most represented transcription factor families expressed in Arabidopsis petals from the data of Wagstaff et al. (2009) whose expression was a up- or b down-regulated. (1) All TFs whose expression changed in petals (2) TFs whose expression only changed in petals (Stage 15 compared to stage 12) but not in senescent leaves (leaf 6: 6 weeks old/leaf 6: 4 weeks old) or senescent siliques (19-23/9-11 DAA), and (3) TFs whose expression changed in all three senescing tissues
The prevalence of the AP2-EREB TFs is not surprising given the established role of ethylene in these species. However the up-regulation of genes encoding AUX-IAA proteins raises again the issue of the role of auxin in petal senescence, which remains to be fully resolved in any species. AUX-IAA proteins are short-lived nuclear proteins forming part of an auxin co-receptor complex (Calderón Villalobos et al. 2012), that bind and repress auxin responsive factor (ARF) TFs. An auxin signal results in degradation of the AUX-IAA protein and de-repression of the ARFs.
Amongst the HB TFs up-regulated in petals was KNAT1, a member of the Class I KNOX family known to modulate cytokinin levels, and four members of the Class II KNOX family most of which are of unknown function (Hay and Tsiantis 2009). A KNAT1 homologue was also up-regulated in senescent carnation petals (Hoeberichts et al. 2007) indicating that the involvement of these genes in the regulation of petal cytokinin levels may be common to different species. Furthermore expression of ARR genes, part of the cytokinin signal transduction chain (To and Kieber 2008), was altered in Arabidopsis senescent petals. Two of these (ARR1 and ARR2) are associated with the vascular tissue and, as noted by Wagstaff et al. (2009), may be important in retaining function of this tissue during senescence-associated nutrient remobilization.
Further work will be needed to establish the role of all these TFs through analysis of over-expression and knock-out mutants, as well as screens to identify their down-stream targets. Work on petal senescence in Arabidopsis will benefit greatly from systems biology approaches to identify hubs in the transcriptional networks regulating senescence and stress responses in leaves (Breeze et al. 2011). However, an analysis of individual TFs has revealed an important role in floral senescence for at least one MADS box TF: AGL42 (FOREVER YOUNG FLOWER; FYF). Chen et al. (2011a) showed that this gene acts as a repressor of ethylene-mediated floral abscission and senescence in Arabidopsis. It acts upstream of BOP2 and IDA, genes involved in floral abscission. It would be interesting to analyse the effects of FYF perturbation on the progression of petal senescence in more detail. It is important to try to clarify whether its role in abscission is distinct from its role in senescence, or whether the two are linked. It will also be interesting to establish whether homologues in other species play a similar role. An orchid (Oncidium sp.) homologue of FYF also delayed petal senescence/abscission when expressed in Arabidopsis (Chen et al. 2011b). This suggests that this gene may indeed be important in other species whose floral senescence is ethylene regulated. Whether it might also play a role in ethylene-independent senescence remains to be tested.
In petunia flowers, the ERF (Ethylene-responsive element binding factor) family of TFs was studied in detail (Liu et al. 2011). This TF family is one of the largest in plants with 122 members in Arabidopsis belonging to 12 groups (Nakano et al. 2006). Thirteen ERF TFs belonging to four of these groups were expressed in petunia corollas, showing distinct patterns of expression. Group VII ERFs, previously associated with fruit ripening and senescence (El-Sharkawy et al. 2009; Yin et al. 2010) were also associated with corolla senescence in petunia. Their expression was up-regulated with ethylene, down-regulated by STS, and up-regulated by MeJA and ABA supporting the link between ethylene and ABA responses. Expression of three of the ERFs, belonging to group IX were down-regulated during corolla senescence and with exogenous ABA. The authors hypothesise that the reduction in expression during senescence may be linked to the rise in endogenous ABA levels (Vardi and Mayak 1989). Expression of several ERFs was also up-regulated by SA treatment indicating an interaction between ethylene and SA signalling in petunia corolla senescence. However, surprisingly none of the ERFs responded to exogenous cytokinin treatment, despite a clear link between cytokinins and ethylene in petunia flowers (Chang et al. 2003).
In other species with ethylene-regulated petal senescence similar TFs have also been identified. In wallflowers expression of genes with homology to WRKY75 and two NAC TFs were up-regulated in both leaf and petal senescence (Price et al. 2008) as they were also in Arabidopsis. In carnation petal senescence, as in Arabidopsis, expression of IAA-AUX TFs (Hoeberichts et al. 2007) was also up-regulated suggesting a link to auxin signaling that is supported by changes in the endogenous levels of this PGR in senescing petals (see above).
Much less is known about the expression of TFs in species where petal senescence is ethylene-insensitive although transcriptomic analyses have revealed a few (Table 1). In Alstroemeria transcripts for 21 TFs were identified from the 2000 ESTs sequenced (Wagstaff et al. 2010). Of these, one C2-H2, one HMG-box, two zinc-finger, one LIM-domain and one homeodomain TF changed in expression during tepal senescence. Expression of several others was also affected by stress treatments indicating shared regulation between these processes. Transcriptomic analyses also identified TFs from daffodil (AP2/ERF and NAC families; Hunter et al. 2002) and Iris (HLH-Zip; van Doorn et al. 2003). It is interesting that WRKY TFs have not been identified from these species so far, although the promoter of a putative E3 ubiquitin ligase that was highly up-regulated in four o’clock senescent petals included putative binding sites for several TF classes including WRKY TFs (Xu et al. 2007b). Cheaper transcriptomic sequencing should help to establish whether WRKY TFs are indeed important in ethylene-independent floral senescence systems.
Another layer of regulation that is becoming increasingly studied is provided by microRNAs (miRNAs). These act as negative regulators and are involved in the regulation of many stages of plant development (Voinnet 2009) including senescence. The leaf-senescence NAC TF ORE1 was found to be regulated by a complex trifurcate feed-forward mechanism involving a miRNA, mi R164 (Kim et al. 2009), and EIN2 from the ethylene-mediated senescence pathway. By negatively regulating ORE1 expression, miR164 may be fine tuning the timing of leaf senescence promoted by EIN2. EIN2 then negatively regulates miR164 as senescence progresses. In fact mathematical modelling showed that persistent rise in EIN2 results in activation of SAG12 supporting this fine tune control. TCP TFs are also miRNA regulated and participate in the regulation of leaf senescence (Schommer et al. 2008). Furthermore some miRNAs are highly conserved across species (Cuperus et al. 2011) providing opportunities for manipulation of developmental programmes through transformation. Thus a fruitful research area is likely to be an investigation of miRNAs regulating floral senescence. It is possible that in petals as well as leaves they may be involved in the fine tuning of senescence progression to orchestrate the complex remobilization processes and delaying cell death as required.
Downstream cell death regulation
Although cell death often starts before visible signs of petal deterioration are evident (Rogers 2006), it is clear from careful analyses of cellular structure (e.g. Wagstaff et al. 2003; Battelli et al. 2011) that cell death is not uniform across the petal. In many species mesophyll cells degenerate completely in older petals while the epidermal cells remain essentially intact. In Iris this differential timing coincides with a closure of plasmodesmata, interrupting the movement of nutrients, PGRs and other small molecules (van Doorn et al. 2003). This may result in effective starvation of the cells and lead to ATP depletion triggering cell death as discussed above.
Furthermore in several species such as Iris, Lilium and Alstroemeria (van Doorn et al. 2003; Battelli et al. 2011; Wagstaff et al. 2003) cell death starts at the petal margins and spreads inwards. The assumption is that this is part of a remobilization programme that favours the survival of the cells nearer the vascular tissue to enable transport of nutrients out of the petal. This presumes a long-range coordination of cellular responses, perhaps involving cytokinins as suggested by the up-regulation of the ARR genes discussed above.
At a cellular level petal PCD resembles closely autophagic (van Doorn and Woltering 2005; Shibuya et al. 2009a) or ‘autolytic’ (van Doorn 2011) cell death. This process is characterised by the formation of small vacuoles that fuse to merge with the central vacuole. The vacuole then ruptures releasing hydrolases which rapidly clear the remaining cytoplasm (van Doorn 2011). Auotophagosomes have been detected in petals (e.g. Japanese morning glory: Shibuya et al. 2009b) and genes encoding components of the conserved autophagic regulatory mechanism are expressed in the petals of several species (e.g. Arabidopsis: Wagstaff et al. 2009; Japanese morning glory: Shibuya et al. 2009b; Alstroemeria: Wagstaff et al. 2010). However to what extent the autophagic mechanism is a requirement for petal cell death is not clear. In Arabidopsis knockouts of autophagy genes, leaf senescence is accelerated, but it has been proposed that this might also be due to an indirect stress effect rather than an indication that autophagy delays senescence (van Doorn and Woltering 2008). A closer investigation of petal senescence phenotype in autophagy gene knockouts would be very useful.
In animal PCD a specific class of proteases, caspases play a key regulatory role both in the initiation and execution of cell death (Sanmartín et al. 2005). Caspase activity has been detected in a number of plant cell death systems (Bonneau et al. 2008), and recently in Lilium (L. longiflorum) petals (Battelli et al. 2011), however caspase genes with homology to those in animals have not been found in plant genomes. In plants, vacuolar processing enzymes (VPEs) are a family of proteases that share structural features with caspases and have caspase-1 activity (Rojo et al. 2004). They localise to the vacuole and are involved in maturation of hydrolases. In tobacco petals expression of a VPE gene increased 100-fold with age, and VPE genes are also up-regulated in senescent petals of carnation (Hoeberichts et al. 2007), daffodil (Hunter et al. 2002), Japanese morning glory (Yamada et al. 2009) and L. longiflorum (Rogers, unpublished). Furthermore of the four VPEs in Arabidopsis, expression of two (β and γ) was up-regulated in senescent petals (Wagstaff et al. 2009). Further work is clearly needed to determine whether perturbation of VPE expression affects petal senescence and to identify VPE substrates in petals. This will shed light on whether VPEs might be playing a regulatory role.
Metacaspase genes also form a large gene family in plants and resemble part of the caspase coding sequence (Tsiatsiani et al. 2011). Moreover, their expression in plants is associated with cell death processes (Coll et al. 2010). A metacaspase gene was expressed in petunia petals (Jones et al. 2009), but its role remains to be established. Expression of two Arabidopsis metacaspase genes (AtMC9 and AtNC3) was up-regulated in senescent petals (Wagstaff et al. 2009) indicating that they may play a role in petal senescence. However as they comprise a large gene family unravelling their roles may take some time.
Petal senescence in many species is also associated with an increase in cysteine protease activity. In some species, such as Iris (Pak and van Doorn 2005) and Sandersonia (Sandersonia aurantiaca; Eason et al. 2002), treatment of flowers with inhibitors of cysteine proteases delayed visible senescence. This indicates a potential regulatory role for these proteases. In several species up-regulation of papain-family cysteine protease genes with a C-terminal KDEL sequence has been reported (E.g. Sandersonia, Eason et al. 2002; day lily, Valpuesta et al. 1995). Expression of some of these genes e.g. Alstroemeria ALSCYP1 was up-regulated before visible signs of senescence (Wagstaff et al. 2002) suggesting that they might participate in a proteolytic cascade activating other down-stream proteases. The KDEL sequence anchors the proteases to the endoplasmic reticulum but they are also associated with ribosome studded vesicles known as ricinosomes (Schmid et al. 1999). Ricinosomes deliver their cargo of protease directly to the cytoplasm following vacuolar rupture and were also detected in senescing day lily petals (Schmid et al. 1999). However, they have not been reported in other species. Identifying the substrates for these early-expressed proteases and defining whether they are indeed released directly into the cytoplasm, are important goals to establish their role in petal cell death. In petunia, down-regulation of cysteine protease genes did not affect senescence progression indicating likely functional redundancy in this gene family (Jones et al. 2009) which will complicate their analysis by a genetic route.
Prospects for understanding and controlling senescence in ornamentals
Our understanding of the genes and processes regulating petal senescence in species where these are coordinated by ethylene is progressing fast. Although many of the pieces are still missing, a tentative model can be assembled to incorporate available data on the genes and processes involved, as discussed above (Fig. 1). Key tools are also becoming available in petunia for this to become a testable model for this group of species. The next step will then be to test and transfer this understanding to other species of commercial relevance and exploit control points to extend vase life. Already several different treatments are available for extending vase life in these species (Rogers and Stead 2011) involving down-regulation of ethylene responses through chemical treatments. Furthermore, prospects seem good for refining chemical approaches e.g. through the use of nanosponges (Seglie et al. 2011). In addition an increasing number of these species can be transformed (Chandler and Tanaka 2007), and transgenic approaches for reducing ethylene signalling can be refined through the use of more specific promoters. Other targets may also be of interest such as FYF or hub TFs driven by tissue specific or senescence induced promoters such as the SAG12 promoter (Chang et al. 2003).
Progress in understanding ethylene-independent floral senescence has lagged behind. Given that the PGR regulation in these species is likely to be very complex, there seems to be much more promise in the manipulation of the signalling and TF intermediaries. It will be crucial to determine whether information from the more tractable ethylene-regulated species can be directly transferred. This will depend on whether the key regulators are shared across the physiological groups. Comprehensive transcriptomic sequencing projects using new high throughput sequencing approaches will be very powerful especially in species such as Lilium where genome sizes of up to 127 000 Mb (Zonneveld et al. 2005) make genome sequencing projects unrealistic. This approach will provide a global view of gene expression changes crucially in up-stream signalling and TFs. This would then enable the use of systems approaches now available for leaf senescence in Arabidopsis. How this information is then used will depend on the species. A major challenge is to develop robust transformation protocols for a transgenic approach in these less tractable species. In some species, however, marker-assisted breeding programmes may be the most immediate way of exploiting the new information.
References
Arrom L, Munné-Bosch S (2010) Tocopherol composition in flower organs of Lilium and its variations during natural and artificial senescence. Plant Sci 179:289–295
Arrom L, Munné-Bosch S (2012a) Sucrose accelerates flower opening and delays senescence through a hormonal effect in cut lily flowers. Science 188–189:41–47
Arrom L, Munné-Bosch S (2012b) Hormonal changes during flower development in floral tissues of Lilium. Planta 236:343–354
Azad AK, Ishikawa T, Sawa Y, Shibata H (2008) Intracellular energy depletion triggers programmed cell death during petal senescence in tulip. J Exp Bot 59:2085–2095
Balbi V, Devoto A (2008) Jasmonate signalling network in Arabidopsis thaliana: crucial regulatory nodes and new physiological scenarios. New Phytol 177:301–318
Battelli R, Lombardi L, Rogers HJ, Picciarelli P, Lorenzi R, Ceccarelli N (2011) Changes in ultrastructure, protease and caspase-like activities during flower senescence in Lilium longiflorum. Plant Sci 180:716–725
Bonneau L, Ge Y, Drury GE, Gallois P (2008) What happened to plant caspases? J Exp Bot 59:491–499
Borohov A, Tirosh T, Halevy AH (1976) Abscisic acid content of senescing petals on cut rose flowers as affected by sucrose and water stress. Plant Physiol 58:175–178
Breeze E, Wagstaff C, Harrison E, Bramke I, Rogers HJ, Stead AD, Thomas B, Buchanan-Wollaston V (2004) Gene expression patterns to define stages of post-harvest senescence in Alstroemeria petals. Plant Biotechnol J 2:155–168
Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim Y, Penfold CA, Jenkins D, Zhang C, Morris K, Jenner C, Jackson S, Thomas B, Tabrett A, Legaie R, Moore JD, Wild DL, Ott S, Rand D, Beynon J, Denby K, Mead A, Buchanan-Wollaston V (2011) High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23:873–894
Calderón Villalobos LIA, Lee S, De Oliveira C, Ivetac A, Brandt W, Armitage L, Sheard LB, Tan X, Parry G, Mao H, Zheng N, Napier R, Kepinski S, Estelle M (2012) A combinatorial TIR1/AFB–Aux/IAAco-receptor system for differential sensing of auxin. Nature Chem Biol 8:477–485
Chanasut U, Rogers HJ, Leverentz MK, Griffiths G, Thomas B, Wagstaff C, Stead AD (2003) Increasing flower longevity in Alstroemeria. Postharvest Biol Technol 29:325–333
Chandler S, Tanaka Y (2007) Genetic modification in floriculture. CRC Crit Rev Plant Sci 26:169–197
Chang H, Jones ML, Banowetz GM, Clark DG (2003) Overproduction of cytokinins in petunia flowers transformed with PSAG12: IPT delays corolla senescence. Plant Physiol 132:2174–2183
Chen J-C, Jiang C-Z, Gookin TE, Hunter DA, Clark DG, Reid MS (2004) Chalcone synthase as a reporter in virus-induced gene silencing studies of flower senescence. Plant Mol Biol 55:521–530
Chen MK, Hsu WH, Lee PF, Thiruvengadam M, Chen HI, Yang CH (2011a) The MADS box gene, FOREVER YOUNG FLOWER, acts as a repressor controlling floral organ senescence and abscission in Arabidopsis. Plant J 68:168–185
Chen MK, Lee PF, Yang CH (2011b) Delay of flower senescence and abscission in Arabidopsis transformed with an FOREVER YOUNG FLOWER homolog from Oncidium orchid. Plant Signal Behav 6:1841–1843
Cho SK, Larue CT, Chevalier D, Wang H, Jinn T-L, Zhang S, Walker JC (2008) Regulation of floral organ abscission in Arabidopsis thaliana. Proc Natl Acad Sci USA 105:15629–15634
Coll NS, Vercammen D, Smidler A, Clover C, Van Breusegem F, Dangl JL, Epple P (2010) Arabidopsis Type I metacaspases control cell death. Science 330:1393–1397
Conner AJ, Albert NW, Deroles SC (2009) Transformation and regeneration of petunia. In: Gerats T, Strommer J (eds) Petunia evolutionary, developmental and physiological genetics. Springer, New York, pp 301–324
Cuperus JT, Fahlgren N, Carrington JC (2011) Evolution and functional diversification of MIRNA genes. Plant Cell 23:431–442
Eason JR, Ryan DJ, Pinkney TT, O’Donoghu EM (2002) Programmed cell death during flower senescence: isolation and characterization of cysteine proteinases from Sandersonia aurantiaca. Funct Plant Biol 29:1055–1064
Edwards D, Batley J (2010) Plant genome sequencing: applications for crop improvement. Plant Biotechnol J 8:2–9
El-Sharkawy I, Sherif S, Mila I, Bouzayen M, Jayasankar S (2009) Molecular characterization of seven genes encoding ethylene responsive transcriptional factors during plum fruit development and ripening. J Exp Bot 60:907–922
Fiil BK, Petersen K, Petersen M, Mundy J (2009) Gene regulation by MAP kinase cascades. Curr Opin Plant Biol 12:615–621
Graham LE, Schippers JHM, Dijkwel PP, Wagstaff C (2012) Ethylene and senescence processes. Annu Plant Rev 44:305–341
Gubrium EK, Clevenger DJ, Clark DG, Barrett JE, Nell TA (2000) Reproduction and horticultural performance of transgenic ethylene-insensitive petunias. J Am Soc Hortic Sci 125:277–281
Guo A, He K, Liu D, Bai S, Gu X, Wei L, Luo J (2005) DATF: a database of Arabidopsis transcription factors. Bioinformatics 21:2568–2569
Hammond JP, Broadley MR, Craigon DJ, Higgins J, Emmerson ZF, Townsend HJ, White PJ, May ST (2005) Using genomic DNA-based probe-selection to improve the sensitivity of high-density oligonucleotide arrays when applied to heterologous species. Plant Method 1:10
Hay A, Tsiantis M (2009) A KNOX family TALE. Curr Opin Plant Biol 12:593–598
Hoeberichts FA, van Doorn WG, Vorst O, Hall RD, van Wordragen MF (2007) Sucrose prevents up-regulation of senescence-associated genes in carnation petals. J Exp Bot 58:2873–2885
Hoekstra FA, Weges R (1986) Lack of control by early pistillate ethylene of the accelerated wilting of Petunia hybrida flowers. Plant Physiol 80:403–408
Hunter DA, Steele BC, Reid MS (2002) Identification of genes associated with perianth senescence in daffodil (Narcissus pseudonarcissus L. ‘Dutch Master’). Plant Sci 163:13–21
Hunter DA, Ferrante A, Vernieri P, Reid MS (2004a) Role of abscisic acid in perianth senescence of daffodil (Narcissus pseudonarcissus ‘Dutch Master’). Physiol Plant 121:313–321
Hunter DA, Yi MF, Xu XJ, Reid MS (2004b) Role of ethylene in perianth senescence of daffodil (Narcissus pseudonarcissus L. ‘Dutch Master’). Postharvest Biol Technol 32:269–280
Iordachescu M, Verlinden S (2005) Transcriptional regulation of three EIN3-like genes of carnation (Dianthus caryophyllus L. cv. Improved White Sim) during flower development and upon wounding, pollination, and ethylene exposure. J Exp Bot 56:2011–2018
Jones ML (2004) Changes in gene expression during senescence. In: Nooden LD (ed) Plant cell death processes. Elsevier, Amsterdam, pp 51–71
Jones ML, Stead AD, Clark DG (2009) Petunia flower senescence. In: Gerats T, Strommer J (eds) Petunia: evolutionary, developmental and physiological genetics. Springer, New York, pp 301–324
Kim JH, Woo HR, Kim J, Lim PO, Lee IC, Choi SH, Hwang D, Nam HG (2009) Trifurcate feed-forward regulation of age-dependent cell death involving mir164 in Arabidopsis. Science 323:1053–1057
Liu J, Li J, Wang H, Fu Z, Liu J, Yu Y (2011) Identification and expression analysis of ERF transcription factor genes in petunia during flower senescence and in response to hormone treatments. J Exp Bot 62:825–840
Ma N, Tan H, Liu X, Xue J, Li Y, Gao J (2006) Transcriptional regulation of ethylene receptor and CTR genes involved in ethylene-induced flower opening in cut rose (Rosa hybrida) cv Samantha. J Exp Bot 57:2763–2773
MAPK Group (2002) Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci 7:301–308
Mayak S, Halevy AH (1970) Cytokinin activity in rose petals and its relation to senescence. Plant Physiol 46:497–499
Miao Y, Zentgraf U (2007) The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell 19:819–830
Miao Y, Laun TM, Smykowski A, Zentgraf U (2007) Arabidopsis MEKK1 can take a short cut: it can directly interact with senescence-related WRKY53 transcription factor on the protein level and can bind to its promoter. Plant Mol Biol 65:63–76
Mor Y, Spiegelstein H, Halevy AH (1983) Inhibition of ethylene biosynthesis in carnation petals by cytokinin. Plant Physiol 71:541–546
Müller R, Stummann BM, Serek M (2000) Characterization of an ethylene receptor family with differential expression in rose (Rosa hybrida L.) flowers. Plant Cell Rep 19:1232–1239. doi:10.1007/s002990000251
Müller R, Owen CA, Xue Z-T, Welander M, Stummann BM (2002) Characterization of two CTR-like protein kinases in Rosa hybrida and their expression during flower senescence and in response to ethylene. J Exp Bot 53:1223–1225
Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140:411–432
Pak C, van Doorn WG (2005) Delay of Iris flower senescence by protease inhibitors. New Phytol 165:473–480
Panavas T, Walker ER, Rubinstein B (1998) Possible role of abscisic acid in senescence of daylily petals. J Exp Bot 49:1987–1997
Picchioni GA, Mackay WA, Valenzuela-Vázquez M (2007) Correlative supply and demand functions in Lupinus havardii: a forgotten side of cut flower physiology? J Am Soc Hortic Sci 132:102–111
Price AM, Aros Orellana DF, Stevens R, Acock R, Buchanan-Wollaston V, Stead AD, Rogers HJ (2008) A comparison of leaf and petal senescence in wallflowers (Erysimum linifolium) reveals common and distinct patterns of gene expression and physiology. Plant Physiol 147:1898–1912
Rogers HJ (2006) Programmed cell death in floral organs: how and why do flowers die? Ann Bot 97:309–315
Rogers HJ (2012) Is there a important role for reactive oxygen species and redox regulation during floral senescence? Plant, Cell Environ 35:217–233
Rogers HJ, Stead AD (2011) Petal abscission: falling to their death or cast out to die? In: Yaish MW (ed) The flowering process and its control in plants: gene expression and hormone interaction. Research Signpost, Kerala, pp 229–258
Rojo E, Martín R, Carter C, Zouhar J, Pan S, Plotnikova J, Jin H, Paneque M, Sánchez-Serrano JJ, Baker B, Ausubel FM, Raikhel NV (2004) VPE gamma exhibits a caspase-like activity that contributes to defense against pathogens. Curr Biol 14:1897–1906
Ronen M, Mayak S (1981) Interrelationship between abscisic acid and ethylene in the control of senescence processes in carnation flowers. J Exp Bot 32:759–765. doi:10.1093/jxb/32.4.759
Sanmartín M, Jaroszewski L, Raikhel NV, Rojo E (2005) Caspases, regulating death since the origin of life? Plant Physiol 137:841–847
Schmid M, Simpson D, Gietl C (1999) Programmed cell death in castor bean endoserm is associated with the accumulation and release of a cysteine endopeptidase from ricinosomes. Proc Natl Acad Sci USA USA 96:14159–14164
Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann J (2005) A gene expression map of Arabidopsis development. Nat Genet 37:501–506
Schommer C, Palatnik JF, Aggarwal P, Chételat A, Cubas P, Farmer EE, Nath U, Weigel D (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6:e230
Seglie L, Martina K, Devecchi M, Roggero C, Trotta F, Scariot V (2011) The effects of 1-MCP in cyclodextrin-based nanosponges to improve the vase life of Dianthus caryophyllus cut flowers. Postharvest Biol Technol 59:200–205
Senthil-Kumar M, Mysore KS (2011) New dimensions for VIGS in plant functional genomics. Trends Plant Sci 16:656–665. doi:10.1016/j.tplants.2011.08.006
Shibuya K, Nagata M, Tanikawa N, Yoshioka T, Hashiba T, Satoh S (2002) Comparison of mRNA levels of three ethylene receptors in senescing flowers of carnation (Dianthus caryophyllus L.). J Exp Bot 53:399–406
Shibuya K, Barry KG, Ciardi JA, Loucas HM, Underwood BA, Nourizadeh S, Ecker JR, Klee HJ, Clark DG (2004) The central role of PhEIN2 in ethylene responses throughout plant development in petunia. Plant Physiol 136:2900–2912
Shibuya K, Yamada T, Ichimura K (2009a) Autophagy regulates progression of programmed cell death during petal senescence in Japanese morning glory. Autophagy 5:546–547
Shibuya K, Yamada T, Suzuki T, Shimizu K, Ichimura K (2009b) InPSR26, a putative membrane protein, regulates programmed cell death during petal senescence in Japanese morning glory. Plant Physiol 149:816–824
Shvarts M, Weiss D, Borochov A (1997) Temperature effects on growth, pigmentation and post-harvest longevity of petunia flowers. Sci Hortic 69:217–227
Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabídopsis. Plant Cell 2:755–767
Stead AD, van Doorn WG (1994) Strategies of flower senescence-a review. In: Scott RJ, Stead AD (eds) Molecular and cellular aspects of plant reproduction. Cambridge University Press, Cambridge, pp 215–238
Taverner EA, Letham DS, Wang J, Cornish E (2000) Inhibition of carnation petal inrolling by growth retardants and cytokinins. Aus J Plant Physiol 27:357–362
ten Have A, Woltering EJ (1997) Ethylene biosynthetic genes are differentially expressed during carnation (Dianthus caryophyllus L.) flower senescence. Plant Mol Biol 34:89–97
Thomas H, Ougham HJ, Wagstaff C, Stead AD (2003) Defining senescence and death. J Exp Bot 54:1127–1132
Tieman DM, Ciardi JA, Taylor MG, Klee HJ (2001) Members of the tomato LeEIL gene family are functionally redundant and regulate ethylene responses throughout plant development. Plant J 26:47–58
To JPC, Kieber JJ (2008) Cytokinin signaling: two-components and more. Trends Plant Sci 13:85–92
Trick M, Cheung F, Drou N, Fraser F, Lobenhofer E K, Hurban P, Magusin A, Town CD and Bancroft I (2009) A newly-developed community microarray resource for transcriptome profiling in Brassica species enables the confirmation of Brassica-specific expressed sequences. BMC Plant Biol: 50
Trivellini A, Ferrante A, Vernieri P, Serra G (2011) Effects of abscisic acid on ethylene biosynthesis andperception in Hibiscus rosa-sinensis L. flower development. J Exp Bot 62:5437–5452
Tsiatsiani L, Van Breusegem F, Gallois P, Zavialov A, Lam E, Bozhkov PV (2011) Metacaspases. Cell Death Diff 18:1279–1288
Valpuesta V, Lange NE, Guerrero C, Reid MS (1995) Upregulation of a cysteine protease accompanies the ethylene-insensitive senescence of daylily (Hemerocallis) flowers. Plant Mol Biol 28:575–582
van Doorn WG (2011) Classes of programmed cell death in plants, compared to those in animals. J Exp Bot 62:4749–4761
van Doorn WG, Woltering EJ (2004) Senescence and programmed cell death: substance or semantics? J Exp Bot 55:2147–2153
van Doorn WG, Woltering EJ (2005) Many ways to exit? Cell death categories in plants. Trends Plant Sci 10:117–122
van Doorn WG, Woltering EJ (2008) Physiology and molecular biology of petal senescence. J Exp Bot 59:453–480
van Doorn WG, Balk PA, van Houwelingen AM, Hoeberichts FA, Hall RD, Vorst O, van der Schoot C, van Wordragen MF (2003) Gene expression during anthesis and senescence in Iris flowers. Plant Mol Biol 53:845–863
van Staden J (1995) Hormonal control of carnation flower senescence. Acta Hortic 405:232–239
van Staden J, Dimalla GG (1980) The effect of silver thiosulphate preservative on the physiology of cut carnations. II. Influence on endogenous cytokinin. Z Pflanzenphysiol 99:19–26
van Staden J, Featonby-Smith BC, Mayak S, Spiegelstein H, Halevy AH (1987) Cytokinins in cut carnation flowers. II. Relation between endogenous ethylene and cytokinin levels in the petals. Plant Growth Regul 5:75–86
Vardi Y, Mayak S (1989) Involvement of abscisic acid during water stress and recovery in petunia flowers. Acta Hortic 261:107–112
Verlinden S (2003) Changes in mineral concentrations in petunia corollas during development and senescence. Hortic Sci 38:71–74
Voinnet O (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136:669–687
Wagstaff C, Leverentz MK, Griffiths G, Thomas B, Chanasut U, Stead AD, Rogers HJ (2002) Protein degradation during senescence of Alstroemeria petals. J Exp Bot 53:233–240
Wagstaff C, Malcolm P, Rafiq A, Leverentz M, Griffiths G, Thomas B, Stead A, Rogers HJ (2003) Programmed cell death (PCD) processes begin extremely early in Alstroemeria petal senescence. New Phytol 160:49–59
Wagstaff C, Yang TJW, Stead AD, Buchanan-Wollaston V, Roberts JA (2009) A molecular and structural characterization of senescing Arabidopsis siliques and comparison of transcriptional profiles with senescing petals and leaves. Plant J 57:690–705
Wagstaff C, Bramke I, Breeze E, Thornber S, Harrison L, Thomas B, Buchanan-Wollaston V, Stead AD, Rogers HJ (2010) A unique group of genes respond to cold drought stress in cut Alstroemeria flowers whereas ambient drought stress accelerates developmental expression patterns. J Exp Bot 61:2905–2921
Weaver LM, Gan S, Quirino B, Amasino RM (1998) A comparison of the expression patterns of several senescence associated genes in response to stress and hormone treatment. Plant Mol Biol 37:455–469
Wilkinson JQ, Lanahan MB, Clark DG, Bleecker AB, Chang C, Meyerowitz EM, Klee HJ (1997) A dominant mutant receptor from Arabidopsis confers ethylene insensitivity in heterologous plants. Nat Biotechnol 15:444–447
Woltering EJ, van Doorn WG (1988) Role of ethylene and senescence of petals: morphological and taxonomical relationships. J Exp Bot 39:1605–1616
Woodson WR, Lawton KA (1988) Ethylene-induced gene expression in carnation petals. Relationship to autocatalytic ethylene production and senescence. Plant Physiol 87:498–503
Wulster G, Sacalis J, Janes HW (1982) Senescence in isolated carnation petals effects of indoleacetic acid and inhibitors of protein synthesis. Plant Physiol 70:1039–1043
Xu X, Gookin T, Jiang C-Z, Reid M (2007a) Genes associated with opening and senescence of Mirabilis jalapa flowers. J Exp Bot 58:2193–2201
Xu X, Jiang C-Z, Donnelly L, Reid MS (2007b) Functional analysis of a RING domain ankyrin repeat protein that is highly expressed during flower senescence. J Exp Bot 58:3623–3630
Yamada T, van Ichimura K, Doorn WG (2007) Relationship between petal abscission and programmed cell death in Prunus yedoensis and Delphinium. Planta 226:1195–1205
Yamada T, Ichimura K, Kanekatsu M, van Doorn WG (2009) Homologues of genes associated with programmed cell death in animal cells are differentially expressed during senescence of Ipomoea nil petals. Plant Cell Physiol 50:610–625
Yang TF, Gonzalez-Caranza ZH, Maunders MJ, Roberts JA (2008) Ethylene and the regulation of senescence processes in transgenic Nicotiana sylvestris plants. Ann Bot 101:301–310
Yin X, Allan AC, Chen K, Ferguson IB (2010) Kiwifruit EIL and ERF genes involved in regulating fruit ripening. Plant Physiol 153:1280–1292
Yoo SD, Cho Y, Sheen J (2009) Emerging connections in the ethylene signaling network. Trends Plant Sci 14:270–279
Zaccai M, Jia G, Chen X, Genis O, Feibin D, Gesua R (2007) Regeneration and transformation system in Mirabilis jalapa. Sci Hortic 111:304–309
Zenoni S, D’Agostino N, Tornielli GB, Quattrocchio F, Chiusano ML, Koes R, Zethof J, Guzzo F, Delledonne M, Frusciante L, Gerats T, Pezzotti M (2011) Revealing impaired pathways in the an11 mutant by high-throughput characterization of Petunia axillaris and Petunia inflata transcriptomes. Plant J 68:11–27
Zhou C, Cai Z, Guo Y, Gan S (2009) An Arabidopsis mitogen-activated protein kinase cascade, MKK9-MPK6, plays a role in leaf senescence. Plant Physiol 150:167–177
Zonneveld BJM, Leitch IJ, Bennett MD (2005) First nuclear DNA amounts in more than 300 angiosperms. Ann Bot 96:229–244
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rogers, H.J. From models to ornamentals: how is flower senescence regulated?. Plant Mol Biol 82, 563–574 (2013). https://doi.org/10.1007/s11103-012-9968-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-012-9968-0