Abstract
Plant heme oxygenase (HO) catalyzes the oxygenation of heme to biliverdin, carbon monoxide (CO), and free iron (Fe2+)—and Arabidopsis and rice (Oryza sativa) HOs are involved in light signaling. Here, we identified that the rice PHOTOPERIOD SENSITIVITY 5 (SE5) gene, which encoded a putative HO with high similarity to HO-1 from Arabidopsis (HY1), exhibited HO activity, and localized in the chloroplasts. Rice RNAi mutants silenced for SE5 were generated and displayed early flowering under long-day conditions, consistent with phenotypes of the null mutation in SE5 gene reported previously (se5 and s73). The herbicide methyl viologen (MV), which produces reactive oxygen species (ROS), was applied to determine whether SE5 regulates oxidative stress response. Compared with wild-type, SE5 RNAi transgenic plants aggravated seedling growth inhibition, chlorophyll loss and ROS overproduction, and decreased the transcripts of some representative antioxidative genes. By contrast, administration of exogenous CO partially rescued corresponding MV hypersensitivity in the SE5 RNAi plants. Alleviation of seed germination inhibition, chlorophyll loss and ROS overproduction, as well as the induction of antioxidant defense were further observed when SE5 or HY1 was overexpressed in transgenic Arabidopsis plants, indicating that SE5 may be useful for molecular breeding designed to improve plant tolerance to oxidative stress.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
It is well established that heme oxygenase (HO; EC 1.14.99.3) catalyzes heme degradation to generate carbon monoxide (CO), biliverdin (BV) and free iron (Fe2+) (Wilks 2002). Genes encoding HOs have been isolated from a wide variety of organisms including mammals, higher plants, red algae, cryptophytes, cyanobacteria and pathogenic bacteria (Wilks and Schmitt 1998; Muramoto et al. 1999; Zhang et al. 2005; Shekhawat and Verma 2010). In addition to well-defined metabolic functions of heme catabolism in animals (e.g. differentiation, hemopoiesis, erythrocyte turnover and Fe trafficking), HO-1, an inducible form of HO, is generally accepted as a mediator of cyto- and tissue protection against a wide variety of injuries and cellular stresses (Ryter et al. 2006).
In higher plants, because BV is the precursor of the phytochrome chromophore, HO is believed to be necessary for proper photomorphogenesis and/or light signaling (Izawa et al. 2000; Davis et al. 2001). Many HO genes in various plant species have been cloned or identified, e.g. Arabidopsis thaliana, rice (Oryza sativa), pea, maize, tomato, pine, sorghum, alfalfa, soybean and rapeseed (Emborg et al. 2006; Cao et al. 2011; Fu et al. 2011). In Arabidopsis, four HO proteins were clustered into two categories: the HO1 and HO2 subfamilies, based on their amino-acid sequence similarity (Davis et al. 2001; Emborg et al. 2006; Gisk et al. 2010). The HO1 subfamily comprises HY1 (HO1), HO3 and HO4, which all contain the canonical HO active site; whereas HO2—distinguished by the lack of a positionally conserved histidine (Davis et al. 2001; Emborg et al. 2006)—is the only member of the HO2 subfamily. Although Arabidopsis HO2 is not considered a true HO and the phenotype of ho2 mutant at least points towards a function within tetrapyrrole metabolism (Gisk et al. 2010), the four Arabidopsis HOs involved in the phytochrome synthesis pathway (Davis et al. 1999; Muramoto et al. 1999, 2002) and the requirement of HY1 in salinity acclimation and UV-C response (Xie et al. 2011, 2012) have also been reported recently. Nevertheless, more detailed evidence for the functionality of the individual members of HO even in Arabidopsis should be investigated in future (Shekhawat and Verma 2010; Shekhawat et al. 2011; Gisk et al. 2012). PHOTOPERIOD SENSITIVITY 5 (SE5) was first presumed to encode a rice HO with high similarity to Arabidopsis HY1, although enzyme activity of SE5 was not confirmed (Izawa et al. 2000). The se5 mutant has a very early flowering phenotype under both short-day and long-day (LD) conditions, and is completely deficient in photoperiodic response (Izawa et al. 2000).
During the last 10 years, increasing attention has been given to demonstrating that, similar to animal responses, plant HO-1 is induced by many factors, including its own substrate heme (Xuan et al. 2008), heavy metals (Noriega et al. 2004, Han et al. 2008), glutathione depletion (Cui et al. 2011), UV radiation (Yannarelli et al. 2006), salinity and osmotic stresses (Liu et al. 2010; Xie et al. 2011), hydrogen peroxide (H2O2) (Chen et al. 2009), nitric oxide (Noriega et al. 2007), auxin (Xuan et al. 2008) and abscisic acid (Cao et al. 2007a; Wu et al. 2011). Thus, the up-regulation of HO-1 in plants can act as an antioxidant barrier against stress-triggered oxidative damage and exhibit hormone-like responses (Shekhawat and Verma 2010). Meanwhile, plant HO and its by-product CO can regulate some developmental processes, such as lateral root formation (Cao et al. 2007b; Xu et al. 2011b) and adventitious rooting (Xuan et al. 2008).
Methyl viologen (1,1′-dimethyl-4,4′-bipyridylium, MV), also known as paraquat, is one of the most widely used herbicides in agriculture. It has a strongly negative redox potential (E0′ = −0.446 V) and is thus capable of accepting electrons from the iron–sulfur cluster Fe–SA/Fe–SB of photosystem I (Lewinsohn and Gressel 1984). This reaction results in a depletion of NADPH, inhibition of CO2 fixation (Dodge 1971; Lewinsohn and Gressel 1984) and the production of bipyridyl radicals that readily react with O2 to produce the superoxide anion (O2 −) and then, through a series of reactions, produce H2O2 and the hydroxyl radical. These reactive oxygen species (ROS) are all very active and cause extensive lipid peroxidation, chlorophyll breakdown, loss of photosynthetic activity and cell membrane integrity (Shaaltiel et al. 1988; Babbs et al. 1989). Thus, MV has been widely used to study oxidative stress (Bowler et al. 1994).
RNA interference (RNAi) using short-interfering RNAs has emerged as a major tool in reverse genetics to demonstrate the functionality of plant genes (Watson et al. 2005). Although previous pharmacological results have shown that hematin, an HO-1 inducer, could protect wheat leaves from oxidative damage triggered by paraquat and H2O2 (Sa et al. 2007), there is no detailed reverse genetic evidence assessing the roles of HY1/CO in Arabidopsis responses to MV treatment. In the present study, we showed that the SE5 protein was located in the chloroplast at least, and exhibited HO activity. Subsequently, RNAi knockdown of the rice SE5 gene results in loss of SE5 mRNA and SE5 protein. We also demonstrated that compared with wild-type, SE5 RNAi plants were more susceptible to MV treatment. As expected in a se5 mutant (Izawa et al. 2000), there was a similar phenotype of early flowering under LD conditions. By contrast, the addition of exogenous CO aqueous solution partially rescued the corresponding MV hypersensitivity. Transgenic Arabidopsis plants overexpressing SE5 and HY1 were generated and their corresponding tolerance phenotypes in the presence of MV were characterized.
Materials and methods
Chemicals
All chemicals were obtained from Sigma-Aldrich unless stated otherwise. Methyl viologen (MV), a redox-active constituent of bipyridyl herbicides, was purchased from Fluka (Buchs, Switzerland). Hemin, was used as an HO-1 inducer, which has been applied in animal and plant researches (Ryter et al. 2006; Xuan et al. 2008). The compound zinc protoporphyrin IX (ZnPP), a specific inhibitor of HO-1(Xuan et al. 2008; Wu et al. 2011), was used at 5 μM. Fresh 50 % CO-saturated aqueous solution was also prepared (Xuan et al. 2008).
Plant materials and growth conditions
Rice (Oryza sativa L., Wuyunjing 7) was kindly provided by Jiangsu Academy of Agricultural Sciences, Jiangsu Province, China. Wild-type (WT) and transgenic seeds were surface-sterilized with 0.1 % HgCl2 for 30 min, washed extensively with distilled water and then germinated in distilled water for 2 days at 28 °C. The seedlings were grown in a growth chamber with 12/12 h (28/25 °C) day/night regimes at 150 μmol m−2 s−1 irradiation for further experiments. To monitor the effect of flowering time, germinated seeds were transplanted into 10 cm soil-containing pots and grown in a greenhouse under LD (14 h light/10 h dark) conditions.
Arabidopsis thaliana (Col-0) seeds, including wild-type (WT), hy1-100 mutant, SE5 and HY1 over-expression line (35S:HY1-3; Xie et al. 2011), were surface-sterilized and rinsed for three times with sterile water, then cultured in Petri dishes on 1/2 Murashige and Skoog (MS, pH 5.8) solid medium containing 1 % (w/v) agar and 1 % (w/v) sucrose in the absence or presence of other indicated chemicals. Seedlings were transferred into a growth chamber with 16/8 h (22/18 °C) day/night regimes with 150 μmol m−2 s−1 irradiation for further experiments.
After various treatments as indicated, corresponding phenotypes of rice and Arabidopsis, including flowering time (Izawa et al. 2000), seedling growth and seeds germination (Xie et al. 2011), and chlorophyll contents (Porra et al. 1989), were measured, and corresponding photographs were taken. Meanwhile, different samples were immediately frozen in liquid nitrogen, and stored at −80 °C until further analysis.
Subcellular localization analysis of SE5
The specific primers 5′-CCATGGCGCCCGCGGCAGCGTCG-3′ (NcoI-SE5-F) and 5′-ACTAGTGGTGAATATGTGACGGAGG-3′ (SpeI-SE5-R) containing the NcoI and SpeI sites (underlined), respectively, were used to amplify the cDNA fragment encoding the full-length SE5 protein. The PCR fragments were inserted into the vector pCAMBIA-1302 at the 5′-terminal of the green fluorescence protein (GFP) gene under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The obtained 35S::SE5-GFP construct, and the 35S::GFP empty vector were transformed into Arabidopsis protoplasts using polyethylene glycol (PEG)-mediated transient gene expression (Yoo et al. 2007), and observed under a TCS-SP2 confocal laser scanning microscope (Leica Lasertechnik GmbH, Heidelberg, Germany) 16 h after transformation.
Expression and purification of recombinant mature SE5
The coding sequence for the mature SE5 (without the predicated chloroplast transit peptide, GenBank accession no. EU781632) was amplified with a pair of primers: 5′-GGATCCGCGGCGGCGACGGCGGCGGAG-3′ containing a BamHI restriction site (underlined) and 5′-GCGGCCGCTTAGGTGAATATGTGACGGAGG-3′ containing a NotI restriction site (underlined). Following digestion of TA cloning with BamHI and NotI, the product was inserted into pET-28a(+) and expressed in Escherichia coli strain Rosetta(DE3)pLysS.
The mSE5 protein was induced by 0.1 mM isopropyl β-d-l-thiogalactopyranoside (IPTG) at 28 °C for 3 h based on the manufacturer’s instructions (Novagen), and purified through Ni-affinity chromatography column. Finally, the purified fusion protein was used for the biochemical experiments.
Enzymatic activities assays
HO activity was assayed as previously described (Xuan et al. 2008). The absorbance changes between 350 and 800 nm were monitored for 20 min at 25 °C. Reaction rates for the formation of BV were determined by measuring absorbance at 665 nm (2-s intervals) for 10 min. Heme concentrations were varied from 0.5 to 20 μM. Values for V max and K m were calculated using Lineweaver–Burk plot. The effects of pH and temperature were determined using the standard assay conditions as described above.
Catalase (CAT) activity was spectrophotometrically measured by monitoring the consumption of H2O2 (ε = 39.4 mM−1 cm−1) at 240 nm for at least 3 min (Xu et al. 2011a). Determination of guaiacol peroxidase (POD) activity was performed by measuring the oxidation of guaiacol (ε = 26.6 mM−1 cm−1) at 470 nm within 2 min (linear phase) after the addition of H2O2 (Xu et al. 2011a). Superoxide dismutase (SOD) activity was measured on the basis of its ability to reduce nitroblue tetrazolium (NBT) by superoxide anion generated by the riboflavin system under illumination. One unit of SOD (U) was defined as the amount of crude enzyme extract required to inhibit the reduction rate of NBT by 50 % (Beauchamp and Fridovich 1971). Protein concentration was determined by the method of Bradford (1976) using bovine serum albumin (BSA) as a standard.
Generation of SE5 RNAi transgenic rice
To make the SE5 RNAi construct, a 392 bp fragment of SE5 cDNA was amplified using primers pSE5-F (5′-AGCAGTAGCAGGAGGATG-3′) and pSE5-R (5′-TATGTAGTGCCGGGAGCA-3′), and subcloned into pMD-19T (TaKaRa). This construct was denoted pSE5-pMD-19T and its sequence was verified. A fragment of pSE5-pMD-19T with EcoRI/KpnI restriction sites was inserted downstream of the atFAD intron, in the sense orientation. Meanwhile, A fragment of pSE5-pMD-19T with BamHI/HindIII restriction sites was inserted upstream of the atFAD intron in the antisense orientation (Fig. 3a). The SE5 RNAi construct was inserted into the BamHI/KpnI sites of pVec8_Ubi between the ubiquitin (Ubi) promoter and the tml terminator to generate pVec8_Ubi::pSE5-RNAi. The SE5 RNAi plasmid was then transformed into rice using Agrobacterium tumefaciens strain LBA4404 (Dai et al. 2001). A cetyltrimethylammonium bromide (CTAB)-based method for genomic DNA extraction from rice seedlings was carried out according to the procedures described by Doyle and Doyle (1987). Afterwards, RNAi plants were genotyped by polymerase chain reaction (PCR) with specific primers (Supplementary Table S1) for SE5 located on the construct vector.
RNA isolation, cDNA synthesis by reverse transcription (RT), and RT-PCR analysis
Total RNA was isolated from 100 mg of fresh-weight tissues using Trizol reagent (Invitrogen, Gaithersburg, MD) according to the user manual. cDNA was synthesized from 2 μg of total RNA using a random primer and avian myeloblastosis virus (AMV) reverse transcriptase XL (TaKaRa). cDNA was then amplified by PCR using specific primers (Supplementary Table S2). To standardize the results, the relative abundance of 18S rRNA and Atactin2 were also determined and used as the internal standard. Amplification products of the expected size were observed, and their identities were confirmed by sequencing (GenScript, Nanjing, China).
Real-time quantitative RT-PCR (qRT-PCR) analysis
qRT-PCR was performed using a Mastercycler® ep realplex real-time PCR system (Eppendorf, http://www.eppendorf.com/) with SYBR® Premix Ex Taq™ (TaKaRa, http://www.takara-bio.com/) according to the manufacturer’s instructions. Using specific primers (Supplementary Table S3 and S4), relative expression levels of corresponding genes are presented as values relative to corresponding control samples at the indicated times or conditions, after normalization to Osactin1 or Actin2 transcript levels.
Western blotting analysis for SE5
Proteins from homogenates were subjected to SDS-PAGE using a 12.5 % acrylamide resolving gel (Xie et al. 2011). The primary antibody used was rabbit polyclonal antibody raised against the recombinant mature SE5 expressed in Escherichia coli with a molecular mass of 25.6 kDa.
Chlorophyll content and lipid peroxidation determination
Chlorophyll a and b contents of aerial tissue and leaf pieces were quantified (Porra et al. 1989). Lipid peroxidation was estimated by measuring the concentrations of thiobarbituric acid reactive substances (TBARS) as described by Liu et al. (2010).
Detection of ROS
Superoxide anion and H2O2 levels were visually detected in the leaves of plants, respectively, with NBT and 3,3′-diaminobenzidine tetrahydrochloride (DAB) as described previously (Fukao et al. 2011). Each experiment was repeated at least five different samples, and representative images were shown.
Generation of transgenic Arabidopsis plants
The coding region of SE5 was amplified by RT-PCR with primers NcoI-SE5-F and SpeI-SE5-R. After verified by sequencing, the fragment was introduced into pCAMBIA-1302 vector, and then transformed into Agrobacterium tumefaciens EHA105. Arabidopsis wild-type plants were transformed by floral dip method. Positive transformants were selected on solid MS media supplemented with 30 mg/L hygromycin. Two independent lines of T3 plants (35S:SE5-1/3) were used for further analysis.
Statistical analysis
Where indicated, results were expressed as the mean ± SE of three independent experiments. Statistical analysis was performed using SPSS 10.0 software. For statistical analysis, Duncan’s multiple range test (P < 0.05 or P < 0.01) was chosen where appropriately.
Results
SE5 was localized to the chloroplast
An 870-bp full length cDNA of SE5 was amplified by RT-PCR from rice seedling leaves. The coding region of SE5 encodes 289 amino acids with a calculated molecular weight of 31.9 kDa. This includes a 64-amino-acid transit peptide identified by the ChloroP algorithm (Emanuelsson et al. 1999; http://www.cbs.dtu.dk/services/ChloroP/), suggesting a mature SE5 protein (mSE5; i.e. without the predicted transit peptide) of 25.6 kDa.
To verify the subcellular localization of SE5, we constructed a vector that constitutively expressed SE5–GFP fusion protein using the CaMV 35S promoter. Then, the resulting construct and GFP control plasmid were transformed into Arabidopsis protoplasts, and the fluorescent signals were observed by a confocal laser scanning microscope. The green fluorescent signal of SE5-GFP fusion protein co-localized with the auto-fluorescence of chlorophylls in chloroplasts (Fig. 1), demonstrating that the fusion protein was efficiently targeted to chloroplasts. By contrast, the protoplast transformed with the empty GFP vector alone has green fluorescent signals in the cytosol and nucleus (Fig. 1). Additionally, non-transformed protoplast (control) for auto-fluorescence with the same acquisition parameters was shown.
Subcellular localization of SE5 protein. SE5-GFP fusion protein or GFP alone expressed under the control of CaMV 35S promoter in Arabidopsis protoplasts was observed under a confocal microscope. Additionally, non-transformed protoplasts for auto-fluorescence with the same acquisition parameters were shown. The photographs were taken in the blue channel (left), in the red channel (middle), and in their combination (right). Scale bars represent 10 μm
Biochemical analysis of the recombinant protein
To confirm that SE5 encodes a HO and to further characterize its properties, the recombinant His-tagged mSE5 protein was induced by IPTG. The recombinant protein was expressed as a soluble protein of 29.1 kDa, approximately corresponding to the molecular weight of the mSE5 protein (25.6 kDa) plus that of 6 × His-tag (0.7 kDa) and the translated vector sequence (2.6 kDa). After purification by Ni-affinity chromatography, it yielded a single band (Fig. 2a, lane 1), which was recognized by the polyclonal antiserum against mSE5 (Fig. 2a, lane 2).
Expression and biochemical characterization of purified recombinant His-tagged mature SE5 (mSE5) protein in E. coli. a Expressed protein purified by Ni-affinity chromatography and its western blotting analysis, 30 μg protein/well. Lane M, marker proteins; lane 1, purified protein; lane 2, western blotting analysis of purified protein developed with the polyclonal antiserum against the mSE5. Arrow indicates the position of the fusion protein of mSE5 with a molecular mass of 29.1 kDa. b Time-course of absorbance changes were determined during the mSE5 reaction with spectra taken at 0, 1, 2, 5, 10 and 20 min after the addition of NADPH. Arrows indicate the direction of the major changes in absorption during the course of the measurements. c Michaelis–Menten plot of the mSE5 reaction for hemin concentrations of 0.5, 2, 5, 10 and 20 μM. Inset: Lineweaver–Burk plot of the same data. Data shown are the mean ± SE from three independent measurements. d pH (left) and temperature (right) dependence of the mSE5 reaction. The relative activity of mSE5 was calculated by BV formation
In a subsequent test, we measured the HO activity of mSE5 by spectrophotometrically examining the conversion of heme to BV (Fig. 2b). Absorbance was monitored between 350 and 800 nm, with bound-heme showing strong absorbance at 405 nm and BV at 665 nm. Over a period of 20 min of incubation, the bound-heme peak decreased substantially, accompanied by a concomitant rise in the BV absorbance maxima (Fig. 2b). The reaction rate for the formation of BV was also determined by monitoring absorbance at 665 nm (Fig. 2c). Using the above data, we determined the kinetic constants for the HO reaction from a Lineweaver–Burk plot (Fig. 2c, insert). Under our experimental conditions, the V max value for the complete reaction was estimated as 62.5 nmol BV h−1 mg protein−1 with an apparent K m value for hemin of 2.9 μM. Furthermore, the rate of the mSE5 reaction increased to a peak value at pH 7.0 and declined thereafter (Fig. 2d, left). In contrast, mSE5 enzyme activity increased with rising temperature within 10–50 °C (Fig. 2d, right), comparable to values obtained from Arabidopsis HY1 (Muramoto et al. 2002, Gisk et al. 2010).
Phenotypic analysis of SE5 RNAi transgenic rice
To investigate the physiological role of SE5 in rice, we obtained two knockdown transgenic lines by an RNAi approach (Fig. 3a). T-DNA insertion of the two T0 transgenic lines (RNAi-1 and RNAi-2) was confirmed by PCR-based analysis (Fig. 3b). Gene-specific RT-PCR showed that about 90 % of SE5 transcripts were specifically decreased by SE5 RNAi, whereas transcripts of OsHO2 were not affected in T2 progeny of transgenic plants (Fig. 3c). The protein levels of SE5 determined by western blotting displayed similar decreasing tendencies (Fig. 3d). The above results clearly indicate that only the transcripts and protein levels of SE5 were specifically reduced in SE5 RNAi transgenic plants.
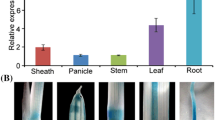
The SE5 RNAi construct and analysis of HO expression in transgenic and wild-type (WT) rice. a Schematic representation of the SE5 RNAi construct and primers (indicated with arrows; Supplementary Table S1) used for genotyping of SE5 mutation. b PCR-based genotyping of T0 progeny of RNAi transgenic and wild-type plants. Ethidium bromide-stained amplicons of different combinations of primers are shown. c RT-PCR-based analyses of SE5 and OsHO2 transcripts. Total RNAs from the T2 progeny of SE5 RNAi transgenic and wild-type plants, were extracted from 14-day-old seedlings. 18 s rRNA expression was used as the loading control. d Corresponding western blotting analysis of SE5 protein level was provided. e Expression profiles of SE5 in various tissues of RNAi transgenic and wild-type plants. The mRNA expression of SE5 in roots (RT), stems (SM), leaves (LF), flowers (FR), and dry seeds (SD) was analysed. 18S rRNA gene was used as a control to show the normalization of the amount of templates in semi RT-PCR assay. Meanwhile, SE5 protein level was determined by western blotting (f). Coomassie Brilliant Blue-stained gels are present to show that equal amounts of proteins were loaded (50 μg protein/well)
Subsequently, RNAi-1 and wild-type plants were selected for further analysis of the expression profiles of SE5 gene and corresponding protein levels in various tissues (Fig. 3e, f). Either SE5 transcripts or SE5 protein were highly expressed in stems and leaves in wild-type, but relatively less in roots and seeds. There was a higher level of SE5 protein in flowers compared to a low abundance of SE5 transcripts. However, the SE5 RNAi plants displayed decreased levels of both SE5 transcripts and SE5 protein. Additionally, except for relatively higher expression in root tissues, there was comparable expression of OsHO2 transcripts in either wild-type or RNAi-1 plants.
In comparison with wild-type, the SE5 RNAi plants had weaker growth, with fewer stems and a yellowish color, when grown under LD conditions. For example, 50-day-old SE5 RNAi plants showed an apparent yellowish phenotype (Fig. 4a) and chlorophyll levels decreased to 47.9 and 54.4 % of the fifth and sixth leaves of wild-type, respectively, while corresponding chlorophyll a to b ratios were 14.5 and 14.0 % higher than those of wild-type plants (Fig. 4b).
Comparison of the leaf and flowering phenotypes of the T2 progeny of SE5 RNAi transgenic and wild-type plants under nature LD conditions (14 h light/10 h dark; 14L:10D). The leaf phenotypes (a) and chlorophyll a/b contents (b) of transgenic and wild-type (WT) plants grown for 50 days. 5, the fifth leaf; 6, the sixth leaf. Scale bar represents 2 cm. c The flowering phenotypes of wild-type (left) and transgenic rice (right). Plants were grown for 80 days. Arrow indicates that SE5 RNAi transgenic plants have spikelets, whereas the wild-type plants were still in the vegetative stage, and showed no signs of flowering. d Flowering time was also recorded. e Comparison of leaf emergence rates between RNAi transgenic and wild-type plants. The leaf number of individual plants was scored on the days indicated until emergence of panicle appearing in RNAi transgenic plants. Data represent mean ± SE (n = 15) from three independent biological replicates
It has been shown that a null mutation in SE5 leads to an early-flowering and photoperiodic-insensitive phenotype (se5 in cultivar Norin 8, Izawa et al. 2000; s73 in cultivar Bahia, Andrés et al. 2009). In our experimental conditions, the SE5 RNAi plants under LD conditions flowered at 71 d after germination, 50 d earlier than wild-type plants (Fig. 4c, d). Leaf emergence reflects the rate of leaf primordia formation in rice—although the leaf numbers of SE5 RNAi plants slightly decreased in the indicated growth time, the rate of leaf emergence in RNAi plants was indistinguishable from that of wild-type until flowering (Fig. 4e).
Knockdown of SE5 gene enhanced sensitivity to MV treatment
To determine whether SE5 regulated plant tolerance to oxidative stress, we incubated wild-type and two SE5 RNAi transgenic seedlings with MV, which stimulates formation of ROS within chloroplasts. Both 2 and 5 μM MV significantly decreased seedling shoot growth in all genotypes, but the reduction was more severe in RNAi plants at the two concentrations tested (P < 0.05; Fig. 5a). Additionally, MV obviously reduced chlorophyll content in RNAi-1, presumably as a secondary consequence of oxidative stress (Fig. 5b, c).
Knockdown of SE5 gene enhances sensitivity to oxidative stress. a Growth inhibition of aerial tissues by MV stress. 7-day-old T2 progeny of SE5 RNAi transgenic and wild-type (WT) seedlings were transferred to MV solution (0, 2, or 5 μM) and incubated for 5 days (14 h light/10 h dark, 150 μmol m−2 s−1 irradiation). Relative fresh weight was calculated by comparison to the nontreated seedlings of individual genotypes. Chlorophyll a (b) and b (c) contents in aerial tissues of plants exposed to MV stress. FW, fresh weight. Data represent mean ± SE (n = 10) from three independent biological replicates. Within each set of experiments, an asterisk indicates a significant difference (P < 0.05) between wild-type and RNAi plants according to Duncan’s multiple range test
Changes of SE5 gene expression and antioxidant defense
In the following experiments, we observed that the amount of rice SE5 mRNA in wild-type was induced within 8 h of exposure to 5 μM MV, followed by a decreasing trend until 24 h. By contrast, knockdown of SE5 obviously kept lower levels of SE5 transcripts (Fig. 6).
Relative mRNA levels of SE5 gene and antioxidative enzyme genes in the T2 progeny of SE5 RNAi transgenic and wild-type (WT) rice seedling leaves exposed to MV. 7-day-old plants were exposed to 5 μM MV for 24 h in the light (150 μmol m−2 s−1 irradiation), and the aerial tissue was subjected to qRT-PCR analysis. The expression levels of the corresponding genes are presented as values relative to the control at 0 h. Data represent mean ± SE from three independent biological replicates, and asterisks indicate a significant difference (* P < 0.05; ** P < 0.01) between wild-type versus RNAi plants according to Duncan’s multiple range test
The abundance of intercellular ROS is tightly regulated through complex antioxidant systems in diverse subcellular compartments. Among ROS-scavenging pathways, major enzymes responsible for selective detoxification of O2 − and H2O2 in plants include SOD, CAT, ascorbate peroxidase (APX) and POD (Apel and Hirt 2004). Because our data indicated a possible link between knockdown of SE5 and MV-triggered lipid peroxidation, we tested the responses of representative antioxidant enzyme genes to MV. Time-dependent analysis showed that both CatA and CatB were oxidative stress inducible and CatB was more prominently induced than CatA in wild-type plants. Comparatively, the inducible pattern of CatA and CatB were delayed or clearly blocked in RNAi plants (Fig. 6). We further noticed that Apx1 or Apx2 mRNA was up-regulated by MV in both wild-type and RNAi transgenic plants, but the induction was more pronounced in the wild-type after 8 h and 12 h of MV exposure. Additionally, SodA1 mRNA was clearly elevated by oxidative stress, with stronger induction in wild-type than RNAi rice seedlings. Based on these directed transcript studies, we suggest that SE5 knockdown may reduce the capability of antioxidant systems, thereby showing sensitivity to oxidative stress.
Knockdown of SE5 gene enhanced sensitivity to oxidative stress
To confirm that knockdown of the SE5 gene enhanced sensitivity to oxidative stress, leaf segments of wild-type and RNAi-1 transgenic plants were treated with 5 μM MV or 10 mM H2O2 (Fig. 7a, b). These treatments decreased the content of chlorophyll a and b in the two genotypes, but to a significantly greater extent in RNAi-1 transgenic plants. To investigate this further, we visualized the accumulation of O2 − and H2O2 using NBT and DAB, respectively, in detached leaves of wild-type and RNAi-1 transgenic plants immediately following MV treatment for 6 h (Fig. 7c). The RNAi transgenic rice seedling leaves exhibited marked blue and brown coloration after MV treatment, suggesting more O2 − and H2O2 accumulation compared to wild-type plants. TBARS content, as a reliable indicator of oxidative damage and lipid peroxidation, was also quantified. Similar to the above histochemical results (Fig. 7c), lipid peroxidation was significantly aggravated in SE5 RNAi transgenic rice after 6 h of MV treatment, compared to wild-type (Fig. 7d).
Knockdown of SE5 gene aggravates chlorophyll degradation triggered by oxidative stresses. Phenotypes (left) and chlorophyll contents (right) of leaf pieces of the T2 progeny of SE5 RNAi-1 transgenic and wild-type (WT) plants exposed to MV (a) and H2O2 (b) treatments. The top leaves of 20-day-old plants were cut into pieces (5 mm × 3.5 mm) and floated on distilled water (Mock), 5 μM MV or 10 mM H2O2 for 24 h at 25 °C in the light (150 μmol m−2 s−1 irradiation). Following treatments, the chlorophyll contents of leaf pieces were assayed spectrometrically. c Accumulation of superoxide anion and H2O2 in leaf blades upon MV treatment. 20-day-old T2 progeny of SE5 RNAi transgenic and wild-type plants were excised at the base with a razor blade and supplied through the cut ends with NBT (1 mg ml−1) or DAB (0.5 mg ml−1) solutions for 8 h, and then exposed to 5 μM MV for 6 h at 25 °C in the light (150 μmol m−2 s−1 irradiation). After that, leaves were decolorized in boiling ethanol (95 %) for 15 min. Scale bar represents 1 cm. d Lipid peroxidation of detached leaves after MV treatment. FW, fresh weight. Data represent mean ± SE (n = 6) from three independent biological replicates, and an asterisk indicates a significant difference (P < 0.05) between wild-type and RNAi plants within each set of experiments according to Duncan’s multiple range test
Up- or down-regulation of SE5 expression contributed to the alleviation or aggravation of chlorophyll loss
The decreases of antioxidant defense in SE5 RNAi transgenic rice suggested that knockdown of SE5 might be related to the sensitivity to oxidative stress. To test this hypothesis, the effects of a potent HO-1 inhibitor (ZnPP) on chlorophyll loss both in the absence and presence of MV were evaluated and compared with that of MV alone. ZnPP aggravated the loss of chlorophyll a (especially) and b contents triggered by MV (Fig. 8a, b). Meanwhile, a ZnPP-inhibited SE5 transcript was observed only in MV-treated wild-type plants (Fig. 8c); however, the ZnPP plus MV-induced chlorophyll loss was markedly recovered by the application of CO aqueous solution. We also noticed that application of CO alone or with other reagents remarkably increased SE5 transcripts, compared to corresponding samples without CO treatment. In addition, the addition of ZnPP alone produced slight but not significant decreases in chlorophyll a content, accompanied by no significant influence on chlorophyll b content and SE5 gene expression.
Effects of hemin, CO, and ZnPP on chlorophyll contents and SE5 transcripts in the wild-type rice upon MV stress. 7-day-old rice seedlings were incubated with water (Con), MV (5 μM), hemin (50 μM), CO (50 % saturation) and ZnPP (5 μM), either alone or in combination for 5 days. Chlorophyll a/b contents were then determined (a, b). SE5 transcripts were analysed after 12 h of various treatments (c). The expression levels of the SE5 transcript are presented as values relative to the corresponding control. Data represent mean ± SE three biological replicates, and different letters above the columns indicate significant different (P < 0.05) according to Duncan’s multiple range test
To further verify the possible role of SE5 in MV-induced oxidative damage, 50 μM hemin was applied. As expected, hemin not only notably reversed the MV-induced chlorophyll loss (Fig. 8a, b), but also up-regulated SE5 transcripts (Fig. 8c). When hemin was used alone, SE5 exhibited a slight but non-significant induction response, and the chlorophyll a content significantly increased. These results suggested that SE5 up-regulation might be required for the alleviation of MV-induced chlorophyll loss.
SE5 knockdown-triggered sensitivity to MV was reversed by CO
Further evidence showed that in the presence of CO, the seedling growth inhibition and the decline in chlorophyll a (but not chlorophyll b) content in MV-treated SE5 RNAi transgenic plants were blocked significantly (Fig. 9a, b). In comparison with the stress alone samples, CatA, Apx2 and SodA1 transcripts were induced significantly when CO was used together with MV in mutant seedling leaves (Fig. 9d). However, there were no obvious differences in the transcripts of SE5, CatB and Apx1. Additionally, the application of CO alone remarkably up-regulated the SodA1 gene expression. Based on these direct transcript studies, combined with changes in chlorophyll content (Fig. 9a–c), we suggest that CO augmented the capability of antioxidant systems, thereby improving tolerance to oxidative stress.
Effects of CO on chlorophyll contents and SE5 transcripts and antioxidative enzyme genes in the T2 progeny of SE5 RNAi transgenic rice seedling leaves exposed to MV. 7-day-old plants were treated with distilled water (Mock), 50 % CO aqueous solution, and 5 μM MV, either alone or in combination. Photographs were taken after 5 days of treatments (a). Scale bar represents 3 cm. Meanwhile, chlorophyll a/b contents in aerial tissue were then determined (b, c). Corresponding gene expression, which are presented as values relative to the corresponding control, were analyzed after 12 h of various treatments (d). Data represent mean ± SE from three independent biological replicates, and different letters above the columns indicate significant different (P < 0.05) according to Duncan’s multiple range test
Overexpression of SE5 and HY1 alleviated seed germination inhibition and chlorophyll loss
To further investigate the function of SE5 in plants, we overexpressed SE5 in transgenic Arabidopsis under the control of a CaMV 35S promoter. Four transformed lines of Arabidopsis were confirmed by hygromycin selection and semi-quantitative RT-PCR. Afterwards, two of them (35S:SE5-1 and 35S:SE5-3) were used for further investigation. The Arabidopsis HY1 mutant hy1-100, which has an AG → AA substitution in the normally conserved sequence at the acceptor site (three boundary) of the first intron (Muramoto et al. 1999), and one overexpression line of HY1 (35S:HY1-3) were also applied (Xie et al. 2011). As expected, HY1 or SE5 were overexpressed in 35S:HY1-3, 35S:SE5-1 and 35S:SE5-3 mutant seedlings, respectively (Fig. 10a).
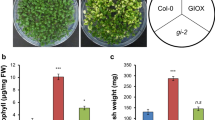
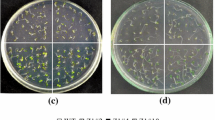
Molecular characterization and the phenotypes of SE5 transgenic Arabidopsis plants. a Semi-quantitative RT-PCR analysis of HY1 and SE5 transcript in the seedlings of wild-type (WT), hy1-100 mutant and T3 progeny of HY1 over-expression line (35S:HY1-3), and T3 progeny of SE5 over-expression line (35S:SE5-1/3). Total RNA was extracted at 5 days under normal growth condition, and Atactin2 was used as an internal control. b Arabidopsis seedlings were grown on 1/2 Murashige and Skoog (MS) solid medium containing the indicated concentrations of MV. Photographs were taken after 5 days of treatments. Scale bar represents 1 cm. Seed germination percentage (c) and chlorophyll content (d) of seedlings were also measured. For each line, chlorophyll contents after various MV treatments were given as percentage values with respect to those in the mock-treated sample. Results represent the mean of three independent experiments. Data represent mean ± SE from three independent biological replicates, and an asterisk was significantly different from wild- type plants at P < 0.05 according to Duncan’s multiple range test
Inhibition of seed germination and seedling growth are two classic responses mediated by MV exposure. In the present study, there was severe inhibition of seed germination and seedling growth induced by MV in wild-type (Fig. 10b, c). For example, upon application of 1 μM MV for 5 d, the root growth of wild-type seedlings was almost totally inhibited and the cotyledon remained small; and treatment with 2 μM MV resulted in even more exaggerated responses. By contrast, seed germination inhibition was significantly attenuated in two SE5 and one HY1 transgenic Arabidopsis lines (P < 0.05). In the hy1-100 mutant there was a contrasting phenomenon: the hy1-100 mutant was more sensitive on germination to MV stress than wild-type plants (21.3 vs. 40.7 % under 1 μM MV, respectively; 6.45 vs. 19.3 % under 2 μM MV, respectively; Fig. 10c). Similar phenomenon were observed in the responses of chlorophyll contents, except for the hy1-100 mutant upon treatment with 2 μM MV, in which there was no significant difference compared with wild-type (Fig. 10d). This could be explained by the fact that the hy1-100 mutant has a yellow-green phenotype itself even under normal growth conditions (Terry 1997).
Overexpression of SE5 and HY1 enhanced oxidative stress tolerance
In comparison with wild-type plants, the hy1-100 mutant upon MV treatment had significant accumulations of O2 − (Fig. 11a) and H2O2 (Fig. 11b) in leaves. By contrast, SE5 and HY1 transgenic Arabidopsis plants subjected to the same treatment exhibited only slight staining, all of which were consistent with the alleviation of seed germination inhibition and chlorophyll loss (Fig. 10). Subsequently, the expression of three antioxidant defense genes (Cat2, Per21 and Csd1) and corresponding enzyme activities were determined. Compared to wild-type, Cat2, Per21 and Csd1 transcripts were induced differentially in SE5 and HY1 transgenic Arabidopsis plants upon MV treatment for 6 h (Fig. 11c). By contrast, Cat2 and Csd1 mRNAs were obviously reduced in the hy1-100 mutant. Comparatively, changes of CAT, POD and especially SOD activities exhibited similar tendencies (Fig. 11d).
O2 − and H2O2 accumulation, and the expression and activities of antioxidative enzymes in wild-type (WT), hy1-100 mutant, SE5 and HY1 transgenic Arabidopsis plants upon MV. 4-week-old plants were sprayed with 5 μM MV for 24 h. Afterwards, leaves were stained with NBT (a) or DAB (b) solutions for 8 h and then decolorized in boiling ethanol (95 %) for 15 min. Scale bar represents 0.5 cm. Meanwhile, corresponding leaves at different times were subjected to qRT-PCR (c) and enzyme activity (d) analysis. The expression levels of the corresponding genes are presented as values relative to the control at 0 h. Data represent mean ± SE from three independent biological replicates, and asterisks were significantly different (* P < 0.05; ** P < 0.01) from wild-type plants according to Duncan’s multiple range test
Discussion
SE5 shared similar subcellular localization and high similarity with its counterpart Arabidopsis HY1
In Arabidopsis, four alleles coding HY1 and HO2/3/4 have been identified. Although this does not exclude the possibility of mitochondrial localization, all Arabidopsis HOs are solely localized to the plastid (Gisk et al. 2010). The amino termini of SE5 deduced from cDNA sequences suggested features of a chloroplast transit peptide according to the ChloroP program. Our experimental results showed chloroplast localization of SE5 in Arabidopsis protoplast (Fig. 1). Furthermore, it was suggested that the purified recombinant mSE5 exhibited HO activity (Fig. 2). Similar to those of HY1 (Muramoto et al. 2002), the degradation activity of the substrate–protein complex of recombinant mSE5 (absorbance peak at 405 nm) concomitant with the generation of BV (absorbance peak at 665 nm), was confirmed in a time-dependent manner (Fig. 2b; Muramoto et al. 2002; Linley et al. 2006; Gisk et al. 2010). The kinetic parameters for the mSE5 reaction were compared with those previously reported for Arabidopsis HY1 (Muramoto et al. 2002; Gisk et al. 2010), pea HO1 (PsHO1) (Linley et al. 2006), alfalfa HO1 (MsHO1) (Fu et al. 2011) and rapeseed HO1 (BnHO1) (Cao et al. 2011). For instance, SE5 had a K m value for hemin of 2.9 μM (Fig. 2c) in comparison with 1.3 μM for HY1. Additionally, SE5 enzyme activity increased with rising temperature (Fig. 2d), similar to results for HY1 (Muramoto et al. 2002).
Davis et al. (2001) discovered that HY1 was actively expressed in the shoot apex, cotyledons, vascular tissue and hypocotyl–root junction. In the present study, using semi-quantitative RT-PCR, we demonstrated that SE5 was very strongly expressed in seedling leaves (Fig. 3e), which was further confirmed by western blotting (Fig. 3f). Similar expression profiles were observed in BnHO1 (Cao et al. 2011). Comparatively, PsHO1 was very strongly expressed in leaves and root tissues (Linley et al. 2006), partially because heme, acting as the cofactor of plant hemoglobins in root nodules (O’Brian 1996), has a crucial role in nodulation and nitrogen fixation in pea plants. Another more interesting feature of SE5 gene and SE5 protein was the strong expression in stem tissue–similar results for PsHO1 were previously reported (Linley et al. 2006).
Although phytochromes have been recently suggested to modulate both biotic and abiotic stresses (Carvalho et al. 2011), there is ample evidence showing that these photoreceptors are involved in a number of processes that control plant growth and development from germination to flowering. For example, mutations in HO-1 lead to a decrease or absence of photochemical activity of functional phytochromes. This conclusion was derived from the phenotypes of HO-1 mutants in rice (se5, Izawa et al. 2000; s73, Andrés et al. 2009), Arabidopsis (hy1, Muramoto et al. 1999) and tomato (yg-2, Terry 1997), such as abnormal stem or hypocotyl elongation, and a yellowish color. Similar to the hy1 mutant in Arabidopsis, and se5 and s73 mutants in rice, we showed that SE5 RNAi plants exhibited a phenotype of reduction in chlorophyll accumulation (Fig. 4a, b), which might be due to reduced 5-aminolaevulinic acid (ALA) formation, one of the two pivotal control points of tetrapyrrole biosynthesis (Davis et al. 2001; Cornah et al. 2003). These results might partially explain why the alleviation or aggravation of chlorophyll loss on exposure to MV was observed in the HY1 and SE5 overexpressing Arabidopsis lines or hy1-100 mutant seedlings, respectively (Fig. 10b, d). Most importantly, as expected (Izawa et al. 2000; Andrés et al. 2009), SE5 RNAi transgenic rice also led to early flowering under LD conditions (Fig. 4c–e), possibly due to the unbalanced expression of Heading date 1 (Hd1) and Early heading date1 (Ehd1), thus resulting in higher levels of Heading date3a (Hd3a) (Andrés et al. 2009).
Knockdown of SE5-triggered MV hypersensitivity impaired antioxidant defense
Similar to Arabidopsis HY1 (Muramoto et al. 1999), previous results demonstrated that SE5 appeared to be the key HO responsible for phytochrome chromophore biosynthesis in rice (Izawa et al. 2000). In the present study, we further discovered that SE5 played a central role, acting as an indispensable endogenous modulator of plant MV tolerance. The following genetic and pharmacological evidence supports this conclusion. Firstly, compared with wild-type plants, the loss of SE5 function in RNAi transgenic plants (especially RNAi-1) increased sensitivity to MV stress, evaluated by the aggravation of seedling shoot growth inhibition and chlorophyll loss (Fig. 5). Secondly, MV induced SE5 gene expression in the first 8 h of treatment in wild-type plants (Fig. 6), which was consistent with the modulation of HO-1 caused by mercury exposure (Han et al. 2007), UV radiation (Yannarelli et al. 2006) and salt stress (Xie et al. 2011). Thirdly, treatment with hemin, CO or ZnPP resulted in increasing or decreasing tendencies in SE5 transcripts in wild-type plants upon MV exposure, consistent with alleviation or exaggeration of MV-induced chlorophyll a and b loss (Fig. 8). However, the above responses of ZnPP were blocked by the addition of CO aqueous solution; and the addition of CO was able to rescue the hypersensitive phenotype of the SE5 RNAi transgenic rice seedlings (Fig. 9a–c). Together, these results clearly suggest that the loss of SE5 function in RNAi transgenic plants is required for MV hypersensitivity.
Previously, HO-1-dependent formation of CO was demonstrated in Arabidopsis and cucumber (Muramoto et al. 2002; Xuan et al. 2008). Recent studies discovered that HO/CO acts as a gaseous signal system in plant ROS signaling (Balestrasse et al. 2006; Xie et al. 2011). Our previous study illustrated that the hy1-100 mutant displayed maximal sensitivity to salinity and no acclimation response, whereas plants overexpressing HY1 (35S:HY1-3/4) showed tolerance characteristics (Xie et al. 2011). Comparatively, both SE5 and HY1 transgenic Arabidopsis lines had significantly decreased MV hypersensitivity and alleviated oxidative damage, in comparison with those of hy1-100 mutant plants (Figs. 10, 11). Given the rescuing effects of CO and hemin, one of the by-products of HO catalytic reactions and the inducer of HO-1, in both wild-type or SE5 RNAi transgenic rice plants (Figs. 8, 9), we deduced that HO-mediated CO may be involved in the alleviation of MV hypersensitivity. The above results further supported the conclusions that SE5 acts as an indispensable endogenous modulator of plant MV tolerance, and strengthens the idea that SE5 shares high similarity with its counterpart Arabidopsis HY1. Certainly, the cytoprotective role of BV, another by-product of HO catalytic reactions, could not be easily ruled out, because it was confirmed previously that BV could act as an antioxidant, alleviating heavy metal stress in soybean plants (Noriega et al. 2004).
It is well established that MV exposure often causes overproduction of ROS and inhibition of some antioxidant enzyme activities in plant cells, and tolerance to MV stress is correlated with a more efficient antioxidant defense (Iturbe-Ormaetxe et al. 1998; Murgia et al. 2004). In the subsequent experiments, four antioxidant enzyme genes, including CatA, CatB, Apx1, and SodA1, significantly decreased in SE5 RNAi transgenic rice seedlings during 24 h after MV treatment, respect to those in the wild-type plants (Fig. 6). These results are consistent with those reported by Willekens et al. (1997), who found that Cat1-deficient tobacco was markedly more sensitive to MV, and also with a previous study showing that knockout-Apx1 Arabidopsis mutant displayed high sensitivity to MV stress (Davletova et al. 2005). In turn, we also noticed that the loss-of-function mutation of SE5 could increase expression of SodB, SodCc1, and Trxh (Supplementary Fig. S1). However, these up-regulations could not fully compensate for the lack of SE5. This deduction was confirmed by the results on leaf segments of both wild-type and knockdown plants treated with MV and H2O2, showing that the RNAi plants seemed more sensitive to oxidative stress, as evaluated by the aggravation of chlorophyll loss, histochemical staining of H2O2 and O2 −, as well as the TBARS overproduction (Fig. 7). We further speculated that MV could block HO activity, a novel antioxidant enzyme proven recently in plants (Shekhawat and Verma 2010; Shekhawat et al. 2011), through the depletion of its electron donor NADPH or ferredoxin (Fd) (Iturbe-Ormaetxe et al. 1998). Furthermore, a reduction of SE5 transcript was observed after 12 and 24 h treatment of 5 μM MV in wild-type plants (Fig. 6).
Genetic evidence showed that overexpression of two rice cytosol APXs (Apx1 and Apx2) in Arabidopsis reduces the accumulation of H2O2, restricts chlorophyll degradation, and enhances survival under salinity stress, and the Apx2 gene has a more functional role than Apx1 in the improvement of salt tolerance in transgenic plants (Lu et al. 2007). Overexpression of Arabidopsis thylakoidal APX gene (tAPX) could increase resistance to MV-induced photooxidative stress (Murgia et al. 2004). It was also reported that mitochondrial SodA1 (mitochondrial Mn-SOD) and SodB (Fe-SOD) were significantly induced by osmotic stress and severe low oxygen levels, respectively (Kaminaka et al. 1999; Magneschi and Perata 2009). In the present study, several antioxidant defense genes (e.g. CatA, Apx2 and especially SodA1), were significantly up-regulated by CO in SE5 RNAi transgenic rice seedlings upon MV treatment. Interestingly, the above changes were consistent with corresponding resistant phenotypes (Fig. 9), suggesting that induction of cytosol APX and Mn-SOD might be the downstream targets of the HO/CO cytoprotective role against various oxidative stresses, by maintaining cellular homeostasis (Sa et al. 2007; Xie et al. 2011).
Together, this study established that the sensitivity of the SE5 knockdown mutant to MV was, at least partially, due to the down-regulation of some representative antioxidant defense. Therefore, future characterization of the direct targets of SE5 in antioxidant defense may reveal the complete pathway of SE5 in oxidative stress signaling. Additionally, combined with the unique role of SE5 in light signaling (Izawa et al. 2000), we further deduced that rice SE5 could not only confer plant tolerance to oxidative stress, but also be useful for molecular breeding.
References
Andrés F, Galbraith DW, Talón M, Domingo C (2009) Analysis of PHOTOPERIOD SENSITIVITY5 sheds light on the role of phytochromes in photoperiodic flowering in rice. Plant Physiol 151:681–690
Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55:373–399
Babbs CF, Pham JA, Coolbaugh RC (1989) Lethal hydroxyl radical production in paraquat-treated plants. Plant Physiol 90:1267–1270
Balestrasse KB, Noriega GO, Batlle A, Tomaro ML (2006) Heme oxygenase activity and oxidative stress signaling in soybean leaves. Plant Sci 170:339–346
Beauchamp C, Fridovich I (1971) Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem 44:276–287
Bowler C, Van Camp W, Van Montagu M, Inzé D (1994) Superoxide dismutase in plants. Crit Rev Plant Sc 13:199–218
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Cao ZY, Huang BK, Wang QY, Xuan W, Ling TF, Zhang B, Chen XY, Nie L, Shen WB (2007a) Involvement of carbon monoxide produced by heme oxygenase in ABA-induced stomatal closure in Vicia faba and its proposed signal transduction pathway. Chin Sci Bull 52:2365–2373
Cao ZY, Xuan W, Liu ZY, Li XN, Zhao N, Xu P, Wang Z, Guan RZ, Shen WB (2007b) Carbon monoxide promotes lateral root formation in rapeseed. J Integr Plant Biol 49:1070–1079
Cao Z, Geng B, Xu S, Xuan W, Nie L, Shen W, Liang Y, Guan R (2011) BnHO1, a haem oxygenase-1 gene from Brassica napus, is required for salinity and osmotic stress-induced lateral root formation. J Exp Bot 62:4675–4689
Carvalho RF, Campos ML, Azevedo RA (2011) The role of phytochrome in stress tolerance. J Integr Plant Biol 53:920–929
Chen XY, Ding X, Xu S, Wang R, Xuan W, Cao ZY, Chen J, Wu HH, Ye MB, Shen WB (2009) Endogenous hydrogen peroxide plays a positive role in the upregulation of heme oxygenase and acclimation to oxidative stress in wheat seedling leaves. J Integr Plant Biol 51:951–960
Cornah JE, Terry MJ, Smith AG (2003) Green or red: what stops the traffic in the tetrapyrrole pathway? Trends Plant Sci 8:224–230
Cui W, Fu G, Wu H, Shen W (2011) Cadmium-induced heme oxygenase-1 gene expression is associated with the depletion of glutathione in the roots of Medicago sativa. Biometals 24:93–103
Dai S, Zheng P, Marmey P, Zhang S, Tian W, Chen S, Beachy RN, Fauquet C (2001) Comparative analysis of transgenic rice plants obtained by Agrobacterium-mediated transformation and particle bombardment. Mol Breed 7:25–33
Davis SJ, Kurepa J, Vierstra RD (1999) The Arabidopsis thaliana HY1 locus, required for phytochrome-chromophore biosynthesis, encodes a protein related to heme oxygenases. Proc Natl Acad Sci USA 96:6541–6546
Davis SJ, Bhoo SH, Durski AM, Walker JM, Vierstra RD (2001) The heme-oxygenase family required for phytochrome chromophore biosynthesis is necessary for proper photomorphogenesis in higher plants. Plant Physiol 126:656–669
Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R (2005) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17:268–281
Dodge AD (1971) The mode of action of the bipyridylium herbicides, paraquat and diquat. Endeavour 30:130–135
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8:978–984
Emborg TJ, Walker JM, Noh B, Vierstra RD (2006) Multiple heme oxygenase family members contribute to the biosynthesis of the phytochrome chromophore in Arabidopsis. Plant Physiol 140:856–868
Fu GQ, Xu S, Xie YJ, Han B, Nie L, Shen WB, Wang R (2011) Molecular cloning, characterization, and expression of an alfalfa (Medicago sativa L.) heme oxygenase-1 gene, MsHO1, which is pro-oxidants-regulated. Plant Physiol Biochem 49:792–799
Fukao T, Yeung E, Bailey-Serres J (2011) The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell 23:412–427
Gisk B, Yasui Y, Kohchi T, Frankenberg-Dinkel N (2010) Characterization of the haem oxygenase protein family in Arabidopsis thaliana reveals a diversity of functions. Biochem J 425:425–434
Gisk B, Molitor B, Frankenberg-Dinkel N, Kötting C (2012) Heme oxygenase from Arabidopsis thaliana reveal different mechanisms of carbon monoxide binding. Spectrochim Acta A Mol Biomol Spectrosc 88:235–240
Han Y, Xuan W, Yu T, Fang WB, Lou TL, Gao Y, Chen XY, Xiao X, Shen WB (2007) Exogenous hematin alleviates mercury-induced oxidative damage in the roots of Medicago sativa. J Integr Plant Biol 49:1703–1713
Han Y, Zhang J, Chen X, Gao Z, Xuan W, Xu S, Ding X, Shen W (2008) Carbon monoxide alleviates cadmium-induced oxidative damage by modulating glutathione metabolism in the roots of Medicago sativa. New Phytol 177:155–166
Iturbe-Ormaetxe I, Escurredo PR, Arrese-Igor C, Becana M (1998) Oxidative damage in pea plants exposed to water deficit or paraquat. Plant Physiol 116:173–181
Izawa T, Oikawa T, Tokutomi S, Okuno K, Shimamoto K (2000) Phytochromes confer the photoperiodic control of flowering in rice (a short-day plant). Plant J 22:391–399
Kaminaka H, Morita S, Tokumoto M, Yokoyama H, Masumura T, Tanaka K (1999) Molecular cloning and characterization of a cDNA for an iron-superoxide dismutase in rice (Oryza sativa L.). Biosci Biotech Biochem 63:302–308
Lewinsohn E, Gressel J (1984) Benzyl viologen-mediated counteraction of diquat and paraquat phytotoxicities. Plant Physiol 76:125–130
Linley PJ, Landsberger M, Kohchi T, Cooper JB, Terry MJ (2006) The molecular basis of heme oxygenase deficiency in the pcd1 mutant of pea. FEBS J 273:2594–2606
Liu Y, Xu S, Ling T, Xu L, Shen W (2010) Heme oxygenase/carbon monoxide system participates in regulating wheat seed germination under osmotic stress involving the nitric oxide pathway. J Plant Physiol 167:1371–1379
Lu Z, Liu D, Liu S (2007) Two rice cytosolic ascorbate peroxidases differentially improve salt tolerance in transgenic Arabidopsis. Plant Cell Rep 26:1909–1917
Magneschi L, Perata P (2009) Rice germination and seedling growth in the absence of oxygen. Ann Bot 103:181–196
Muramoto T, Kohchi T, Yokota A, Hwang I, Goodman HM (1999) The Arabidopsis photomorphogenic mutant hy1 is deficient in phytochrome chromophore biosynthesis as a result of a mutation in a plastid heme oxygenase. Plant Cell 11:335–348
Muramoto T, Tsurui N, Terry MJ, Yokota A, Kohchi T (2002) Expression and biochemical properties of a ferredoxin-dependent heme oxygenase required for phytochrome chromophore synthesis. Plant Physiol 130:1958–1966
Murgia I, Tarantino D, Vannini C, Bracale M, Carravieri S, Soave C (2004) Arabidopsis thaliana plants overexpressing thylakoidal ascorbate peroxidase show increased resistance to Paraquat-induced photooxidative stress and to nitric oxide-induced cell death. Plant J 38:940–953
Noriega GO, Balestrasse KB, Batlle A, Tomaro ML (2004) Heme oxygenase exerts a protective role against oxidative stress in soybean leaves. Biochem Biophys Res Commun 323:1003–1008
Noriega GO, Yannarelli GG, Balestrasse KB, Batlle A, Tomaro ML (2007) The effect of nitric oxide on heme oxygenase gene expression in soybean leaves. Planta 226:1155–1163
O’Brian MR (1996) Heme synthesis in the rhizobium-legume symbiosis: a palette for bacterial and eukaryotic pigments. J Bacteriol 178:2471–2478
Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975:384–394
Ryter SW, Alam J, Choi AMK (2006) Heme oxygenase-1/carbon monoxide: from basic science to therapeutic applications. Physiol Rev 86:583–650
Sa ZS, Huang LQ, Wu GL, Ding JP, Chen XY, Yu T, Shi C, Shen WB (2007) Carbon monoxide: a novel antioxidant against oxidative stress in wheat seedling leaves. J Integr Plant Biol 49:638–645
Shaaltiel Y, Glazer A, Bocion PF, Gressel J (1988) Cross tolerance to herbicidal and environmental oxidants of plant biotypes tolerant to paraquat, sulfur dioxide and ozone. Pestic Biochem Physiol 31:13–23
Shekhawat GS, Verma K (2010) Haem oxygenase (HO): an overlooked enzyme of plant metabolism and defence. J Exp Bot 61:2255–2270
Shekhawat GS, Dixit S, Verma K, Nasybullina EI, Kosmachevskaya OV, Topunov AF (2011) Heme oxygenase: enzyme with functional diversity. J Stress Physiol Biochem 7:88–94
Terry MJ (1997) Phytochrome chromophore-deficient mutants. Plant Cell Environ 20:740–745
Watson JM, Fusaro AF, Wang M, Waterhouse PM (2005) RNAi silencing platforms in plants. FEBS Lett 579:5982–5987
Wilks A (2002) Heme oxygenase: evolution, structure, and mechanism. Antioxid Redox Sign 4:603–614
Wilks A, Schmitt MP (1998) Expression and characterization of a heme oxygenase (Hmu O) from Corynebacterium diphtheriae. Iron acquisition requires oxidative cleavage of the heme macrocycle. J Biol Chem 273:837–841
Willekens H, Chamnongpol S, Davey M, Schraudner M, Langebartels C, Van Montagu M, Inzé D, Van Camp W (1997) Catalase is a sink for H2O2 and is indispensable for stress defence in C3 plants. EMBO J 16:4806–4816
Wu M, Huang J, Xu S, Ling T, Xie Y, Shen W (2011) Haem oxygenase delays programmed cell death in wheat aleurone layers by modulation of hydrogen peroxide metabolism. J Exp Bot 62:235–248
Xie YJ, Xu S, Han B, Wu MZ, Yuan XX, Han Y, Gu Q, Xu DK, Yang Q, Shen WB (2011) Evidence of Arabidopsis salt acclimation induced by up-regulation of HY1 and the regulatory role of RbohD-derived reactive oxygen species synthesis. Plant J 66:280–292
Xie Y, Xu D, Cui W, Shen W (2012) Mutation of Arabidopsis HY1 causes UV-C hypersensitivity by impairing carotenoid and flavonoid biosynthesis and the down-regulation of antioxidant defence. J Exp Bot 63:3869–3884
Xu S, Lou T, Zhao N, Gao Y, Dong L, Jiang D, Shen W, Huang L, Wang R (2011a) Presoaking with hemin improves salinity tolerance during wheat seed germination. Acta Physiol Plant 33:1173–1183
Xu S, Zhang B, Cao ZY, Ling TF, Shen WB (2011b) Heme oxygenase is involved in cobalt chloride-induced lateral root development in tomato. Biometals 24:181–191
Xuan W, Zhu FY, Xu S, Huang BK, Ling TF, Qi JY, Ye MB, Shen WB (2008) The heme oxygenase/carbon monoxide system is involved in the auxin-induced cucumber adventitious rooting process. Plant Physiol 148:881–893
Yannarelli GG, Noriega GO, Batlle A, Tomaro ML (2006) Heme oxygenase up-regulation in ultraviolet-B irradiated soybean plants involves reactive oxygen species. Planta 224:1154–1162
Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Prot 2:1565–1572
Zhang X, Migita CT, Sato M, Sasahara M, Yoshida T (2005) Protein expressed by the ho2 gene of the Cyanobacterium synechocystis sp. PCC 6803 is a true heme oxygenase. FEBS J 272:1012–1022
Acknowledgments
We thank Dr M. Wang (CSIRO Plant Industry, Canberra, Australia) for kindly providing the binary vector pVec8_Ubi. This work was supported by the Priority Academic Program Development of Jiangsu Higher Education Institutions, the 111 project, the Fundamental Research Funds for the Central Universities (KYZ200905), and the National Natural Science Foundation of China (30960182).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xu, S., Wang, L., Zhang, B. et al. RNAi knockdown of rice SE5 gene is sensitive to the herbicide methyl viologen by the down-regulation of antioxidant defense. Plant Mol Biol 80, 219–235 (2012). https://doi.org/10.1007/s11103-012-9945-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-012-9945-7