Abstract
Agronomic traits controlling the formation, architecture and physiology of source and sink organs are main determinants of rice productivity. Semi-dwarf rice varieties with low tiller formation but high seed production per panicle and dark green and thick leaves with prolonged source activity are among the desirable traits to further increase the yield potential of rice. Here, we report the functional characterization of a zinc finger transcription factor, OsGATA12, whose overexpression causes increased leaf greenness, reduction of leaf and tiller number, and affects yield parameters. Reduced tillering allowed testing the transgenic plants under high density which resulted in significantly increased yield per area and higher harvest index compared to wild-type. We show that delayed senescence of transgenic plants and the corresponding longer stay-green phenotype is mainly due to increased chlorophyll and chloroplast number. Further, our work postulates that the increased greenness observed in the transgenic plants is due to more chlorophyll synthesis but most significantly to decreased chlorophyll degradation, which is supported by the reduced expression of genes involved in the chlorophyll degradation pathway. In particular we show evidence for the down-regulation of the STAY GREEN RICE gene and in vivo repression of its promoter by OsGATA12, which suggests a transcriptional repression function for a GATA transcription factor for prolonging the onset of senescence in cereals.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Grain yield in cereals is principally determined by agronomic traits controlling the formation and architecture of source and sink organs and by the physiology of the plant affecting source-sink relationships. Alteration of plant architecture characteristics in cereals has resulted in successfully boosting yield potential, especially in rice and wheat (Khush 1999). In the past, reduction of plant height by the manipulation of gibberellin metabolic pathways and achieving plants with greener leaves, sufficient tillers and strong stems, for example, has contributed to double the yield of traditional tall rice cultivars (from 4 t ha−1 to 8 t ha−1) and also increase harvest indices (Khush 2003; Sasaki et al. 2002). Despite this, semidwarf rice cultivars produce a large number of unproductive tillers and excessive leaves that reduce light capture for photosynthesis. Consequently, current research to further increase the yield potential of rice includes development of semidwarf rice varieties with low tiller formation while keeping high seed production per panicle and dark green and thick leaves plus a deep root system (Ikeda et al. 2013).
In rice, grain yield is mainly produced by primary tillers and early secondary tillers. Therefore, whereas increased tillering for high biomass production may be advantageous, reducing the formation of late secondary and tertiary tillers which rarely contribute to yield but still compete for plant nutrients is also a desirable trait (Xing and Zhang 2010). For example, expression of OsSPL14, a member of the SQUAMOSA PROMOTER BINDING PROTEIN-LIKE transcription factor, modifies rice plant architecture resulting in fewer tillers but more productive panicles and consequently higher grain yields (Jiao et al. 2010; Miura et al. 2010). Similar efforts to reduce axillary buds to increase yield potential have been pursued through manipulating the expression of genes involved in the biosynthesis or signaling of strigolactones (SLs) (Umehara et al. 2008). A reduced tiller formation also brings the possibility for cultivars to be planted at higher density (hence higher yields per area) or the advantage of presenting enhanced light capture for photosynthesis and nitrogen storage for grain filling that could also benefit grain yield in rice.
On the other hand, sink strength (high photosynthesis and nutrient remobilization) and source activity (import and reuse of photoassimilates and nitrogen) also have a decisive role in grain filling which is another determining factor for high productivity in rice. Coordination between these organs is also critical. For instance, any carbon imbalance caused by a slow sugar transport or insufficient sink strength in source organs often results in feedback down-regulation of photosynthetic efficiency in leaves (Paul and Foyer 2001). During the rice reproductive stage, panicles are the main sinks whereas the top two leaves and the flag leaf in particular are the primary sources producing over 80% of the total carbohydrate accumulated in the grains (Gladun and Karpov 1993; Yu et al. 2015). Hence, delaying leaf senescence and maintaining photosynthetic activity for longer periods also improves C assimilation, and are among the current approaches for conferring higher yields. The onset of leaf senescence (transition to N remobilization) and the phase of post-anthesis development can be influenced by environmental conditions (Yang and Zhang 2006) but breeders have taken advantage of the genetic regulation of the stay-green trait in crops to prolong canopy photosynthesis and obtain higher yields. In rice, for example, a functional stay-green genotype, SNU-SG1, presents high grain yield (Yoo et al. 2007). In addition, endogenous factors including leaf age and phytohormones have been associated with the control of stay green phenotypes and to the regulation of source-sink activities (Albacete et al. 2014; Wilkinson et al. 2012). Further, cytokinin (CK) levels and signaling pathways mediate or influence leaf longevity (Lim et al. 2007), however regulation of CK-mediated leaf senescence at the level of gene expression has not been elucidated.
A group of transcription factors (TFs), namely GATA TFs, represents a possible link between the regulation of source-sink activities and hormonal signals. The two best characterized GATA TFs are GNC (GATA, NITRATE-INDUCIBLE, CARBON-METABOLISM INVOLVED) and GNL/CGA1 (GNC-LIKE/CYTOKININ-RESPONSIVE GATA FACTOR1) from Arabidopsis thaliana (Bi et al. 2005; Richter et al. 2010). GNC and CGA1 are induced by CKs and are thought to participate in regulating the balance between GA and CK signaling pathways (Hudson et al. 2011) as well as CK and auxin (Richter et al. 2013). This has also been evidenced by phenotypic analysis of gnc and cga1 mutants which, among other characteristics, present reduced chlorophyll levels (Hudson et al. 2011; Richter et al. 2010). In rice, manipulation of OsCGA1 expression affects chloroplast formation, chlorophyll content, senescence, and tiller and biomass production (Hudson et al. 2013), suggesting that expression of GATA factors can have potential agricultural applications.
In this work, we describe the functional characterization of OsGATA12, a new GATA TF whose expression regulates chlorophyll levels, senescence and yield parameters in rice. We provide evidence to propose a molecular mechanism for the control of chlorophyll degradation by this TF. The delayed senescence and modified plant architecture with low tiller formation of transgenic overexpression plants translated in a high harvest index, thus presenting an option for increased rice yield potential under high density planting.
Materials and methods
Plant growth conditions
Wild-type (WT) plants and single copy homozygous transgenic lines (OE) (Oryza sativa ssp. japonica, cultivar Kaybonnet) were grown in pots with a 1:4 mixture of peat moss:vermiculite and 1 g of Nutricote® 13-13-13 slow-release fertilizer with micronutrients (Plant Products). Plants were grown in growth chambers with 12 h of light (500 µmol m−2 s 1) at 30 °C, 12 h of dark at 25 °C, and constant 65% relative humidity. For high and low density growing assays, WT and OE seeds were sown in water-soaked sand for germination and after 10 days, seedlings were transferred into 75 l plastic containers with the same substrate mix. Plants were kept in a greenhouse with natural light (April to November crop season; 43°N) and controlled constant temperature of approximately 29 °C. For low density conditions, 24 uniformly spaced plants were placed per container (equivalent to 100 plants m−2), while for high density conditions, 96 plants were transplanted in each container (or 400 plants m−2). In both cases plants were able to complete their life cycle. Plants were kept watered with a fertilizer solution.
Plant transformation
Generation of overexpression and RNAi lines was performed by Syngenta. A genetic construct to overexpress OsGATA12 in rice was generated by cloning the full-length cDNA into an ubiquitin-driven vector. Agrobacterium tumefaciens-mediated transformation was performed according to standard protocols. Single-copy transgenic lines were identified examining the selfed progeny of independent T1 plants presenting 3:1 co-segregation of the hygromycin gene and by mapping one T-DNA insertion according to a TAIL-PCR method described by Liu and Chen (2007). T2 plants producing 100% of transgenic progeny were selected for the experiments. For generating the RNAi lines, a conserved and specific cDNA segment (319 bp) was amplified and cloned into the pANDA-RNAi vector using the GATEWAY™ Cloning System. The ProOsGATA12::GUS construct was generated by fusing a 1848 bp upstream region from the OsGATA12 start codon into the pMDC163 target vector. Primers used for cloning are listed in Supplementary Table S1.
Gene expression analysis
Extraction of total RNA, cDNA synthesis, real-time PCR and relative quantification for each target gene were performed according to Lu et al. (2015). Primer sequences for each gene analyzed and for the OsACTIN2 internal reference gene were designed with the Primer Express 2.0 software (Applied Biosystems) and are listed in Supplementary Table S2.
Tissue staining and microscopy
Longitudinal and cross-sections were taken from similar positions near the middle part of shoots or leaf blades in 4-week-old plants, stained with 0.05% toluidine blue for 2–3 min and then rinsed with water before observation using a Leica DM LS2 microscope.
Subcellular localization of OsGATA12
A C-terminus fusion of the coding sequence of OsGATA12 was generated with the N-terminus of the green fluorescent protein (GFP) and inserted into the pMDC32 vector to generate the 35S::OsGATA12-GFP construct. A vector carrying GFP alone was used as control. The vectors were transformed into rice protoplasts according to the protocol described previously (Chen et al. 2006). Fluorescence signals were imaged using a confocal laser-scanning microscope (Leica TCS SP5).
Transcriptional activation in yeast
The OsGATA12 coding sequence was amplified and cloned directional and in-frame into the pBGKT7-DB vector between the NdeI and BamHI restriction sites. Positive clones were transformed into the Y2HGold strain using the LiAc yeast transformation method according to Yeastmaker™ Yeast Transformation System 2 instructions (Clontech) and then grown on plates containing the minimal synthetic defined medium without tryptophan (SD/-Trp), or SD/-Trp supplemented with 40 µg/ml X-α-Gal and 125 ng/ml Aureobasidin A (SD/-Trp/X/A) or SD/-His plates. Plates were incubated at 30 °C for 2 days until photographed. Lack of autoactivation was indicated by white colonies on SD/-Trp and absence of colony growth on SD/-Trp/X/A and SD/-His plates. A pBGKT7-DB empty vector was used as a negative control.
Determination of chlorophyll content
Leaf tissues were ground in liquid nitrogen and 100 mg of leaf powder was used to extract pigments using 80% acetone. Chlorophyll absorbance was analyzed by spectrophotometry at 645 and 663 nm and the contents determined by the method described by Arnon (1949).
Amino acid quantification
Total soluble amino acids were extracted from 100 mg fresh weight of frozen leaf tissue at room temperature successively with 80, 50 and 0% ethanol in 10 mM HEPES–KOH (pH 7.4). Supernatants were pooled and assayed according to Rosen (1957).
Starch and sugar analysis
For starch quantification, leaves (100 mg fresh weight) were flash frozen in liquid nitrogen, ground to a fine powder and extracted with 1 ml 100% methanol by shaking at 70 °C for 15 min. The extraction was repeated twice and insoluble residue was freeze dried overnight and weighed. 20 mg of the extracted sample were used to determine the content of starch and total sugars using a Megazyme Assay kit according to the manufacturer’s instructions (Megazyme International Ireland Ltd).
Root phenotyping
Ten-day-old seedlings were transferred to a hydroponic system to continue growth for eight additional weeks. For each sample, an intact root system was evenly spread apart in a water layer on a transparent tray and imaged with a scanner. Images were analyzed using the WinRHIZO software (V5.0, Regent Instruments) to calculate the total sum length of the root system, total root surface area, average diameter and number of root tips.
Microarray and GO analyses
Eight-week-old plants were used for microarray analysis. Five microgram of total RNA from each sample was used to synthesize double-stranded cDNAs. Labeled complementary RNA, synthesized from the cDNA, was hybridized to the NSF Rice 45 K oligonucleotide array (http://www.ricearray.org). Hybridization signal of the arrays was obtained with a GeneChip scanner 3000 and quantified using MAS 5.0 (Affymetrix). Intensity data of all probe sets was analyzed using GeneSpring software (Agilent). The data was normalized with a default setting of the program, followed by gene filtering which required that each transcript must have either a present (P) or marginal (M) flag in the three replicate samples. Differentially expressed genes in the transgenic line with at least 1.5-fold change were identified first, and then ANOVA was used to detect significance (Welch’s t test; P value cut-off at 0.05). Gene Ontology (GO) term enrichment analysis was performed with the AgriGO tool (http://bioinfo.cau.edu.cn/agriGO; Du et al. 2010). Lists of up- and down-regulated genes were exported to AgriGO and significant GO terms were identified by Singular Enrichment Analysis (SEA) using locus IDs and available GO term annotation from the Rice TIGR Genome Reference. Fisher and FDR statistical test methods were used considering a 0.05 significance level.
Transient assay for promoter repression analysis
A DNA sequence (2137 bp upstream the ATG start codon of the cDNA) corresponding to the SGR promoter was amplified from rice genomic DNA using the OsSGR-prom-F/R primer pair and cloned in the pCAMBIA1305.1 vector to generate a GUS reporter construct. For the effector construct, the coding region of OsGATA12 was inserted into the pMDC32 vector between the DNA recombination sequences (att sites) and behind the cauliflower mosaic virus (CaMV) 35S promoter. Primers used for cloning are listed in Supplementary Table S1. An empty vector with CaMV35S was used as negative control. Reporter and effector plasmids were introduced separately into the Agrobacterium tumefaciens EHA105 strain by electroporation. Transient expression assays were performed using an agroinfiltration method described by Yang et al. (2000) in fully expanded leaves of 7-week-old Nicotiana benthamiana plants and with equal ratio of effector:reporter for co-infiltration. After infiltration, plants were maintained in a growth chamber at 22 °C and 16 h light for 48 h or at constant darkness to activate the SGR promoter. Experiments were done in triplicate; in each experiment six 12 mm discs from infiltrated areas of three independently infiltrated plants were frozen in liquid nitrogen and total protein was extracted to measure GUS activity. Six discs were taken from infiltrated areas from three independent plants. Samples were frozen in liquid nitrogen and total protein was extracted to measure GUS activity. Each experiment was repeated at least twice. Transient GUS expression was evaluated by quantification of the GUS activity using the fluorometric method described by Jefferson et al. (1987) and enzyme activity was recorded as concentration of the 4-methylumbelliferone product over total protein content. Protein content was determined according to the Bradford protein assay using bovine serum albumin as a standard.
Statistical analysis
Data were analysed using the Student’s t-test. Significant differences were determined based on P < 0.05 or P < 0.01. All analysis was performed with the Statistical Package for Social Sciences software (SPSS ver. 11.0 for Windows).
Results
OsGATA12 is a member of the Zn-GATA family of transcription factors
Because chlorophyll content (greenness) is an indicator of plant health and can be used to optimize nitrogen (N) fertilizer application, we have focused our investigation in characterizing TFs whose expression affects chlorophyll levels and also responds to changes in N conditions (Bi et al. 2005). Based on the phenotype of Arabidopsis GATA T-DNA insertion mutants (Bi et al. 2005) and the homology between Arabidopsis and rice GATA genes, we identified few rice genes encoding GATA TFs with potential role in chlorophyll accumulation or N response. One of them is Os03g08312000/LOC_Os03g61570, which is among the 28 different loci in the rice genome encoding proteins containing GATA-like zinc fingers (Reyes et al. 2004). It belongs to the Group B or Subfamily II of rice GATAs characterized for being short proteins with a conserved LLM-(leucine–leucine–methionine) domain at their very C-terminus and a conserved zinc finger DNA-binding domain (Behringer and Schwechheimer 2015). A BLAST analysis indicated that its sequence overlaps with a gene encoding a 136 amino acid protein designated as OsGATA12 by Reyes et al. (2004).
Expression of OsGATA12 was examined by real-time qPCR in different tissues. The gene was expressed in vegetative and reproductive tissues including sheaths, panicles, stems, leaves and roots (Fig. 1a). Expression of the gene was also monitored using transgenic lines bearing a genetic construct with the GUS reporter gene fused to the OsGATA12 promoter. GUS staining was detected in the same tissues and, consistent with the qPCR data, the OsGATA12 promoter activity was most intense in roots (Fig. 1a).
Alteration of OsGATA12 expression affects greenness and biomass production
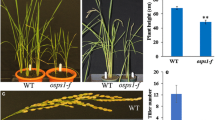
To study the function of OsGATA12, transgenic plants representing 12 overexpression (OE) and six RNAi knockdown lines were generated. Within each class, all single-copy transgenic lines displayed similar phenotypes and two genetically stable homozygous lines of each category were chosen for detailed functional analysis (Supplementary Fig. S1). Compared to WT plants, OsGATA12 OE lines presented obvious phenotypic differences in terms of biomass and greenness throughout plant growth (Fig. 2a–d). A reduction of the number of leaves and tillers and total biomass was observed in OE lines during vegetative and reproductive stages (Fig. 2a–g) however no occurrence of stem bending (lodging) was observed. Flowering time (days to heading) was also delayed ca. 10 days in the same transgenic lines compared to WT (Fig. 2h). In particular, leaves in the overexpression lines were greener than in WT throughout the vegetative and reproductive phases (Fig. 2a–d). The greener appearance of the OE lines was corroborated by quantifying the total chlorophyll content at 35, 70, 105 and 140 days after germination. Although leaves from OE lines presented higher chlorophyll content at the vegetative stages, bigger differences were evident in the reproductive and mature stages compared to WT plants, suggesting also a delayed senescence phenotype in the transgenic lines (Fig. 2i). This increased chlorophyll content was positively linked to significantly higher content of amino acids during all developmental stages (Supplementary Fig. S2a) and higher starch and soluble sugar concentrations maintained at maturity in leaves of OE plants compared to their WT counterparts (Supplementary Fig. S2b, c). In contrast to OE lines, the phenotype of RNAi knockdown lines targeting OsGATA12 did not show differences in terms of plant architecture and yield parameters compared to WT plants (Supplementary Fig. S3), but presented less greenness in aerial tissues (Fig. 2j). The reduced greenness of RNAi plants compared to WT correlated with a reduction in total chlorophyll content which was quantified in the leaves (Fig. 2k).
Phenotype of OsGATA12 transgenic lines. a–d Wild-type (WT) and transgenic lines overexpressing (OE) OsGATA12 (O13, O16) at a 35 d, b 70 d, c 105 d and d 140 d after germination. e–h Growth parameters of WT and OE lines at 35 d showing: e number of leaves, f number of tillers, g biomass (as total dry weight) and h days to heading. i Total chlorophyll content in WT and OE lines during the same developmental stages. j Comparison of WT and RNAi lines for OsGATA12 (R3, R5) at 140 d. k Chlorophyll content in WT and RNAi lines at the same mature stage (140 d). Bars represent means ± SD (n = 12; Student’s t-test P-values, *P < 0.05, **P < 0.01)
Based on the phenotypic changes observed, the OE plants were used for the further examination of morphological characteristics. At the root level, overexpression of OsGATA12 also resulted in decreased root growth that translated into reduced total length of the root system and fewer root tips but not significant less surface area (Supplementary Fig. S4a-f). Plant height was also affected by overexpression of OsGATA12. Reduction of plant height was more significant during the vegetative phase than during the mature stage (Fig. 2a–d, Supplementary Fig. S5a). Close examination of stem epidermal cells showed that the length of stem epidermal cells was not reduced in OE lines compared to WT (Supplementary Fig. S5b–d), suggesting that any differences in height at this stage may be due to a disparity in cell division. In addition, transversal sections of leaves and sheaths revealed a larger number of chloroplasts in OE lines than in WT that accumulated mainly in mesophyll cells (Supplementary Fig. S5e), which accounts for the pronounced greenness and increased chlorophyll content in these transgenic plants. All these observations indicate that overexpression of OsGATA12 affects plant characteristics during vegetative and reproductive growth but particularly indicates a specific role of OsGATA12 associated with chlorophyll accumulation.
OsGATA12 OE lines exhibit increased seed yield when planted at high density
The reduced tiller number and biomass of OE transgenic plants translated in declining yield parameters compared to WT plants (Supplementary Fig. S6). Panicle length, number of seeds and seed yield per panicle and seed size (i.e., 1000 grain weight) in OE lines were not different from WT plants (Supplementary Fig. S6a–d), indicating a similar seed setting rate between the genotypes. However total seed number and total yield per plant in the OE lines were lower than in WT (Supplementary Fig. S6e, f). This was essentially due to the fewer tillers formed in the transgenic plants (Fig. 2f). As one of the focuses for breeding high yielding rice is to maximize the number of panicles per unit area rather than panicles per plant, we tested the effect of increasing the planting density four-fold and compared the biomass production and yield of the transgenic lines with those when they were grown under normal low planting density (LD) of 100 plants per m2. At LD planting, OsGATA12 OE lines presented around half the number of tillers per unit area of that of WT, whereas when planted at high density (HD) the transgenic plants had almost threefold the total number of tillers per unit area while WT plants only increased 30% (Fig. 3a). Tillering and yield per panicle was affected in WT plants when grown at HD. Even when OE plants exhibited fewer tillers than WT, the yield per panicle in OE plants grown at HD did not diminish and was higher than in WT. This distinction translated in differences in seed production per unit area. Seed yield per area was reduced ca. 15% in WT plants when planted at HD, meanwhile OsGATA12 OE lines presented around 100% increase in seed yield compared to LD planting and around 30% more seed yield/area than WT plants (Fig. 3b). Although the reduced biomass and tillering in OsGATA12 OE plants led to a reduced yield, under LD conditions their harvest index (seed mass/total biomass) was similar to that of WT plants. However, when planted at HD, OsGATA12 OE lines could maintain an elevated harvest index while in WT plants the harvest index was reduced approximately 50% (Fig. 3c).
Growth and yield performance of OsGATA12 OE lines under different density planting systems. WT and OsGATA12 OE lines where planted at low density (LD, 100 plants/m2 or high density (HD, 400 plants/m2). Measurements were performed at 140 d after sowing. a Number of tillers. b Yield per area. c Harvest index. Bars represent means ± SD (n = 12; Student’s t-test P-values, *P < 0.05, **P < 0.01)
OsGATA12 acts as a transcriptional repressor of chlorophyll degradation
Analysis of OsGATA12 amino acid sequence showed that there are strong putative nuclear localization signals between positions 55 and 84 and from 107 to 115, implying that as TF, GATA12 should be localized in the nucleus. To verify this, subcellular localization of OsGATA12 was examined by monitoring the GFP fluorescence in rice protoplasts transformed with the fusion construct OsGATA12-GFP. Green fluorescence signals were exclusively observed in the nucleus of cells transformed with the fusion construct (Supplementary Fig. S7a), indicating that OsGATA12 is indeed a nuclear protein.
To investigate if OsGATA12 functions as transcriptional activator, a yeast one-hybrid analysis was followed. Yeast cells containing the control vector pGBKT7 or the fusion plasmid with OsGATA12 fused to the GAL4 DNA-binding domain grew well on selective SD medium without tryptophane (-Trp). However, growth was inhibited when cells were streaked on SD medium lacking tryptophane and supplemented with X-Gal and Aureobasidin A (-Trp/X/A) or histidine (-His), indicating that OsGATA12 could not function as a transcriptional activator. A NAC TF (ANAC032) used as positive control grew well in these media (Supplementary Fig. S7b).
A microarray experiment was conducted to analyze the potential target genes and pathways affected by OsGATA12 overexpression. 533 probe sets (333 up-regulated and 200 down-regulated) were identified as exhibiting differential expression (greater than 2–fold change, P < 0.05) in leaves between a transgenic line and WT (Supplementary Table S3). Gene Ontology (GO) functional enrichment analysis was performed with the gene lists of up- and down-regulated genes in the transgenic line compared to WT. GO terms for biological processes are shown in Supplementary Fig. S8 and are plotted as a heat map to visualize distinct and common processes enriched in the two categories of differentially expressed genes (DEG). Out of 49 terms, only 11 were commonly enriched between both up- and down-regulated genes. 24 and 36 different GO terms were significantly enriched between the up- and down-regulated genes, respectively (Supplementary Fig. S8). Those represented diverse ontology terms associated with metabolism, signaling, regulation, transport, defense and stress. Among the up-regulated terms, the most significantly enriched were those related to transcription, DNA replication and integration, oxidation–reduction, carbohydrate metabolism and proteolysis and flower development. With regard to genes involved in carbohydrate metabolic process, we focused our attention to genes encoding sugar transporters whose function can affect reserve mobilization and yield. We evaluated the expression of genes that respond to cytokinin and gibberellin signaling and participate in sink/source sugar mobilization. Six out of ten genes encoding for a cell wall invertase (OsCIN1), sucrose transporters (OsSUT2, OsSUT5), sucrose phosphate synthases (OsSPS1, OsSPS11) and a SWEET sugar efflux carrier (OsSWEET3a) were upregulated in OsGATA12 OE plants (Fig. S9). On the other hand, enriched down-regulated genes included diverse GO terms related to transport and metabolism. Interestingly, two of the significant enriched terms in this category were associated with chlorophyll and light-harvesting complex II catabolic processes (Supplementary Fig. S8). Annotation of genes in this group was performed using the PlantGDB database and additional BLAST searches for unknown genes. Those were represented by Os09g0532000, encoding the senescence-inducible chloroplastic STAY-GREEN PROTEIN 1 (SGR); Os01g0227100, encoding NON-YELLOW COLORING 1 (NYC1), a short chain dehydrogenase/reductase and Os06g0354700 and Os02g0705100, encoding the hydrolases NON-YELLOW COLORING 3 (NYC3) and NYC3-like 1 (NHL1). As these genes could be potential targets of GATA12, we evaluated their expression in both WT and OE plants along with the expression of other genes known to be implicated in chlorophyll degradation. Among the eight genes evaluated (listed in Supplementary Table S2), expression of NYC3, SGR and genes encoding a red chlorophyll catabolite reductase (RCCR) and a pheophorbide a oxygenase (PAO) was significantly reduced in OE lines compared to WT (Fig. 4a). Since GATA factors also affect chlorophyll formation, expression of seven important chlorophyll biosynthesis genes was also analyzed. Of those, only expression of HEMA1 (encoding a NADPH-dependent glutamyl-tRNA reductase), PORA (protochlorophyllide oxidoreductase a) and CAO1 (chlorophyllide a oxygenase 1) was up-regulated less than two-fold in OE plants (Fig. 4b). In addition, besides SGR, other senescence associated genes (SAGs) showed down-regulated in the array experiment. Thus, we selected four reported rice SAGs: H36 (encoding a senescence-induced aminotransferase; Lee et al. 2001), OsNAP/PS1 (premature senescence-1 NAC TF; Liang et al. 2014a), OsAP10 (aspartic proteinase oryzasin-1; Lee et al. 2001) and CatB (Catalase B; Liu et al. 2008) to evaluate their transcript levels. Significant down-regulation of OsNAP, OsAP10 and CatB was detected in the transgenic plants (Fig. 4c). In the array data, transcription signals of these three genes were also reduced in the OE line (Supplementary Table S3).
Expression analysis of genes associated with chlorophyll metabolism and senescence. a Expression levels of chlorophyll degradation-related genes. b Expression of chlorophyll biosynthesis genes. c Expression of senescence associated genes. qPCR (three biological replicates and three technical replicates) was performed with samples from 70-day-old WT and OsGATA12 OE plants. Name of genes are detailed in Supplementary Table S2. Bars represent means ± SD (n = 9; Student’s t-test P-values, *P < 0.05, **P < 0.01)
Because down-regulation of SGR expression was the most drastic in the OE transgenic lines, the possibility that OsGATA12 may in fact act by repressing SGR transcription was evaluated by testing the activity of the SGR promoter in an in vivo transactivation assay. Co-transfection with an effector construct for constitutive expression of OsGATA12 (Fig. 5a) caused a significant reduction of GUS activity driven by the SGR promoter compared to the assay with an empty effector vector used as control (Fig. 5b). Similar substantial repression of the SGR promoter activity was observed when the transfected tissue was incubated in the dark to induce SGR activity (Fig. 5b). These data indicates that OsGATA12 may be acting as a repressor of chlorophyll degradation in rice which concurs with the phenotype observed in the OE lines. In this respect, our observations suggest that OsGATA12 functions primarily in delaying chlorophyll degradation and leaf senescence.
In vivo repression of the SGR promoter by OsGATA12. a Representation of the reporter and effector constructs used in the transient expression assays. b Agrobacterium cells harboring the reporter construct SGRprom::GUS was co-infiltrated into N. benthamiana leaves with (+) or without (−) bacteria cells carrying the effector construct 35S::OsGATA12. Transformed plants were then incubated under constant light or dark for 48 h. Bars indicate GUS activities means ± SD (n = 9). Statistically differences are shown between activity with and without effector construct (Student’s t-test P-values, **P < 0.01)
Discussion
GATA transcription factors play different roles in growth, development, and cell proliferation (Patient and McGhee 2002; Scazzocchio 2000). In plants, GATA factors have been principally linked to responses to the environment, mainly light- and circadian-regulated gene expression (Behringer and Schwechheimer 2015) and other biological functions are not well understood. Here, we report that expression of OsGATA12 affects aspects of rice plant architecture and prolongs source leaf activity.
Effect of OsGATA12 on greenness and source activity
Recent studies of overexpression and cga1 and gnc loss-of-function lines have substantiated a role for CGA1 and GNC as transcriptional regulators of chloroplast biogenesis, acting downstream of cytokinin (CK) and enhancing chloroplast growth and division as well as chlorophyll synthesis (Chiang et al. 2012; Hudson et al. 2011). Similar to CGA1 and OsCGA1 (Hudson et al. 2013), OsGATA12 promotes chloroplast development in rice (Supplementary Fig. S5d). In the case of rice overexpressing OsCGA1, up-regulation of the rice FtsZ chloroplast division gene by CGA1 offers a potential mechanism for controlling chloroplast proliferation (Hudson et al. 2013). Although FtsZ was not detected in the OsGATA12 microarray analysis, because of the comparable characteristics of leaf cross sections and chloroplast abundance in OsGATA12 OE and OsCGA1 OE transgenic plants, we can speculate that stimulation of similar steps of chloroplast biogenesis may be occurring in both of these genotypes.
GNC and CGA1/GNL were also identified as repressors of auxin and gibberellins (GA) signaling influencing plant growth (Richter et al. 2013), raising the possibility that besides transcriptional activators they may also act as repressors. Even then, the mechanisms by which these plant GATA factors are able to repress transcription have remained for the most part unexplored. Some evidence comes from fungal GATA transcription factors, which are also engaged in nitrogen metabolism and light induction (Scazzocchio 2000). Interestingly, the mechanisms by which they regulate nitrogen catabolic genes and biosynthesis of certain secondary nitrogen metabolites in response to nitrogen availability in fungi also involve repression of transcription (Tudzynski 2014).
In our study, OsGATA12 was unable to activate transcription in a yeast transactivation assay, suggesting that it may act as a repressor. According to our transcriptomic analysis, several genes associated with senescence and chlorophyll metabolism were down-regulated in OsGATA12 OE plants (Supplementary Table S3). These included two senescence-induced receptor-like protein kinases (Os09g0359500 and Os09g0355400), one leaf senescence related protein (Os03g0817900) and a chloroplast early light-induced protein (ELIP; Os01g0246400). ELIPs have a photoprotective role of thylakoid membranes against oxidative stress, for example during natural senescence (Binyamin et al. 2001); thus, down-regulation of ELIP expression in GATA12 OE plants might result from a lower requirement for this protein when senescence is delayed. The senescence associated genes CatB, OsNAP and OsAP10 (Liu et al. 2008; Liang et al. 2014a; Lee et al. 2001) also appeared in the array data and down-regulation of their expression was validated through qPCR (Fig. 4c). Cysteine proteases are also usually induced during senescence (Malgorzata and Barbara 2004). In the array, three Cys proteinases 1 (Os03g0752500, Os04g0311400 and Os09g0442300) were down-regulated 1.6- to 4.5-fold in the OE line. All these data supports the perception that the senescence process is delayed in OE plants. In addition, four genes involved in chlorophyll degradation had reduced expression in OE lines (Fig. 4a). NYC3, encoding a plastid-localizing a/b hydrolase family protein, was down-regulated about 40% compared to WT. Expression of NYC3 increases during leaf senescence in rice and its loss-of-function mutant displays a stay-green phenotype (Morita et al. 2009). Though no enzymatic activity has yet been proven for NYC3, it is ortholog to pheophytin pheophorbide hydrolase (PPH; Hörtensteiner 2013) and a mutation in NYC3 affects the function of NYC1/Chl b reductase and chlorophyllase, the enzymes that catalyze the first steps of chlorophyll b and a degradation (Morita et al. 2009). In the transgenic line, expression of PAO and RCCR, encoding enzymes that degrade chlorophyll(ide) to a non-photoreactive chlorophyll catabolite (Hörtensteiner 2013), were about half of that in WT (Fig. 4a). Finally, and most noteworthy, the expression levels of SGR (also a senescence associated gene) in the OsGATA12 OE lines were ca. 20% of those in WT (Fig. 4a).
SGR encodes a chloroplast protein required to trigger chlorophyll degradation during natural and dark-induced leaf senescence (Park et al. 2007). Although it is not completely clear how SGR activates chlorophyll degradation during leaf senescence, it is known that this protein interacts in vivo with subunits of the light-harvesting complex of photosystem II (LHCII) and with five plastid-localized chlorophyll catabolic enzymes (including NYC1, PPH, PAO and RCCR) all of which form a protein complex for chlorophyll degradation (Sakuraba et al. 2015). Another rice homolog, SGRL (for SGR-like), may have overlapping roles but, because of its expression pattern, it is considered to be involved in chlorophyll turnover or degradation in pre-senescent leaves and in the degradation of LHCI proteins (Rong et al. 2013). Thus, these two SGR proteins are involved in a disassembling mechanism of intact LHCs in the thylakoid membranes and their expression would be a precondition for chlorophyll breakdown (Hörtensteiner 2013). Similar to OsGATA12 OE lines, rice sgr mutants maintain greenness during leaf senescence while transgenic rice overexpressing SGR produces yellowish-brown leaves (Park et al. 2007). Though we do not discard the option that OsGATA12 also affects chlorophyll biosynthesis (Fig. 4b), our gene expression analysis suggests that one way by which OsGATA12 affects greening is through down-regulation of genes involved in chlorophyll stability. Four putative GATA binding sites (with the A/TGATAA/G consensus) are present in the rice SGR promoter (within 2 kb upstream the transcription start site). Whether OsGATA12 binds directly to one of these sites or interferes with a transcriptional complex, remains to be investigated. Changes in the phenotype of OsGATA12 RNAi plants were less severe that in OE lines suggesting a possible functional redundancy exists among B-GATA proteins. In addition, the fact that B-GATA factors can interact with each other could suggest that they may act as dimeric complexes having varied DNA-binding specificities that can modulate their function as transcriptional repressors or activators (Behringer et al. 2014; Zhang et al. 2013). Interestingly, in Arabidopsis, the SGR1 gene is induced by the bHLH transcription factors phytochrome interacting factor 4 (PIF4) and PIF5 during senescence and by transcription factors mediating ABA signaling (Sakuraba et al. 2015), suggesting that the role of ABA during senescence involves SGR1-mediated chlorophyll degradation. Several reports identified PIFs as key participants in the expression of genes regulated by GA, brassinosteroids (BR) and auxin to modulate plant growth and development (reviewed in de Lucas and Prat 2014), thus, it is possible that SGR is among the target genes of several hormone signals where GATA TFs such as OsGATA12 participate.
Ethylene, ABA, JA and SA are also internal factors known to promote leaf senescence, whereas CK, auxin and GA can postpone it. Modification of hormone signaling pathways also leads to altered leaf senescence (Li et al. 2012). Positive correlation has been found between CK production and delayed leaf senescence (Gan and Amasino 1995), suggesting that CK delays senescence by promoting the phase of active photosynthesis in green tissues. This has direct implications on source-sink relations that can result in a morphological adjustment to optimize growth and hence can impact crop productivity. Likewise, evidence exists to support positive effects of GA on plant growth and yield by increasing source strength and promoting cell division, growth and carbohydrate import thus increasing sink activities (Iqbal et al. 2011; Roitsch and González 2004). However it is still unclear whether GATA TFs have a role in CK or GA signaling controlling source-sink relations. Likewise, any possible interaction between GATA TFs and other hormonal pathways possible involved in late rice senescence (e.g., ethylene or jasmonate) cannot be excluded prematurely.
The molecular mechanisms by which leaf greenness is combined with effective photosynthetic capacity in stay-green genotypes remain unclear (Gregersen et al. 2013). It is plausible that the delayed senescence phenotype of OsGATA OE lines is not only a result of down-regulation of genes associated with the catabolism of chlorophyll and LHCII but also of higher protection of PSII and thylakoid membranes and increased antioxidative capacity, as suggested by enrichment of GO terms related to responses to oxidative stress (Supplementary Fig. S8). This could facilitate the regeneration of chloroplast ultrastructures by maintaining protein synthesis (Luo et al. 2013). In effect, according to the microarray data, photosynthesis-related genes encoding chlorophyll a-b binding proteins, PSII polypeptides and rbc small chain showed higher expression in the OE line (Supplementary Table S3).
Delayed leaf senescence in OsGATA12 lines can also be explained by the availability of excess nitrogen relative to low levels of carbon after anthesis (Supplementary Fig. S2; Wingler et al. 2006). Thus nitrogen stored during pre-anthesis in vegetative tissues and carbon reserve buildup after anthesis facilitated transfer of reserves to the grain to contribute to grain-filling. Genetic factors determining extended periods of grain filling to increase yield in functional stay-green genotypes have been valuable for breeding approaches (Gregersen et al. 2013). The OsGATA12 OE lines presented not only delayed chlorophyll degradation but perhaps also extended carbon assimilation as suggested by more soluble sugars and more starch in leaves than WT plants at mature stages (i.e., 140 DAG; Fig. S2). This observation also agrees with the fact that overexpression of OsCGA1 results in more starch content in seeds (Hudson et al. 2013). In addition, GO terms for carbohydrate metabolic process and carbohydrate transport were enriched among up-regulated genes in OsGATA12 OE (Supplementary Fig. S8), implying that an adjustment of carbon resources to secure yield may also be influenced at the transcriptional level. Related to this, we found that genes encoding proteins involved in source/sink sucrose mobilization were upregulated in OsGATA12 OE plants (Fig. S9). Expression of genes encoding for cell wall invertases, sucrose transporters, sucrose phosphate synthases and SWEET sugar efflux carriers or their enzymatic activities have been reported to be regulated by CK and GA in rice and other cereals (Chávez-Bárcenas et al. 2000; Yuan et al. 2014; Wang et al. 2016), which concurs with the notion that GATA TFs mediate CK and GA signals and may be facilitating resource mobilization.
Effect of OsGATA12 on plant architecture and yield
Though growth and heading (flowering) time was somewhat delayed in OsGATA12 OE plants, mature plants presented similar height and fully productive panicles compared to non transgenic controls (Supplementary Figs. S5, S6). On the other hand, reduced expression of OsGATA12 did not cause any pleiotropic phenotype in plant growth or yield besides an obvious reduction of chlorophyll content (Fig. 2j, k). Notably, overexpression of OsGATA12 also caused reduction of the number of tillers (Fig. 2). Several low or high tiller mutants of rice have been characterized (Liang et al. 2014b) and most of them turned out to be defective in steps controlling hormone levels or hormone signaling pathways. It is plausible that OsGATA12 may also affect hormonal signals controlling branching. Auxin, brassinosteroid and strigolactone pathways are involved in determining rice plant architecture as has been shown recently (Liang et al. 2014b; Xu et al. 2015). Among the genes differentially expressed in OsGATA12 OE plants, there were six coding for AUX/IAA factors that were up-regulated and two coding for auxin efflux carriers that were down-regulated (Supplementary Table S3). Though most BR-related dwarf mutants present more tillers than wild-type (Morinaka et al. 2006), a rice BR mutant, dlt, with reduced sensitivity to BR has erect leaves and, similarly to OSGATA12 OE plants, has a semi-dwarf phenotype with fewer tillers and dark-green leaves (Tong et al. 2009). It has been suggested that BR affects rice tillering by regulating DLT expression through OsBZR1 (Tong et al. 2009). Interestingly, one gene encoding a BR receptor, three BR receptor-associated kinases and one gene encoding a BES/BZR1 homolog (a negative regulator of BR signaling) were differentially expressed in OsGATA12 OE compared to WT (Supplementary Table S3). Although this mechanism for controlling tiller formation has not been completely elucidated, it has been postulated that it could be a result of a complex interaction between BR and other hormones (Zhang et al. 2014). For instance, strigolactones influence auxin flux and distribution (Koltai 2014) which can also alter the number of tiller in rice plants (Zhang et al. 2010). Whether OsGATA12 is involved in an interaction between BR, auxin and other signaling pathways remains to be explored. In addition, besides tiller formation, transgenic rice plants expressing genes that control BR hormone levels in vegetative tissues showed enhanced carbon assimilation, increased glucose content in flag leaves, more starch in seeds and yield more seeds than wild-type plants (Wu et al. 2008), which suggest that BRs also have a positive effect in loading photoassimilates to sinks. It is possible that the extended carbon accumulation in OsGATA12 OE lines could also be a result of adjusted hormonal signaling pathway(s).
Reduced tillering allowed us to test the OsGATA12 OE plants under high density. From a yield perspective, the optimal planting for WT plants was at low density whereas OsGATA12 OE plants grew well at higher density. Results showed that these transgenic plants retained their yield per panicle and this was especially significant for maintaining the harvest index and to produced increased yield per area (Fig. 3) which has significant agronomic implications for improving field productivity. Likewise, other studies have also shown that while some rice BR biosynthesis mutants have lower yield than wild-type at harvest under conventional planting density, they present higher biomass and yield at high planting density (Morinaka et al. 2006). The same research group noted that the osdwarf4 and d11 mutants, with limited defects in BR biosynthesis, present erect leaf phenotype which was associated with enhanced grain yields under dense planting conditions, even without extra fertilizer (Sakamoto et al. 2006). In these examples, it is reasoned that the increased grain yields of these cultivars are due to their erect leaves that offer better light capture for photosynthesis and nitrogen storage for grain filling (Morinaka et al. 2006; Sakamoto et al. 2006). Besides the dominant effect of BR on leaf angle, signaling of auxin (IAA) and gibberellins (GA) also plays roles in the regulation of lamina joint bending in rice (Luo et al. 2016). Even though OsGATA12 OE lines could be grown at high densities, we did not observe significant differences in leaf or tiller angle affected by OsGATA12, yet any interaction between OsGATA12 and the hormone signals affecting such architectural traits should be matter of future investigations. Our observations indicate that OE plants assimilate biomass in reproductive organ development more effectively at high planting density compared to WT. Even though OsGATA12 OE lines could be grown at high densities, we did not observe significant differences in leaf or tiller angle affected by OsGATA12, yet any interaction between OsGATA12 and hormone signals affecting such architectural traits should be matter of future investigations. Our results with OsGATA12 OE plants suggest that their lower tiller formation and delayed senescence resulted in effective assimilate buildup in reproductive organs at high planting density. Thus, OsGATA12 represents an example of a practical gene for developing rice cultivars with optimal plant architecture and yield performance for sustainable and efficient farming practices.
References
Albacete AA, Martínez-Andújar C, Pérez-Alfocea F (2014) Hormonal and metabolic regulation of source-sink relations under salinity and drought: from plant survival to crop yield stability. Biotechnol Adv 32:12–30
Arnon DI (1949) Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol 24:1–15
Behringer C, Schwechheimer C (2015) B-GATA transcription factors - insights into their structure, regulation, and role in plant development. Front Plant Sci 6:90
Behringer C, Bastakis E, Ranftl QL, Mayer KF, Schwechheimer C (2014) Functional diversification within the family of B-GATA transcription factors through the leucine-leucine-methionine domain. Plant Physiol 166:293–305
Bi YM, Zhang Y, Signorelli T, Zhao R, Zhu T, Rothstein SJ (2005) Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J 44:680–692
Binyamin L, Falah M, Portnoy V, Soudry E, Gepstein S (2001) The early light-induced protein is also produced during leaf senescence of Nicotiana tabacum. Planta 212:591–597
Chávez-Bárcenas AT, Valdez-Alarcón JJ, Martínez-Trujillo M, Chen L, Xoconostle-Cázares B, Lucas WJ, Herrera-Estrella L (2000) Tissue-specific and developmental pattern of expression of the rice sps1 gene. Plant Physiol 124:641–654
Chen S, Tao L, Zeng L, Vega-Sanchez ME, Umemura K, Wang GL (2006) A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol Plant Pathol 7:417–427
Chiang YH, Zubo YO, Tapken W, Kim HJ, Lavanway AM, Howard L et al (2012) Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis. Plant Physiol 160:332–348
de Lucas M, Prat S (2014) PIFs get BRright: PHYTOCHROME INTERACTING FACTORs as integrators of light and hormonal signals. New Phytol 202:1126–1141
Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38:W64–W70
Gan S, Amasino RM (1995) Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270:1986–1988
Gladun IV, Karpov EA (1993) Distribution of assimilates from the flag leaf of rice during the reproductive period of development. Russ J Plant Physiol 40:215–219
Gregersen PL, Culetic A, Boschian L, Krupinska K (2013) Plant senescence and crop productivity. Plant Mol Biol 82:603–622
Hörtensteiner S (2013) Update on the biochemistry of chlorophyll breakdown. Plant Mol Biol 82:505–517
Hudson D, Guevara D, Yaish MW, Hannam C, Long N, Clarke JD et al (2011) GNC and CGA1 modulate chlorophyll biosynthesis and glutamate synthase (GLU1/Fd-GOGAT) expression in Arabidopsis. PLoS ONE 6:e26765
Hudson D, Guevara DR, Hand AJ, Xu Z, Hao L, Chen X et al (2013) Rice cytokinin GATA transcription Factor1 regulates chloroplast development and plant architecture. Plant Physiol 162:132–144
Ikeda M, Miura K, Aya K, Kitano H, Matsuoka M (2013) Genes offering the potential for designing yield-related traits in rice. Curr Opin Plant Biol 16:213–220
Iqbal N, Nazar R, Khan MIR, Masood A, Khan NA (2011) Role of gibberellins in regulation of source–sink relations under optimal and limiting environmental conditions. Curr Sci 100:998–1007
Jefferson R, Kavanagh T, Bevan M (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G et al (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet 42:541–544
Khush GS (1999) Green revolution: preparing for the 21st century. Genome 42:646–655
Khush GS (2003) Productivity improvements in rice. Nutr Rev 61:S114–S116
Koltai H (2014) Receptors, repressors, PINs: a playground for strigolactone signaling. Trends Plant Sci 19:727–733
Lee RH, Wang CH, Huang LT, Chen SC (2001) Leaf senescence in rice plants: cloning and characterization of senescence up-regulated genes. J Exp Bot 52:1117–1121
Li Z, Peng J, Wen X, Guo H (2012) Gene network analysis and functional studies of senescence-associated genes reveal novel regulators of Arabidopsis leaf senescence. J Integr Plant Biol 54:526–539
Liang C, Wang Y, Zhu Y, Tang J, Hu B, Liu L et al (2014a) OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc Natl Acad Sci USA 111:10013–10018
Liang WH, Shang F, Lin QT, Lou C, Zhang J (2014b) Tillering and panicle branching genes in rice. Gene 537:1–5
Lim PO, Kim HJ, Nam HG (2007) Leaf senescence. Annu Rev Plant Biol 58:115–136
Liu YG, Chen Y (2007) High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 43:649–656
Liu L, Zhou Y, Zhou G, Ye R, Zhao L, Li X, Lin Y (2008) Identification of early senescence-associated genes in rice flag leaves. Plant Mol Biol 67:37–55
Lu G, Coneva V, Casaretto JA, Ying S, Mahmood K, Liu F et al (2015) OsPIN5b modulates rice (Oryza sativa) plant architecture and yield by changing auxin homeostasis, transport and distribution. Plant J 83:913–925
Luo PG, Deng KJ, Hu XY, Li LQ, Li X, Chen JB et al (2013) Chloroplast ultrastructure regeneration with protection of photosystem II is responsible for the functional ‘stay-green’ trait in wheat. Plant Cell Environ 36:683–696
Luo X, Zheng J, Huang R, Huang Y, Wang H, Jiang L, Fang X (2016) Phytohormones signaling and crosstalk regulating leaf angle in rice. Plant Cell Rep 35:2423–2433
Małgorzata G, Barbara Z (2004) Multifunctional role of plant cysteine proteinases. Acta Biochim Pol 51:609–624
Miura K, Ikeda M, Matsubara A, Song XJ, Ito M, Asano K et al (2010) OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet 42:545–549
Morinaka Y, Sakamoto T, Inukai Y, Agetsuma M, Kitano H, Ashikari M, Matsuoka M (2006) Morphological alteration caused by brassinosteroid insensitivity increases the biomass and grain production of rice. Plant Physiol 141:924–931
Morita R, Sato Y, Masuda Y, Nishimura M, Kusaba M (2009) Defect in non-yellow coloring 3, an alpha/beta hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J 59:940–952
Park S-Y, Yu J-W, Park J-S, Li J, Yoo S-C, Lee N-Y et al (2007) The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 19:1649–1664
Patient RK, McGhee JD (2002) The GATA family (vertebrates and invertebrates). Curr Opin Genet Dev 12:416–422
Paul MJ, Foyer CH (2001) Sink regulation of photosynthesis. J Exp Bot 52:1383–1400
Reyes JC, Muro-Pastor MI, Florencio FJ (2004) The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol 134:1718–1732
Richter R, Behringer C, Müller IK, Schwechheimer C (2010) The GATA type transcription factors GNC and GNL/CGA1 repress gibberellins signaling downstream from DELLA proteins and PHYTOCHROME INTERACTING FACTORS. Genes Dev 24:2093–2104
Richter R, Behringer C, Zourelidou M, Schwechheimer C (2013) Convergence of auxin and gibberellin signaling on the regulation of the GATA transcription factors GNC and GNL in Arabidopsis thaliana. Proc Natl Acad Sci USA 110:13192–13197
Roitsch T, González MC (2004) Function and regulation of plant invertases: sweet sensations. Trends Plant Sci 9:606–613
Rong H, Tang Y, Zhang H, Wu P, Chen Y, Li M et al (2013) The Stay-Green Rice like (SGRL) gene regulates chlorophyll degradation in rice. J Plant Physiol 170:1367–1373
Rosen H (1957) A modified ninhydrin colorimetric analysis for amino acids. Arch Biochem Biophys 67:10–15
Sakamoto T, Morinaka Y, Ohnishi T, Sunohara H, Fujioka S, Ueguchi-Tanaka M et al (2006) Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat Biotechnol 24:105–109
Sakuraba Y, Park SY, Paek NC (2015) The divergent roles of STAYGREEN (SGR) homologs in chlorophyll degradation. Mol Cells 38:390–395
Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D et al (2002) Green revolution: a mutant gibberellin-synthesis gene in rice. Nature 416:701–702
Scazzocchio C (2000) The fungal GATA factors. Curr Opin Microbiol 3:126–131
Tong H, Jin Y, Liu W, Li F, Fang J, Yin Y et al (2009) DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J 58:803–816
Tudzynski B (2014) Nitrogen regulation of fungal secondary metabolism in fungi. Front Microbiol 5:656
Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N et al (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455:195–200
Wang W, Hao Q, Tian F, Li Q, Wang W (2016) Cytokinin-regulated sucrose metabolism in stay-green wheat phenotype. PLoS ONE 11:e0161351
Wilkinson S, Kudoyarova GR, Veselov DS, Arkhipova TN, Davies WJ (2012) Plant hormone interactions: innovative targets for crop breeding and management. J Exp Bot 63:3499–3509
Wingler A, Purdy S, MacLean JA, Pourtau N (2006) The role of sugars in integrating environmental signals during the regulation of leaf senescence. J Exp Bot 57:391–399
Wu CY, Trieu A, Radhakrishnan P, Kwok SF, Harris S, Zhang K et al (2008) Brassinosteroids regulate grain filling in rice. Plant Cell 20:2130–2145
Xing Y, Zhang Q (2010) Genetic and molecular bases of rice yield. Annu Rev Plant Biol 61:421–442
Xu J, Zha M, Li Y, Ding Y, Chen L, Ding C, Wang S (2015) The interaction between nitrogen availability and auxin, cytokinin, and strigolactone in the control of shoot branching in rice (Oryza sativa L.). Plant Cell Rep 34:1647–1662
Yang J, Zhang J (2006) Grain filling of cereals under soil drying. New Phytol 169:223–236
Yang Y, Li R, Qi M (2000) In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J 22:543–551
Yoo SC, Cho SH, Zhang H, Paik HC, Lee CH, Li J et al (2007) Quantitative trait loci associated with functional stay-green SNU-SG1 in rice. Mol Cells 24:83–94
Yu SM, Lo SF, Ho THD (2015) Source–sink communication: regulated by hormone, nutrient, and stress cross-signaling. Trends Plant Sci 20:844–857
Yuan M, Zhao J, Huang R, Li X, Xiao J, Wang S (2014) Rice MtN3/saliva/SWEET gene family: evolution, expression profiling, and sugar transport. J Integr Plant Biol 56:559–570
Zhang SY, Li G, Fang J, Chen W, Jiang H, Zou J et al (2010) The interactions among DWARF10, auxin and cytokinin underlie lateral bud outgrowth in rice. J Integr Plant Biol 52:626–638
Zhang X, Zhou Y, Ding L, Wu Z, Liu R, Meyerowitz EM (2013) Transcription repressor HANABA TARANU controls flower development by integrating the actions of multiple hormones, floral organ specification genes, and GATA3 family genes in Arabidopsis. Plant Cell 25:83–101
Zhang C, Bai MY, Chong K (2014) Brassinosteroid-mediated regulation of agronomic traits in rice. Plant Cell Rep 33:683–696
Acknowledgements
This work was supported in part by the Natural Sciences and Engineering Research Council of Canada to SJR, and funding from Syngenta Biotechnology, Inc.
Authors contributions
GL designed and performed the cloning experiments, generation of transgenics, collected phenotypic data and tissues, worked on gene expression analysis and contributed to manuscript writing. JAC helped with density experiments, performed microarray and qRT-PCR data analyses, interpreted the data and wrote the manuscript. SY helped with the generation of transgenic lines and qRT-PCR analysis. KM performed the transcriptional activation in yeast. FL performed the promoter repression assay. YMB and SJR helped conceiving the study, participated in its coordination and contributed to manuscript editing. All authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Guangwen Lu and José A. Casaretto have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lu, G., Casaretto, J.A., Ying, S. et al. Overexpression of OsGATA12 regulates chlorophyll content, delays plant senescence and improves rice yield under high density planting. Plant Mol Biol 94, 215–227 (2017). https://doi.org/10.1007/s11103-017-0604-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-017-0604-x