Abstract
Mutations in H3.3-ATRX-DAXX chromatin remodeling pathway have been reported in pediatric GBMs. H3.3 (H3F3A) mutations may affect transcriptional regulation by altered global histone-methylation. Therefore, we analyzed yet partly understood global histone code (H3K-4/9/27/36) trimethylation pattern in H3F3A-ATRX mutants and wild-type. H3F3A, HIST1H3B, IDH1, ATRX, DAXX and Tp53 mutations were identified by sequencing/immunohistochemistry in 27 pediatric GBMs. Global histone-methylation H3K-4/9/27/36me3 and Polycomb-protein EZH2 expression were evaluated by immunohistochemistry. H3F3A-ATRX mutation was observed in 66.7 % (18/27) of pediatric GBMs. K27M and G34R-H3F3A mutations were found in 37 % (10/27) and 14.8 % (4/27) patients respectively. G34V-H3F3A, HIST1H3B and IDH1 mutations were absent. Notably, commonest global histone-methylation mark lost was H3K27me3 (17/25, 68 %) followed by H3K4me3 (45.5 %, 10/22) and H3K9me3 (18.2 %, 4/22). Global H3K36me3 showed no loss. Most significant observation was loss of one or more histone-trimethylation mark in 80 % (20/25) pediatric GBMs. Notably, simultaneous loss of H3K27me3 and H3K4me3 were present in 7/22 (31.8 %) of pediatric GBMs. Low expression of EZH2 was found in 12/24 (50 %) of cases. However no significant correlation of loss of histone-marks or EZH2 expression with H3F3A-ATRX mutants (loss of at least one histone-marks in 87.5 % (14/16) cases) versus wild-types (loss of at least one histone-marks in 75 % (6/8) cases) was seen. The present study highlights for the first time combinatorial loss of one or more histone-trimethylation marks associated with majority of pediatric GBMs and the finding suggests significant role of histone-code in the molecular biology that underlies pediatric GBMs. Hence therapies for patients with particular combinations of histone modifications present opportunity to design innovative patient-tailored treatment protocols.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pediatric glioblastomas (pGBMs) constituting approximately 20 % of all brain tumors in children, are biologically aggressive and associated with poor outcome despite aggressive therapeutic approaches [1]. Previous studies including ours indicate that the molecular pathogenesis of pGBMs is largely different from that of adults [2–6]. Recently, two recurrent mutations were identified in histone H3.3 (H3F3A gene) at two critical positions within the histone tail namely K27M and G34V/R [5]. These two mutations were found to be highly specific to pGBMs, pediatric diffuse intrinsic pontine gliomas (DIPGs) and very small proportion (3 %) of young adult GBMs [5, 7]. Additionally, in DIPGs, mutation of HIST1H3B was also described [7, 8]. Further, H3F3A (especially G34R/V) mutations were seen to be significantly associated with mutations in ATRX(α thalassemia/mental retardation syndrome X-linked) and DAXX(death-domain–associated protein), part of chromatin remodeling complex required for H3.3 incorporation at pericentric heterochromatin and telomeres [5, 9]. Differences have also been reported in the clinical features (site, age), gene expression profile, methylation profile and prognostic outcome of H3.3 mutant versus wild types as well as between K27M versus G34R/V mutant types [5, 10, 11]. Therefore, the finding of recurrent gain of function mutations for the first time in H3.3, a regulatory histone, triggered interest in understanding how this affects various histone post translational modifications (PTMs) which play a significant role in regulation of gene expression. The most widely studied histone modification is histone lysine (H3K) methylation which can be an activator or repressor of transcription depending on the particular histone residue methylated. Thus, methylation of H3K9 and H3K27 is generally associated with silencing of transcription, while methylation of H3K4 and H3K36 are associated with activation of transcription [12]. There exist also numerous methyltransferases and demethylases regulating the level of methylation of histone lysine residues [13]. One such important histone methyltransferase is EZH2 which is the catalytic subunit of the polycomb repressive complex 2(PRC2) and involved in repressing gene expression through methylation of histone H3 on lysine 27(H3K27me3) [14]. Overexpression of EZH2 has been shown in many solid tumors including GBMs [15–18]. A few recent studies reported significant decrease in global H3K27me3 expression and/or temporal change in H3K27 trimethylation mark at a set of genes in the H3F3A-K27M mutant pediatric GBM and DIPG group, without significant change in H3K4me3 and H3K36me3 [19–21].
Recent lines of evidence suggest combinatorial action of histone-methylation marks and indeed in many cancers multiple histone methylation marks have been shown to be altered without any known H3.1 or H3.3 histone mutation. These alterations have also been shown to correlate with recurrence and survival [22–30]. However, despite of known histone mutation, no such study characterizing histone code trimethylation is available in pediatric GBMs. Hence, first, we studied the H3F3A/ATRX/DAXX mutation status to genetically characterize pediatric GBMs. Further, to get a comprehensive view of histone modifications, we evaluated expression of all four major histone tri-methylation marks (H3K27me3, H3K9me3, H3K4me3, and H3K36me3) using immunohistochemistry (IHC) in pGBMs and correlated these histone-methylation expression patterns with H3F3A/ATRX/DAXX mutation status and EZH2 expression. Interestingly we observed a loss of histone-trimethylation marks, commonest being H3K27me3 and H3K4me3 in majority of pGBMs. Notably, neither of global histone-trimethylation mark or EZH2 expression was exclusively associated with H3F3A-ATRX mutants compared to H3F3A-wildtype.
Materials and methods
Patients and tissue samples
A retrospective study was conducted with the approval of institutional ethics committee for study on human subject samples. 27 pediatric (≤18 years) cases of GBMs diagnosed over a period of 7 years (2006–2012) in which fresh frozen tumor samples and paraffin blocks with adequate tumor tissue available were included. Fresh H & E-stained slides of these cases were reviewed for the reconfirmation of the diagnosis between three trained neuropathologists (CS, MCS, VS), based on the WHO classification (2007).
H3F3A (H3.3), HIST1H3B (H3.1), IDH1 and TP53 gene sequencing
Genomic DNA was extracted from freshly frozen tumor tissue using DNeasy Blood and Tissue Kit (M/s. Qiagen) according to the manufacturer’s protocol. Genotyping of all three H3F3A-(K27 and G34R/V) and one HIST1H3B-(K27) Single nucleotide variations were analyzed using single base primer extension assay (SNaPshot multiplex reaction mix) following the manufacturer’s protocol (ABI, UK) on ABI Prism 3130xl Genetic Analyzer. Primers were designed by Primer3, a web based tool (Supplementary Table 1). IDH1 in all the cases and TP53 gene sequencing (Exon 5-8) in nine cases were performed as described elsewhere [31].
Immunohistochemistry
Immunohistochemical analysis was performed as described before [3]. The details of the primary antibodies and their dilution were enlisted in Supplementary Table 2. As negative control; we used paraffin sections from the same tissues without application of the primary antibody. Positive controls included: breast carcinoma for EZH2 (normal brain: negative), normal brain and breast carcinoma for H3K4me3, H3K9me3, H3K27me3, H3K36me3, ATRX and DAXX.
Quantification of immunohistochemistry
The labeling indices for all antibodies were calculated as a percentage of the positively stained nuclei. Normal adjacent brain tissue was excluded from analysis. One thousand tumor cells were counted in the areas with highest density of positive nuclei (at least ten representative microscopic fields) under ×400 magnification. Care was taken to exclude the vascular endothelial cells and hematogenous cells in the counts. For H3K27me3, H3K4me3, H3K9me3, H3K36me3 and EZH2, tumors containing ≥10 % stained cells were considered positive [22]. For ATRX and DAXX complete negative staining pattern was considered as evidence of mutation. Nuclear accumulation of p53 protein suggestive of possible presence of TP53 mutation (positive cut off ≥10 % of positively stained nuclei) was considered as TP53 mutants.
Statistical analysis
Analysis of data was performed using SPSS 16.0 version. Associations were analyzed with two-sided Fisher exact test or Chi square. A P value of <0.05 was considered significant.
Results
H3F3A, HIST1H3B and IDH mutation analysis
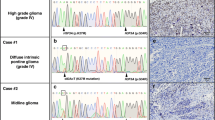
Of the 27 pGBMs, 14 (52 %) tumors showed H3F3A mutation. Most commonly H3F3A mutation was found at K27M, (10 cases; 71.4 %) while four cases harboured G34R mutation (28.5 %). Individually K27M and G34R accounted for 33 % (10/27) and 14.8 (4/27 cases) respectively. G34V mutation of H3F3A and HIST1H3B mutation were absent in our series. No case of pGBM showed IDH1 mutation (Figs. 1, 2). K27M mutation occurred in 3/10 (30 %) thalamic, 3/10 (30 %) cases in cerebral hemisphere, 2/10 (20 %) cases in posterior fossa and 2/10 (20 %) spinal region tumors. However G34R was mostly, 3/4 (75 %) confined to cerebral hemispheric tumors (Fig. 1, Supplementary Table 3).
Genotyping of all three H3F3A mutations was carried out using the SNaPshot method based on the single base extension principle. SNaPshot electropherograms showing representative H3F3A heterozygous mutations encoding a p.Lys27 Met (c83 A>T) and H3F3A heterozygous mutation (lower) encoding a p.Gly34Arg (c103 G>A) substitution. Mutations are shown in the indicated tumor compared to matched normal DNA
Analysis of ATRX, DAXX and TP53 mutation
Loss of immunopositivity for ATRX which is a surrogate marker for gene mutation was seen in 48 % (13/27) of pGBMs. Notably, 6/10 (60 %) cases with K27M mutation had associated ATRX loss while 3/4 (75 %) cases with G34R mutation had ATRX loss (P > 0.05). No case showed loss of DAXX expression. TP53 mutation was found in 61 % (14/23) of pGBMs. However the 60 % (9/15) patients harbouring either of K27M, G34R or ATRX mutation showed TP53 mutation (Figs. 1, 3).
Immunohistochemical staining in normal brain and tumor tissues of pediatric GBM. All the immunostaining were nuclear. a, d, g and j are showing H3K27me3, H3K9me3, H3K4me3 and H3K36me3 immunopositivity in normal brain. b, e and h are showing H3K27me3, H3K9me3 and H3K4me3 immunonegativity and c, f, i and k showed H3K27me3, H3K9me3, H3K4me3 and H3K36me3 immunopositivity in a representative case of pediatric GBM. l showing EZH2 low expression and m with EZH2 high expression in a representative case of pediatric GBM. n showing ATRX immunonegativity (Endothelial cells are positive (internal control)) and o with ATRX Immunopositivity in a representative case of pediatric GBM. p showing H&E of a representative case of pediatric H3F3AK27M mutant GBM with q H3K27me3 immunnopositivity shown by H3K27me3 specific abcam and r with Cell signaling antibody respectively. All images are reported at a ×200 magnification
Global analysis of histone-trimethylation expression in H3F3A mutants versus wild-type
Global H3K27me3 expression
Among pGBMs, the commonest histone mark lost was H3K27me3 in 68 % (17/25) cases and majority of H3K27me3 immunonegative cases were associated with mutation. Thus, of the 17 H3K27me3 immunonegative pediatric GBMs, interestingly significant difference was found as 11 (65 %) had H3F3A-ATRX mutations while six (33.3 %) had no mutation (P = 0.03). However, no significant difference was noted in the loss of H3K27me3 expression between the K27M versus G34R mutant and K27M versus wild type (P > 0.05) (Figs. 1, 3, 4).
H3K4me3 expression
Loss of H3K4me3 was noted in 45.5 % (10/22) of pGBMs. Similar to H3K27me3 and its association with mutation in chromatin remodelling genes, most of the H3K4me3 immononegative cases were significantly associated with H3F3A–ATRX mutation (70 % in mutants vs. 30 % wild-type, P = 0.03). Akin to H3K27me3, no significant correlation was noted within different types of mutation with loss of H3K4me3 expression (P > 0.05) (Figs. 1, 3, 4).
H3K9me3 and H3K36me3 expression
Loss of H3K9me3 expression was less frequent and seen in 4/22 (18.2 %) cases. It was noted that, unlike H3K27me3 and H3K4me3, H3K9me3 immunonegativity was more closely associated with H3F3-K27M mutation. Thus, three among four H3K9me3 histone mark negative cases had K27M mutations and remaining one was wild-type. Moreover all H3K9me3 negative cases essentially had H3K27me3 mark loss. In contrary to other histone-trimethylation marks, immunohistochemical analysis of H3K36me3 showed no loss in any of the pGBM cases. In fact all the cases showed diffusely nuclear positivity of H3K36me3 (Figs. 1, 3, 4).
Combinatorial histone-methylation expression analysis in pediatric GBMs
Since H3K36me3 was positive in all cases, we noted combinatorial losses in H3-K27, −K4 and −K9me3. The most striking and remarkable observation was that 80 % (20/25) pGBMs had loss of one or more histone-trimethylation marks. Of the ten cases with only one histone-trimethylation marks loss, commonest was H3K27me3 loss (7 of 10 pGBMs). Another significantly notable feature was loss of at least two histone-trimethylation marks in 8/22 (36.4 %) pGBMs (Fig. 1).
Among the combinatiorial histone mark losses, most marked was simultaneous loss of H3K27me3 and H3K4me3 in 7/22 (31.8 %) pediatric cases. Further, 3/22 (13.6 %) pGBMs showed loss of all three histone tri-methylation marks. Thus, it was found that 7/8 (87.5 %) pGBM with two histone mark loss were all important H3K27me3 and H3K4me3. Again it was seen that loss of 1, 2 or 3 histone-trimethylation marks was frequently associated with H3F3-ATRX (14/20, 70 %) mutation than without mutation (6/20, 30 %) (P = 0.005). However, there was no correlation of combinatorial histone-trimethylation mark loss with type of mutation (K27M vs. G34R vs. WT or ATRX only) (P > 0.05) (Figs. 1, 5).
Correlation of H3F3A-ATRX mutation and histone-trimethylation with EZH2
Low expression of EZH2 was found in 12/24 (50 %) of pGBMs. Notably, 6/9 (66.6 %) H3F3A−K27M mutants and 4/5 (80 %) of K27M+ATRX showed low expression of EZH2. Conversely, 1/4 (25 %) H3F3A−G34R mutants and 3/3 (100 %) of G34R+ATRX showed high expression of EZH2. Further, there was no significant correlation between H3K27me3 and EZH2 expression. Among 17 of H3K27me3 immunonegative cases, 9/17 (53 %) showed low expression of EZH2 and remaining were high (P < 0.05). Similar to the correlation of EZH2 and H3K27me3, low expression of EZH2 was observed in about half of H3K4me3 and H3K9me3 immunonegative cases. However most of the EZH2 immunonegative cases like H3K27me3 were significantly associated with H3F3A–ATRX mutation (P = 0.007). (Figs. 1, 4).
Discussion
The present study revealed H3F3A mutation in 52 % of pGBMs, which was higher as compared to the findings of Schwartzentruber et al. (32 %). K27M mutation was commonest representing 37 %, G34R in 14.8 % cases compared to 18.5 % of K27M and 10.4 % of G34R reported earlier [5]. A sample bias in our study could be the reason of higher frequency of histone mutation. No case with H3F3A-G34V or HIST1H3B mutation was noted as G34V is rare and HIST1H3B mutation is mostly found in DIPGs. Unlike findings of Schwartzentruber et al. there was no significant difference between the age groups of patient with G34R (mean age = 13 year) and K27M (mean age = 12.6) mutation. Sturm et al. reported that K27M mutation exclusively occurs in midline and rarely in spinal cord. However apart from midline, we also found K27M mutation in 30 % of cerebral hemisphere and 20 % posterior fossa. G34R mutation was located in 75 % of cerebral hemispheric region as reported before in literature (Supplementary Table 3, 5). However, first time we found G34R mutation in basal ganglia in a patient. Therefore our observation further extends the knowledge of H3F3A mutation and its location in pGBMs.
Out of the 14 cases with H3F3A mutation, 35.7 % (5/14) had no ATRX mutation (4 of K27M and 1 of G34R). The remaining half of the cases was associated with ATRX loss (six of K27M and three of G34R). As a corollary, 60 % of K27M cases and 75 % of G34R mutant cases had ATRX loss. Schwartzentruber et al. noted that K27M mutants were associated with ATRX mutations in about a third of cases. However, 100 % of the G34R mutants had associated ATRX loss, slightly different to only 75 % in our series.
H3K4me3 in general is associated with active transcription and H3K27me3 with repression [32, 33]. Several lines of evidence now indicate that alterations in global levels of histone methylation have a role in initiation and progression of cancer and correlate with clinical outcome and survival (Supplementary Table 4). In several cancers, multiple methyl-marks have been shown to change in concert, highlighting the importance of combinatorial modification effect on biological function [22–30]. Notably, it has been shown that combined histone methylation marks can have different roles to the same methylation marks appearing in isolation [24].
Very little information is available regarding histone code in gliomas. A recent study by Venetti et al. noted that H3K9-trimethylation was significantly associated with IDH1 mutation in oligodendroglioma and grade II astrocytomas but not in grade-III and IV astrocytomas [34]. Lewis et al. reported that DIPGs containing H3K27M mutation exhibited significantly lower overall H3K27me3 levels. They also found that H3K27M inhibits the enzymatic activity of PRC2 through interaction with the EZH2 subunit. They noted that H3K4me3 and H3K36me3 were similar in K27M genotype arguing that global changes in PTMs were specific to H3K27 [20]. Venneti et al. reported absent/lowered expression of H3K27me3 in six K27M mutants [19].
Our study is the first to look at four histone marks by immunohistochemistry in pGBMs, with the mutation of H3F3A-ATRX. We found that the histone code of pGBMs is distinct and characterized by loss of one or more histone marks. To our surprise, H3K27me3 loss in non H3F3A-K27M was also observed in present study. For H3K27me3, we used two different antibodies to further validate our result in representative cases with and without K27M mutation and found similar loss of H3K27me3 in some of H3F3A wild-type and normal expression in some of K27M mutants. There are other studies evaluating histone-trimethylation level that mechanistically support our findings viz, H3K27me3 loss in several other cancers like breast, pancreas, ovary etc. with no reported histone H3 mutation. Likewise loss of H3K4me3 and H3K9me3 were also reported not only in glioma but also in other tumors (Supplementary Table 4).
Loss of trimethylation marks of histones H3K4, H3K9 and H3K27 (alone or in combination) were more frequently associated with mutation (H3F3A alone and H3F3A+ATRX, and/or ATRX alone) than without mutations. However, no marked difference was noted in histone-trimethylation loss of either type between K27M and G34R variants or with wild type. No correlation was noted of loss of histone-trimethylation with EZH2 status which is similar to the observation of Venetti et al. [19]. Although, the underlying pathology that lead to alteration in global H3K4, H3K27 (in wild types) and H3K9 trimethylation levels remained unclear. Several histone lysine methylases and demethylases are described whose expression in brain tumors remains largely unknown. However, recently, a mutation in SETD2, a H3K36 trimethyltransferase was also identified in 15 % of pediatric high grade gliomas with substantial decrease in H3K36me3 [35]. Hence, it is possible that many other histone methylases and demethylases are deregulated in GBMs which results in the aberrant histone code in pGBMs.
One drawback of our study is that we have not been able to directly establish the prognostic significance of the loss of histone-methylation marks because of lack of follow up. However, now it is well established from several previous studies that G34R has greater overall survival (OS) versus wild-types and K27M. [10, 36, 37–39]. Further, combinatorial loss of H3K4me3 and H3K27me3 may have a prognostic significance in pGBMs, since similar observations have been reported in medulloblastoma wherein histone, H3-(K4−/K27−) marks had been shown to be associated with poor and H3-(K4+/K27−) mark with better survival outcome respectively [25]. Overlaying the known survival outcome of G34R, K27M and Wild-types in pGBMs, interestingly we found comparable downstream events (pattern of histone code trimethylation and polycomb EZH2) in these three subgroups (Supplementary Table 6). K27M which shows poor OS than G34R patients, also harbored more frequent loss of isolated H3-K27/4/9 trimethylation, combined loss of H3-(K4 and K27)me3, combined loss of H3-(K4/K27/K9)me3 and EZH2 than G34R in our study. Further, H3-(K4+/K27−)me3, associated with better OS was also found more in G34R than K27M. Notably, frequency of alteration in the isolated or combined histone trimethylation and polycomb EZH2 expression in our study was positioned in between G34R and K27M (except H3K27me3 which was almost similar to K27M and H3K4me3 which was less than G34R). Therefore, in the light of known report that wild-types and K27M show a marginal difference in OS (Strum D et al.), our findings fits well into the ambit of prognostication [10]. Hence our results mechanistically revealed the underlying biology of variable survival outcome of different subgroup of pediatric GBMs. Most significantly about one third of pGBMs showed H3-(K4−/K27−) mark loss. Therefore it is important to study the prognostic significance of H3-(K4−/K27−) in larger series of pGBMs to further illustrate clinical significance of these observations.
In conclusion, our study for the first time highlights that the combinatorial loss of histone marks are enriched with H3F3A-ATRX mutants in pGBMs. Therefore, we further advanced the knowledge of chromatin machinery related genetic alteration while taking into account of both H3F3A and ATRX mutation together and its correlation with loss of histone-trimethylation marks so that the entire chromatin machinery is examined. The major outcome of this study was that (1) loss of global H3K27me3 was present in H3F3A-ATRX mutants but also not restricted to H3F3A-K27M mutants only, (2) global H3K4/K9me3 loss represented more than half of pediatric GBMs and (3) Combined loss of H3K27me3-H3K4me3 and H3K4me3 positive but H3K27me3 negative patients, probably with poor and better survival outcome respectively offers a hope to clinicians to prognosticate. Importantly histone mutation was reported in one third of pediatric GBMs and about two third of DIPGs. Therefore not only the pathogenesis of pediatric GBMs lies under the chromatin remodeling pathway but also the great promise of therapeutic option. Thus our findings demonstrating global loss of one or more histone trimethylation in about 80 % of pGBMs, and combinatorial histone code loss mechanistically linked to overall survival and specific to particular mutant subgroup augurs a phenomenon not only restricted to H3F3A-ATRX mutants but also beyond it. However, overall these alterations suggest development of therapies for the reversal of epigenetic changes in a customized manner for patients with particular combinations of histone modifications.
References
Broniscer A, Gajjar A (2004) Supratentorial high-grade astrocytoma and diffuse brainstem glioma: two challenges for the pediatric oncologist. Oncologist 9:197–206
Tamber MS, Rutka JT (2003) Pediatric supratentorial high-grade gliomas. Neurosurg Focus 14:e1 Review
Suri V, Das P, Pathak P, Jain A, Sharma MC et al (2009) Pediatric glioblastomas: a histopathological and molecular genetic study. Neuro Oncol 11:274–280
Gilheeney SW, Kieran MW (2012) Differences in molecular genetics between pediatric and adult malignant astrocytomas: age matters. Future Oncol 8:549–558
Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E et al (2012) Driver mutations in histone H3.3 and chromatin remodeling genes in pediatric glioblastoma. Nature 482:226–231
Appin CL, Brat DJ (2014) Molecular genetics of gliomas. Cancer J 20(1):66–72
Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS et al (2012) Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and nonbrainstem glioblastomas. Nat Genet 44:251–253
Saratsis AM, Kambhampati M, Snyder K, Yadavilli S, Devaney JM et al (2014) Comparative multidimensional molecular analyses of pediatric diffuse intrinsic pontine glioma reveals distinct molecular subtypes. Acta Neuropathol 127:881–895
Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD (2010) Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci USA 107:14075–14080
Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT et al (2012) Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22:425–437
Bender S, Tang Y, Lindroth AM, Hovestadt V, Jones DT et al (2013) Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell 24:660–672
Martin C, Zhang Y (2005) The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 6:838–849
klose RJ, Zhang Y (2007) Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol 8:307–318
Margueron R, Reinberg D (2011) The Polycomb complex PRC2 and its mark in life. Nature 469:343–349
Liu L, Xu Z, Zhong L, Wang H, Jiang S et al (2013) Prognostic value of EZH2 expression and activity in renal cell carcinoma: a prospective study. PLoS ONE 8:e81484
Holm K, Grabau D, Lövgren K, Aradottir S, Gruvberger-Saal S et al (2012) Global H3K27 trimethylation and EZH2 abundance in breast tumor subtypes. Mol Oncol 6:494–506
Shen L, Cui J, Liang S, Pang Y, Liu P (2013) Update of research on the role of EZH2 in cancer progression. Onco Targets Ther 6:321–324
Orzan F, Pellegatta S, Poliani PL, Pisati F, Caldera V et al (2011) Enhancer of Zeste 2 (EZH2) is up-regulated in malignant gliomas and in glioma stem-like cells. Neuropathol Appl Neurobiol 37:381–394
Venneti S, Garimella MT, Sullivan LM, Martinez D, Huse JT et al (2013) Evaluation of histone 3 lysine 27 trimethylation (H3K27me3) and enhancer of Zest 2 (EZH2) in pediatric glial and glioneuronal tumors shows decreased H3K27me3 in H3F3A K27M mutant glioblastomas. Brain Pathol 23:558–564
Chan KM, Fang D, Gan H, Hashizume R, Yu C et al (2013) The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev 27:985–990
Lewis PW, Müller MM, Koletsky MS, Cordero F, Lin S et al (2013) Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340:857–861
Zhang A, Xu B, Sun Y, Lu X, Gu R et al (2012) Dynamic changes of histone H3 trimethylated at positions K4 and K27 in human oocytes and preimplantation embryos. Fertil Steril 98:1009–1016
Nakazawa T, Kondo T, Ma D, Niu D, Mochizuki K et al (2012) Global histone modification of histone H3 in colorectal cancer and its precursor lesions. Hum Pathol 43:834–842
Rogenhofer S, Kahl P, Mertens C, Hauser S, Hartmann W et al (2012) Global histone H3 lysine 27 (H3K27) methylation levels and their prognostic relevance in renal cell carcinoma. BJU Int 109:459–465
Dubuc AM, Remke M, Korshunov A, Northcott PA, Zhan SH et al (2013) Aberrant patterns of H3K4 and H3K27 histone lysine methylation occur across subgroups in medulloblastoma. Acta Neuropathol 125:373–384
Watanabe T, Morinaga S, Akaike M, Numata M, Tamagawa H et al (2012) The cellular level of histone H3 lysine 4 dimethylation correlates with response to adjuvant gemcitabine in Japanese pancreatic cancer patients treated with surgery. Eur J Surg Oncol 38:1051–1057
Tamagawa H, Oshima T, Numata M, Yamamoto N, Shiozawa M et al (2013) Global histone modification of H3K27 correlates with the outcomes in patients with metachronous liver metastasis of colorectal cancer. Eur J Surg Oncol 39:655–661
Chen YW, Kao SY, Wang HJ, Yang MH (2013) Histone modification patterns correlate with patient outcome in oral squamous cell carcinoma. Cancer 119:4259–4267
Chapman-Rothe N, Curry E, Zeller C, Liber D, Stronach E et al (2013) Chromatin H3K27me3/H3K4me3 histone marks define gene sets in high-grade serous ovarian cancer that distinguish malignant, tumour-sustaining and chemo-resistant ovarian tumour cells. Oncogene 32:4586–4592
Rogenhofer S, Miersch H, Göke F, Kahl P, Wieland WF et al (2013) Histone methylation defines an epigenetic entity in penile squamous cell carcinoma. J Urol 189:1117–1122
Jha P, Suri V, Sharma V, Singh G, Sharma MC et al (2011) IDH1 mutations in gliomas: first series from a tertiary care centre in India with comprehensive review of literature. Exp Mol Pathol 91:385–393
Zhang Y, Reinberg D (2001) Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev 15:2343–2360
Greer EL, Shi Y (2012) Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet 13:343–357
Venneti S, Felicella MM, Coyne T, Phillips JJ, Gorovets D et al (2013) Histone 3 lysine 9 trimethylation is differentially associated with isocitrate dehydrogenase mutations in oligodendrogliomas and high-grade astrocytomas. J Neuropathol Exp Neurol 72:298–306
Fontebasso AM, Schwartzentruberentruber J, Khuong-Quang DA, Liu XY, Sturm D et al (2013) Mutations in SETD2 and genes affecting histone H3K36 methylation target hemispheric high-grade gliomas. Acta Neuropathol 125:659–669
Khuong-Quang DA, Buczkowicz P, Rakopoulos P, Liu XY, Fontebasso AM et al (2012) K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol 124:439–447
Fontebasso AM, Liu XY, Sturm D, Jabado N (2013) Chromatin remodeling defects in pediatric and young adult glioblastoma: a tale of a variant histone 3 tail. Brain Pathol 23:210–216
Venneti S, Santi M, Felicella MM, Yarilin D, Phillips JJ et al (2014) A sensitive and specific histopathologic prognostic marker for H3F3A K27M mutant pediatric glioblastomas. Acta Neuropathol 128(5):743–753
Buczkowicz P, Bartels U, Bouffet E, Becher O, Hawkins C (2014) Histopathological spectrum of paediatric diffuse intrinsic pontine glioma: diagnostic and therapeutic implications. Acta Neuropathol. doi:10.1007/s00401-014-1319-6
Acknowledgements
The authors are thankful to Indian Council of Medical Research (ICMR), Neuro Sciences Centre and Department of Pathology, All India Institute of Medical Sciences, New Delhi, India for funding. The authors are also thankful to Dr. Supriya mallick, Department of Radiation Oncology, All India Institute of Medical Sciences, New Delhi, for assistance in patient follow up and Dr. Mitali Mukerji, Genomics and Molecular Medicine, (CSIR-IGIB), New Delhi for providing genomics facility for this work.
Conflict of interest
The authors declare that there is no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Pankaj Pathak and Prerana Jha have equally contributed to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pathak, P., Jha, P., Purkait, S. et al. Altered global histone-trimethylation code and H3F3A-ATRX mutation in pediatric GBM. J Neurooncol 121, 489–497 (2015). https://doi.org/10.1007/s11060-014-1675-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-014-1675-z