Abstract
Histones form chromatin and play a key role in the regulation of gene expression. As an epigenetic information form, histone modifications such as methylation, phosphorylation, acetylation, and ubiquitination are closely related to the regulation of genes. In the last two decades, cancer scientists discovered that some histone modifications, including acetylation and methylation, are perturbed in cancer diseases. Recurrent histone mutations, which hinder histone methylation and are implicated in oncogenesis, are recently identified in several cancer disease and called oncohistones. Well-known oncohistones, with mutations on both H3.1 and H3.3, include H3K36M in chondroblastoma, H3K27M in glioma, and H3G34 mutations that exist in bone cancers and gliomas. Oncohistone expression can lead to epigenome/transcriptome reprogramming and eventually to oncogenesis. The H3K27M, H3G34V/R, and H3K36M histone mutations can lead to the substitution of amino acid(s) at or near a lysine residue, which is a methylation target. H3K27M characteristically exists in diffuse intrinsic pontine glioma (pediatric DIPG), and its expression can cause a global decrease of the methylation of histone at the lysine residue. Uncovering the molecular mechanisms of H3K27M-driven tumorigenesis has recently led to the identification of some potential therapeutic targets in diffuse intrinsic pontine glioma. In this chapter, we will review and summarize recent studies on the H3K27M-driven tumorigenic mechanisms and properties and the role of H3.1K27M and H3.3K27M oncohistones in brain tumors.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
3.1 Introduction
As alkaline proteins, histones exist in the nuclei of eukaryotic cells. They mainly function to control the appropriate DNA package/order the DNA into the nucleosomes units. In addition, histones form chromatin and play a key role in the regulation of gene expression. In the nucleosomes, histone proteins can be modified dynamically by an epigenetic writer(s). Methylation, phosphorylation, and acetylation as well as ubiquitination are key modification processes of histone proteins. Indeed, the posttranslational modification processes of histone proteins are one form of epigenetic information and play significant roles in regulating the gene expression [1,2,–3].
The formation of cancer diseases is widely known to be related to the misregulation or mutation of specific gene expression in cancer cells. However, in the last two decades, cancer scientists have discovered that modification processes of histone proteins, including acetylation and methylation, are perturbed in cancer diseases. Chromatin-remodeling protein mutations often lead to these perturbations in cancer cells. Therefore, much attention has been recently paid for investigating the role of aberrant modifications of histone proteins in cancer development and pathology.
Recently, recurrent histone mutations, which are also implicated in oncogenesis, are identified in several cancer disease and called oncohistones. Well-known oncohistones, with mutations on both H3.1 and H3.3, include H3K36M in chondroblastoma and H3K27M in glioma, as well as H3G34 mutations that exist in bone cancers and gliomas. Both H3.1 and H3.3 have conserved major lysine residues, including K9, K4, K36, and K27, despite their slight differences in the amino acid sequences. Notably, while H3.3 is a variant histone existing in both the gene bodies and promoters of genes that are actively transcribed, H3.1 can be found throughout the genome [4]. In addition, certain histone chaperone complexes, including DAXX–ATRX and HIRA, mediate the incorporation of H3.3 histone onto the chromatin that is cell cycle independent [4, 5]. Notably, all well-documented oncohistones have mutation on or near these residues that are conserved and act as sites for the modifications of histone. Oncohistone expression can lead to transcriptome reprogramming and eventually to oncogenesis [3]. Other types of mutations, such as those on glycine 34 of histone H3, are also found in brain tumors, like H3K27M [6]. These mutations are characterized with the substitution of glycine with valine/arginine (H3G34V/R) and, together with H3K27M, exist in different brain regions [6].
3.2 Role of Oncohistones H3.1K27M and H3.3K27M in Brain Tumors
The oncohistones H3.1K27M and H3.3K27M play important role in brain tumors. One of the most studied brain tumors is the diffuse intrinsic pontine glioma (DIPG). DIPG is an aggressive, glial cell-derived brain tumor that is common in children [7]. DIPG children have a limited median rate of survival (1 year only). Histone H3 somatic mutations, which result in a substitution of lysine with methionine specifically on position 27 (H3K27M), have been identified in 60% of DIPG patients and up to 30% of patients with glioblastoma [6, 8, 9]. Several studies have showed that these somatic mutations occur mostly on the genes that encode histones H3.1 and H3.3, which are HIST1H3B and H3F3A genes, respectively [10, 11].
In DIPG, H3.1K27M and H3.3K27M tumors represent two different subgroups with a characteristic clinical manifestation(s). For example, while H3.3K27M tumors exist along the brain midline, tumors with H3.1K27M mutations are only restricted to the pons [10,11,12]. In addition, H3.1K27M and H3.3K27M tumors have characteristic associating mutations of DNA such as H3.1K27M-associated ACVR1 mutations [13], and H3.3K27M-associated amplified mutations in CCND2, PDGFRA, TP53, and MYC [12, 14]. Moreover, H3.1K27M/ H3.3K27M subgroups have characteristic expression profiles. Thus, H3.3K27M tumors have a characteristic oligodendroglial phenotype, while H3.1K27M tumors have a remarkable mesenchymal phenotype. The clinical outcomes also vary between H3.1K27M and H3.3K27M tumors. Thus, patients with H3.1K27M have a longer median survival of 15 months, while patients with H3.3K27M have 11 months’ median survival only [10]. In addition, H3.3K27M tumor types display more metastatic relapses and show more radiotherapy resistance [10]. In contrast, expressing both H3.1K27M and H3.3K27M can result in similar epigenomic changes. Notably, a recent immunohistochemical study has detected histone H3 K27M mutation, H3K27me3 (but not H3K27Ac), and posttranslational modifications in pediatric DIPG and suggested H3K27M and H3K27me3 staining for pediatric DIPG diagnosis [15].
3.3 Role of H3K27me3 Landscape Changes in Reprogramed Epigenome and Gene Expression in Brain Tumors
Both trimethylation at histone H3 lysine 27 (H3K27me3) and H3K27ac are mutually exclusive and important in gene regulation. For instance, as a repressive histone mark, H3K27me3 is important for several processes such as the inactivation of X-chromosome, silencing of HOX genes, and genome imprinting [16]. The deposition of H3K27me3 relies on the histone methyl-transferase (HMT), named as EZH2, which represents a subunit of the polycomb-repressive complex 2 (PRC2). H3K27 acetylation (H3K27ac) antagonizes the activity of PRC2 and plays an important role in gene regulation due to its association with both enhancer regions and active gene transcription [17]. Notably, H3K27M expression can influence both the acetylation and methylation of lysine 27 (of Histone H3 protein; [3]).
Despite the general low incidence of H3K27M mutation, which takes place on 1 out of 16 genes that encode histone H3, it can clearly change the reported epigenome(s) of H3K27M-DIPG cells.
In addition, in diffuse intrinsic pontine glioma (DIPG) patient cells, recent data have shown that H3K27M represents 3–17% only of total H3 [18]. However, H3K27M expression in these cells is enough to reprogram the landscape of H3K27me3 [18]. Notably, a remarkable decrease of trimethylated H3K27 has been reported in different types of H3.3K27M brain tumors, including glioblastomas and gliomas [18,19,20,21], with an associated gain of H3K27ac in some reports [18, 22, 23]. However, other studies have reported an enhancement of H3K27me3 in a large number of gene loci [20, 24]. Interestingly, the promoter-associated regions loss H3K27me3, while the intergenic regions are associated with gaining the mark [19].
Several studies have demonstrated that H3K27M expression can lead to H3K27me3 loss, which is cell-type independent. In addition, the decrease of H3K27me3 that is observed in DIPG cells can be recapitulated by expressing H3.1K27M/H3.3K27M in 293T, human astrocyte, and murine embryonic fibroblast cells [20, 21]. Remarkably, incubating H3-WT nucleosome with H3K27M peptide can significantly decrease methylation that is mediated by PRC2 (by almost sixfold), suggesting that H3K27M can clearly impede the activity of PRC2 in trans [18]. To exert these activities, H3K27M interacts with the SET-domain (of EZH2), which has a characteristic active site lined with highly conserved residues of tyrosine and plays a key role in EZH2 methyl-transferase activity. Indeed, a substitution of one of these tyrosine residues (Y641N) that exist in the SET-domain can lead to sensitivity loss to the inhibition of H3K27M. This suggests a hypothesis that the aromatic tyrosine-hydrophobic methionine interactions are important for H3K27M inhibitory effect [3]. A recent structural study by Justin and colleagues that focuses mainly on the crystal structure of K27M peptide-bound EZH2 has confirmed this important hypothesis [25]. Indeed, there are many configuration and charge property similarities between both SET-bound lysine and methionine side chain, and these similarities enable methionine to occupy the “lysine channel” that is lined with the aromatic residues [25]. Interestingly, a 16-fold tighter binding was observed between PRC2 and H3K27M peptide(s) when compared to the wild-type H3 peptide(s) [25]. Moreover, a more recent study has demonstrated that both DIPGs and posterior fossa type A (PFA) ependymomas are partially driven by the activities of peptidyl PRC2 inhibitors (the EZHIP and K27M oncohistone), which lead to promoting tumorigenesis by dysregulating gene silencing [26].
3.4 Involvement of H3K27ac Gain in the Epigenetic Landscape Changes in Brain Tumors
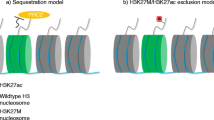
Currently, there are two suggested models that explain H3K27me3 loss in H3K27M tumors/expressing cells: the sequestration model and the H3K27M/H3K27ac exclusion model. Figure 3.1 shows a sequestration model that is supported by several studies and describes both the recruitments and local retaining of EZH2 and the stop of PRC2 complex spreading by H3K27M, and these activities can result in H3K27me3 loss globally [3, 18, 25]. This sequestration model has been opposed recently by a study on H3K27M genome-wide distributions in diffuse intrinsic pontine gliomas [23]. This study has shown that the two subunits of the PRC2 complex, EZH2 and SUZ12, are clearly eliminated from the H3K27M-containing nucleosomes [23]. Notably, H3K27M can be detected at the actively transcribed region(s) and coincides with both RNA polymerase II and H3K27ac in diffuse intrinsic pontine gliomas [23]. This H3K27M- H3K27ac colocalization agrees with previous studies by Lewis and co-workers in pediatric glioblastoma, in which an increased H3K27ac level was detected in the H3K27M-containing oligo-nucleosome array [18]. Interestingly, the study by Lewis and co-workers has suggested another model, H3K27M- H3K27ac exclusion model (Fig. 3.2), in which the H3K27me3 loss is not due to the recruitments of PRC2 by H3K27M but because of the H3K27M/H3K27ac nucleosome-dependent exclusion of PRC2 from the chromatin [18].
The H3K27M/H3K27ac exclusion model explaining H3K27me3 loss in H3K27M tumors/expressing cells. The H3K27M/H3K27ac exclusion model is based on Piunti et al. [23] studies that demonstrated that H3K27M rather than PRC2 subunits can colocalize with H3K27ac. PRC2 is, therefore, excluded in this model from H3K27M/H3K27ac heterotypic nucleosomes
3.5 Influence of H3K27M and H3K27me3 Alterations on the Gene Expression in Brain Tumors
Several studies have demonstrated the importance of changes in H3K27me3 occupancy in long-term gene repression in different biological processes. Thus, the expression of H3K27M in cells can lead to a concomitant change in the expression of several differentially expressed genes that are key players of the embryonic morphogenesis, activity of transcription factors, cancer pathways, and differentiation on neurons [19, 20]. These differentially expressed genes include MHC class I polypeptide-related sequence A (MICA) and platelet-derived growth factor receptor-α (PDGFRA) as well as cyclin-dependent kinase inhibitor 2A (CDKN2A) [24, 27, 28]. While MICA downregulation has been suggested in glioma as an important mechanism for the immune evasion, the upregulation of PDGFRA is involved in gliomagenesis [27, 28]. Moreover, the repression of CDKN2A has recently been reported to be involved in H3K27M-driven tumorigenesis [24]. Interestingly, a new model was suggested by which H3K27me2/3 can facilitate both the genome stability maintenance and the nonhomologous end joining (NHEJ) capability in more recent researches [29].
3.6 Different H3K27M-Driven Tumorigenic Mechanisms and Their Impacts on the Epigenome
The repressive histone mark, H3K27me3, is globally lost in cells expressing H3K27M. However, some H3K27me3 still existed and/or enriched at some loci in the cells expressing H3K27M [20, 24]. This was supported by a recent study on pediatric gliomas that used Pdgfβ overexpressing murine model and found in wild-type H3 mouse that H3K27me3-enriched CpG islands (CGIs) can gain and retain H3K27me3 in the H3K27M counterparts [24]. These data have led to the suggestion that the H3K27me3 histone mark-retaining loci are strong and important targets for polycomb in gliomas [24], since it is well-known that polycomb complexes target CGIs [30]. Consequently, these data have led to a conclusion that H3K27M incorporations at its strong and important polycomb targets are not apparently sufficient for inhibiting the overall activity of PRC2, and, therefore, the levels of H3K27me3 remain unchanged in this study on pediatric gliomas [24] (Fig. 3.3a, b). Importantly, in H3K27M-DIPG, H3K27me3-gaining genes play important roles in the maintenance of the DIPG cell identity. Yet, the exact mechanisms by which some loci can gain H3K27me3 are still unclear (Fig. 3.3c).
Three models of the landscape of H3K27me3 at different loci in H3K27M-expressing cells. (a) At the H3K27me3-enriched strong polycomb target(s), the trimethylation mark persists even when H3K27M nucleosome is present. (b) At weak polycomb targets with a scarce H3K27me3, the loss of H3K27me3 is due to H3K27M incorporations. (c) The expression of H3K27M can lead to H3K27me3 gain at some loci, in which PRC2 complex is recruited by still- unidentified repressor, as suggested by Mohammad et al. [24]. Adapted from Wan et al. [3]
The remarkable gene ontology analyses of H3K27me3-enriched loci in the recent study by Mohammad and colleagues have demonstrated a clear and remarkable enrichment of polycomb targets that play important regulatory roles of key biological/developmental processes, including pattern specification, and transcription regulation as well as embryonic development [24]. Notably, wild-type H3 mice, genes that are associated with H3K27me3 in neural stem cells, can completely overlap with that of H3K27M neural stem cells [24]. This has led to a conclusion that some de novo polycomb targets exist in the H3K27M/ PDGFβ model, suggesting that H3K27M-derived DIPG oncogenesis is due to H3K27M supports of a specific transcriptional pattern that is characteristic for the cell-of-origin of DIPG [24]. Nevertheless, it was reported in H3K27M mice that 20 genes that gain H3K27me3, including cyclin-dependent kinase inhibitor 2A (CDKN2A) that is importantly involved in the development of H3K27M-driven DIPG, did not apparently show an enrichment of H3K27me3 in gliomas [24]. This suggests that the oncogenic transformation process involves the selected silencing of these 20 genes [24].
The cyclin-dependent kinase inhibitor 2A (CDKN2A) plays important roles in both cell stress and oncogenic activation by encoding p16 tumor suppressor protein, which can terminate the cell cycle during both the cell stress and oncogenic activation [31]. Notably, the homozygous CDKN2A deletion is rare in pediatric high-grade glioma, despite the findings by Brennan and co-workers that it can be detected in >55% of adult high-grade glioma [32]. These findings have led to the conclusion that the gene repression mediated by PRC2 could be considered as an alternative pathway for silencing the expression of CDKN2A [33]. Indeed, a recent study using H3.3K27M murine model by Cordero et al. [34] has shown that CDKN2A repression could lead to the acceleration of gliomagenesis [34]. In addition, the DIPG growth relies on CDKN2A repression since inducing p16 expression in DIPG cell lines can lead to the growth arrest of these cells [24].
3.7 Other H3K27M-Driven Tumorigenic Mechanisms in the Brain
There are other H3K27M-driven tumorigenic mechanisms, in addition to the silencing of CDKN2A. It is well reported that H3K27M is localized to the transcriptionally active regions, together with RNA polymerase II and H3K27ac. It also coincides with that of the members of the family of bromodomain and extra-terminal domain (BET)-containing proteins such as BRD4 and BRD2.
Both the BET domain-containing proteins, BRD2 and BRD4, are also important H3K27M-driven oncogenic mechanisms, and this was confirmed by inhibiting both BRD2 and BRD4 using JQ1 in H3K27M-DIPG cells. This approach helps with dissecting the functional roles of these BET domain-containing proteins, which also play important roles in the transcription elongation, in H3K27M-DIPG cells. Because of their importance in the transcription elongation, Piunti and co-workers found that inhibiting both BRD2 and BRD4 using JQ1 leads to antitumor effects without CDKN2A derepression and can effectively suppress the activity of well-known transcribed genes that are also direct targets since they are occupied by H3K27M and BRD2/4. These data suggest other oncogenic pathway and H3K27M-driven tumorigenic mechanism in the brain that are independent of p16 [3, 23]. In addition, the study by Piunti et al. [23] has shown evidences that H3K27M can occupy the active enhancer region, which is characteristically marked by H3K27ac/H3K4me1. This recent data suggests another oncogenic-driven mechanism, in which H3K27M can contribute to the super enhancers’ formation [23]. In addition, since almost 25% of DIPGs harbor active ACVR1 mutations, which are associated with H3.1K27M mutations, a remarkable recent study has shown that both H3.1K27M and ACVR1 R206H can promote the initiation of tumors and increase gliomagenesis, through activating the signal transducer and activator of transcription 3 (Stat3) [35].
Furthermore, other co-occurring mutations were shown recently to contribute to H3K27M-driven tumorigenesis. For example, in H3.3K27M-expressing mice used to study high-grade gliomas, ATRX knockdown can lead to the formation of focal tumor, while the overexpression of PDGFRA results in the reduction of tumor latency [36].
3.8 H3K27M-DIPG Therapeutic Approaches and Strategies
Currently, the precise and detailed H3K27M-driven tumorigenic mechanisms in the brain are still not clear. However, recent studies on H3K27M-DIPG have led to the identification of some important druggable targets, including small molecules that are effective against H3K27M-DIPG in vivo and in culture, using different approaches such as the reverse of the H3K27ac gain, the rescue of H3K27me3, and targeting the activities of residual EZH2.
3.8.1 The Reverse of H3K27ac Gain Approach
Several studies have provided evidences that the trimethylation loss at K27 can consequently lead to the acetylation that is gained at K27 also [10, 11]. This suggests a possible therapeutic strategy/approach that aims to reverse these epigenetic alternations. For instance, Grasso and colleagues have provided evidences that the HDAC inhibitor, panobinostat, is an effective therapy in H3K27M-DIPGs [37].Thus, treatment of K27M-DIPG cell lines with panobinostat has led to increased cell apoptosis, reduced cell viability, downregulated MYC target genes, and enhanced H3K27ac as well as the rescue of H3K27me3 levels [37]. This important study has also shown that panobinostat can decrease H3K27M-DIPG cell viability by acting synergistically with GSKJ4, which is an inhibitor of the demethylases of H3K27me2/3, which are responsible for catalyzing the H3K27me2/3 demethylation [37]. In addition, recent studies on the bromodomain and extra-terminal domain (BET) inhibitor, JQ1, have demonstrated its anti-tumorigenic activity in DIPG cell lines, probably by decreasing the levels of H3K27ac [23]. Other effects of JQ1 on H3K27M-DIPG cells include the promotion of both certain neuronal-like morphological change(s) and anti-proliferation activities by stimulating the differentiation markers of mature neurons, the tubulin beta 3 (TUBB3) and microtubule-associated protein 2 (MAP2), and the cell cycle arrest marker p21, respectively, in these cells [3, 23].
3.8.2 The Rescue of H3K27me3 Approach
Several recent studies show evidences that the rescue of H3K27me3 is an important approach
to develop an effective K27M-DIPGs therapy since H3K27M expression leads to decreasing H3K27me3 [3]. Studies using the demethylase inhibitor GSKJ4, which has remarkable anti-tumorigenic activities in vivo/ in culture [38], represent a good example for this rescue H3K27me3 approach. For example, the treatment of human H3K27M-DIPG cell lines with GSKJ4 increased cell apoptosis but decreased both cell viability and clonogenic activities [3, 39]. Mechanistically, the GSKJ4 anti-tumorigenic activities in H3K27M-DIPG cell lines are probably due to the inhibition of the well-known H3K27me3 demethylase, JMJD3 [38, 40]. Further analyses of the GSKJ4-treated cell transcriptome are needed to uncover GSKJ4 effects in H3K27M-DIPG cells.
3.8.3 Targeting the Activities of Residual EZH2 Approach
Since residual EZH2 activities play an important role in DIPG cell growth, both targeting and inhibiting EZH2 activities are considered as important approaches for the treatment of diffuse intrinsic pontine glioma (DIPG) [23, 24]. This was supported by a recent study showing decreased proliferation, probably by derepressing p16, and reduced colony-forming abilities of H3K27M-DIPG cell lines after treatment with the highly selective EZH2 inhibitor, EPZ6438 [24]. The EPZ6438 anti-proliferative effects on H3K27M-DIPG cell lines may be also p16-independent effects [23]. Interestingly, treatments of DIPG cells with EPZ6438 cause an increase of p16, while the treatment with JQ1 upregulates the other key regulator of the cell cycle, p21. Further in vivo and in vitro research studies are needed to explore whether treatments with EPZ6438 and JQ1 together can synergistically cause the suppression of DIPG cell growth.
3.9 Conclusion, Perspective, and Future Direction
Since 2012, several studies on the recurrent mutation of H3K27M in gliomas have highlighted the importance of histone mutation-driven tumorigenesis [6, 8, 9]. This was well exemplified in the role of H3.3K27M oncohistone in the diffuse intrinsic pontine glioma (DIPG) that is well-investigated. Indeed, better understanding of H3.3K27M-driven oncogenic mechanisms has recently helped with the identification of some potential DIPG therapeutic targets. For example, a recent study on ACVR1 and H3.1K27M mutations has identified LDN212854 compound as a potential treatment for DIPGs [35]. In addition, the identification and characterization of pediatric glioma-exclusive H3K27M have led to the conclusion that both types of gliomas (pediatric and adult) are different on the molecular levels and, therefore, require different therapeutic strategies and approaches.
Despite accumulated data on the role of oncohistones in brain tumors, more studies are still needed. For example, more research is still needed to uncover the details of the occurrence of oncohistones in specific tumor types that is currently mostly unknown. H3K27M, for example, exists on both H3.3 and H3.1. H3.1 is characteristic in its ubiquitous expression and unformal distribution in the genome. Nevertheless, current research studies have only identified H3.1K27M in the diffuse intrinsic pontine glioma (DIPG). Identification of factors that play a role in the H3.1K27M tumor-type specificity is needed to better understand the H3K27M-driven tumors in the brain.
It is well-known that cancer development is associated with perturbations of histone methylations and changes of histone methyltransferases. Morin and colleagues have, for instance, identified EZH2-inactivating mutations in two types of lymphomas (diffuse large B cell lymphoma and follicular lymphoma; [41]). In contrast, studies by Kanu and co-workers have not detected H3K36M in renal cancer cells that contain inactivating mutations of SETD2 [42], while Lehnertz et al. [43] have recently reported a low frequency (1/415) of H3K27M in follicular lymphomas. Remarkably, inactivating SETD2, together with EZH2, was found to cause methylation abnormalities at histone H3 lysine residues. More studies are, therefore, still needed to clarify and identify the reasons of infrequency of H3K36M and H3K27M in these cancer types.
Furthermore, the posttranslational modifications of histone do not only include the methylation of lysine on histone H3 but extend to include the acetylation of lysine and serine/threonine phosphorylation as well as ubiquitination. Notably studies have related cancers with many newly discovered writers and reader/eraser mutations of these histone tail modifications [44]. Importantly, further studies are needed to identify the functional roles of more oncohistones in brain tumors.
Abbreviations
- BET:
-
Bromodomain and extra-terminal domain
- CGIs:
-
CpG islands
- CDKN2A:
-
Cyclin-dependent kinase Inhibitor 2A
- DIPG:
-
Diffuse intrinsic pontine glioma
- H3K27ac:
-
Histone H3 lysine 27 acetylation
- H3K27me3:
-
Histone H3 lysine 27 trimethylation
- HMT:
-
Histone methyl-transferase
- MAP 2:
-
Microtubule-associated protein 2
- MICA:
-
MHC class I polypeptide-related sequence A
- NHEJ:
-
Nonhomologous end joining
- PDGFRA:
-
Platelet-derived growth factor receptor-α
- PRC2:
-
Polycomb-repressive complex 2
- PFA:
-
Posterior fossa type A
- Stat3:
-
Signal transducer and activator of transcription 3
- TUBB3:
-
Tubulin beta 3
References
Cox M, Nelson DR, Lehninger AL (2005) Lehninger principles of biochemistry. W.H. Freeman, San Francisco
Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W (2002) Histone H2A variants H2AX and H2AZ. Curr Opin Genet Dev 12(2):162–169
Wan YCE, Liu J, Kui Ming Chan KM (2018) Histone H3 mutations in cancer. Curr Pharmacol Rep 4:292–300
Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S et al (2010) Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140(5):678–691
Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y (2004) Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116(1):51–61
Sturm D, Witt H, Hovestadt V, Khuong-Quang D-A, Jones DTW, Konermann C et al (2012) Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22(4):425–437
Jones C, Perryman L, Hargrave D (2012) Paediatric and adult malignant glioma: close relatives or distant cousins? Nat Rev Clin Oncol Nat Publ Group 9(7):400–413
Schwartzentruber J, Korshunov A, Liu X-Y, Jones DTW, Pfaff E, Jacob K et al (2012) Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482(7384):226–231
Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J et al (2012) Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet 44(3):251–253
Castel D, Philippe C, Calmon R, Le Dret L, Truffaux N, Boddaert N et al (2015) Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropathol 130(6):815–827
Fontebasso AM, Papillon-Cavanagh S, Schwartzentruber J, Nikbakht H, Gerges N, Fiset PO et al (2014) Recurrent somatic mutations in ACVR1 in pediatric midline high-grade astrocytoma. Nat Genet Nat Publ Group 46(5):462–466
Mackay A, Burford A, Carvalho D, Izquierdo E, Fazal-Salom J, Taylor KR et al (2017) Integrated molecular meta-analysis of 1,000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell 32(4):520–537.e5
Taylor KR, Mackay A, Truffaux N, Butterfield YS, Morozova O, Philippe C et al (2014) Recurrent activatingACVR1mutations in diffuse intrinsic pontine glioma. Nat Genet Nat Publ Group 46(5):457–461
Wu G, Diaz AK, Paugh BS, Rankin SL, Ju B, Li Y et al (2014) The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet 46(5):444–450
Huang T, Garcia R, Qi J, Lulla R, Horbinski C, Behdad A, Wadhwani N, Shilatifard A, James C, Saratsis AM (2018) Detection of histone H3 K27M mutation and post-translational modifications in pediatric diffuse midline glioma via tissue immunohistochemistry informs diagnosis and clinical outcomes. Oncotarget 9(98):37112–37124
Cao R, Zhang Y (2004) The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev 14(2):155–164
Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A et al (2009) CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila polycomb silencing. Development 136(18):3131–3141
Lewis PW, Müller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA et al (2013) Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340(6134):857–861
Bender S, Tang Y, Lindroth AM, Hovestadt V, Jones DTW, Kool M et al (2013) Reduced H3K27me3 and DNA Hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell 24(5):660–672
Chan K, Fang D, Gan H, Dev G, Chan K, Fang D et al (2013) The histone H3.3K27M mutation in pediatric glioma reprogramsH3K27 methylation and gene expression. Genes Dev 27:985–990
Chan KM, Han J, Fang D, Gan H, Zhang Z (2013) A lesson learned from theH3.3K27 Mutation found in pediatric glioma. Anew approach to the study of the function of histone modifications in vivo? Cell Cycle 12(16):2546–2552
Herz HM, Morgan M, Gao X, Jackson J, Rickels R, Swanson SK et al (2014) Histone H3 lysine-to-methionine mutants as a paradigm to study chromatin signaling. Science 345(6200):1065–1070
Piunti A, Hashizume R, Morgan MA, Bartom ET, Horbinski CM, Marshall SA et al (2017) Therapeutic targeting of polycomb and BET bromodomain proteins in diffuse intrinsic pontine gliomas. Nat Med 23(4):493–500
Mohammad F, Weissmann S, Leblanc B, Pandey DP, Hojfeldt JW, Comet I et al (2017) EZH2 is a potential therapeutic target for H3K27M mutant pediatric gliomas. Nat Med 23(4):483–492
Justin N, Zhang Y, Tarricone C, Martin SR, Chen S, Underwood E et al (2016) Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat Commun 7:11316
Jain SU, Do TJ, Lund PJ et al (2019) PFA ependymoma-associated protein EZHIP inhibits PRC2 activity through a H3 K27M-like mechanism. Nat Commun 10(1):2146
Verhaak RGW, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD et al (2010) Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17(1):98–110
Zarghooni M, Bartels U, Lee E, Buczkowicz P, Morrison A, Huang A et al (2010) Whole-genome profiling of pediatric diffuse intrinsic pontine gliomas highlights platelet-derived growth factor receptor α and poly (ADP-ribose) polymerase as potential therapeutic targets. J Clin Oncol 28(8):1337–1344
Zhang Y, Chang JF, Sun J, Chen L, Yang XM, Tang HY, Jing YY, Kang X, He ZM, Wu JY, Wei HM, Wang DL, Xu RG, Zhu RB, Shen Y, Zeng SY, Wang C, Liu KN, Zhang Y, Mao ZY, Jiang CZ, Sun FL (2018) Histone H3K27 methylation modulates the dynamics of FANCD2 on chromatin to facilitate NHEJ and genome stability. J Cell Sci 131:jcs215525
Deaton A, Bird A (2011) CpG islands and the regulation of transcription. Genes Dev 25(10):1010–1022
Okamoto A, Demetrick DJ, Spillare EA, Hagiwara K, Hussain SP, Bennett WP et al (1994) Mutations and altered expression of p16INK4 in human cancer. Proc Natl Acad Sci U S A 91(23):11045
Brennan CW, Verhaak RGW, McKenna A, Campos B, Noushmehr H, Salama SR et al (2013) The somatic genomic landscape of glioblastoma. Cell 155(2):462–477
Mohammad F, Helin K (2017) Oncohistones: drivers of pediatric cancers. Genes Dev 31(23–24):2313–2324
Cordero FJ, Huang Z, Grenier C, He X, Hu G, McLendon RE et al (2017) Histone H3.3K27M represses p16 to accelerate gliomagenesis in a murine model of DIPG. Mol Cancer Res 15(9):1243–1254
Hoeman CM, Cordero FJ, Hu G et al (2019) ACVR1 R206H cooperates with H3.1K27M in promoting diffuse intrinsic pontine glioma pathogenesis. Nat Commun 10(1):1023
Pathania M, De Jay N, Maestro N, Harutyunyan AS, Nitarska J, Pahlavan P et al (2017) H3.3 K27M cooperates with Trp53 loss and PDGFRA gain in mouse embryonic neural progenitor cells to induce invasive high-grade gliomas. Cancer Cell 32(5):684–700.e9
Grasso CS, Tang Y, Truffaux N, Berlow NE, Liu L, Debily MA et al (2015) Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat Med 21(6):555–559
Hashizume R, Andor N, Ihara Y, Lerner R, Gan H, Chen X et al (2014) Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat Med 20(12):1394–1396
Heinemann B, Nielsen JM, Hudlebusch HR, Lees MJ, Larsen DV, Boesen T et al (2014) Inhibition of demethylases by GSK-J1/J4. Nature 514(7520):E1–E2
Agger K, Cloos PAC, Christensen J, Pasini D, Rose S, Rappsilber J et al (2007) UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449(7163):731–734
Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R et al (2010) Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet 42(2):181–185
Kanu N, Grönroos E, Martinez P, Burrell RA, Yi Goh X, Bartkova J et al (2015) SETD2 loss-of-function promotes renal cancer branched evolution through replication stress and impaired DNA repair. Oncogene 34(46):5699–5708
Lehnertz B, Zhang YW, Boivin I, Mayotte N, Tomellini E, Chagraoui J et al (2017) H3K27M/Imutations promote context dependent transformation in acute myeloid leukemia with RUNX1 alterations. Blood 130(20):2204–2214
Chi P, Allis CD, Wang GG (2010) Covalent histone modifications — miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer 10(7):457–469
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
El-Hashash, A.H.K. (2021). Histone H3K27M Mutation in Brain Tumors. In: Fang, D., Han, J. (eds) Histone Mutations and Cancer. Advances in Experimental Medicine and Biology, vol 1283. Springer, Singapore. https://doi.org/10.1007/978-981-15-8104-5_3
Download citation
DOI: https://doi.org/10.1007/978-981-15-8104-5_3
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-8103-8
Online ISBN: 978-981-15-8104-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)