Abstract
Seed dormancy is an adaptive trait that widely exists in angiosperms and gymnosperms. The mechanisms for the release of seed dormancy have been less well studied. Using smoke tree (Cotinus coggygria var. Cinerea Engler) seeds, the effect of cold moist stratification (5 °C, 18.5 % humidity and 0–75 days) on dormancy release, changes of respiration rate, ABA and GA3 content, and the differentially expressed proteins during dormancy release were investigated. Seed dormancy was released during cold moist stratification, seed respiration rate was increased while both ABA and GA3 concentrations were decreased. A total of 28 protein spots with significant changes in relative expression abundance were detected by two-dimensional electrophoresis. Among these protein spots, four proteins of ATPase β subunit, heat-shock cognate protein 70, aspartic proteinase 1 and actin were successfully identified by the matrix assisted laser desorption/ionization time-of-flight/time-of-flight mass spectrometry. These four proteins were all down-expressed during dormancy release, and their possible implications for dormancy release of C. coggygria seeds was discussed. A better understanding of seed dormancy release and germination has practical benefits to seedling production for forest regeneration purposes.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Seed dormancy is defined as the absence of germination of a viable seed in a specified period of time, under the presence of all favorably environmental factors (temperature, light/dark, etc.) (Baskin and Baskin 1998, 2004; Bewley and Black 1994; Hilhorst 2007). Seed dormancy is an adaptive trait that contributes to the survival, dispersion and establishment of the next generation in time or space, especially for temperate plants (Baskin and Baskin 1998, 2004; Bewley and Black 1994). Under natural conditions, dormancy release of seeds is usually triggered by after-ripening (the exposure to relative dry and hot conditions) and stratification (imbibition at low temperature) (Gubler et al. 2005).
The release of seed dormancy is controlled by interactions between external environmental factors and internal genetic characteristics that are less understood (Gubler et al. 2005). A phytohormone equilibrium theory on seed dormancy was suggested as abscisic acid (ABA) and gibberellic acid (GA) function at different times in plant life cycle (Karssen and Laçka 1986). However, ABA and GA in Sorghum bicolor and Zea mays may function at the same time in seed dormancy and germination (Steinbach et al. 1997; White and Rivin 2000; White et al. 2000). On the one hand, in general, ABA stimulates dormancy development during seed maturation while GA promotes the release of seed dormancy and germination, particularly when the ABA biosynthesis was inhibited (Page-Degivry et al. 1996). On the other hand, concentration of ABA was transiently increased after exogenous GA was applied to the dormant Arabidopsis thaliana seeds (Ali-Rachedi et al. 2004). The release of seed dormancy of A. thaliana was therefore related to the increase of both GA biosynthesis and ABA hydrolysis (Ali-Rachedi et al. 2004; Cadman et al. 2006). These results indicated that it was the ABA/GA ratio, not their absolute concentration or content, which controls seed germination.
Proteomic approaches could elucidate the involved proteins in regulating seed dormancy and germination (Kermode 2005). However, the protein expression profiles are not consistent during dormancy release of seeds from different plant species (Chibani et al. 2006; Lee et al. 2006; Pawłowski 2007, 2009). These inconsistencies indicate that the mechanisms of seed dormancy release and germination may be species dependent if different plants have distinctive seed proteins. Proteomic approaches could be important in elucidating the role of protein expression during seed dormancy release.
Smoke tree (Cotinus coggygria var. Cinerea Engler, Anacardiaceae) is a pioneer shrub growing in temperate regions and is used for afforestation in desert mountains because of its tolerance to drought, poor or saline and alkaline soils (Liu 2000). It is also a widely used landscape plant species because of its beautiful red leaves in autumn and is an economically valued tree species for its yellowish wood as well as providing yellow dye, tannin and fragrant oil extractives from leaves and woods (Wu et al. 2008). However, the typically low germination rates for this species adversely affects the economics of nursery seedling production. A better understanding of seed dormancy release and germination thus has practical benefits to seedling production. The results of previous studies suggested that the C. coggygria seed displayed a combinational dormancy (physical dormancy plus type 3 non-deep physiological dormancy), and its dormancy release was stronger under stratification (5 °C) than under cool (15 °C) germination conditions (Deng et al. 2010). Germination of C. coggygria seeds is negative photoblastic, and the optimum germination temperature is 10 °C (Deng et al. 2010). In the present study we investigated the effect of cold moist stratification on seed dormancy release, ABA and GA3 concentrations, and protein expression during dormancy release to better understand the mechanism of nondeep physiological dormancy release in C. coggygria seeds.
Materials and methods
Plant seeds
Mature C. coggygria seeds were collected between May and June 2008 in Xiangshan mountain (39°59′N, 116°13′E, 73–452 m above sea level), Beijing, China (seeds get mature in May–August of year in nature, Flora of China, 2008). The annual mean temperature is 11.8 °C (1.1 °C in winter and 24.4 °C in summer). Annual rainfall ranges from 600 to 700 mm (>70 % in June–August). The mature and filled seeds were separated from the unfilled ones using water flotation and dried for 1 week at 20 °C and 46 % relative humidity. The seeds were then subjected to water content determination, initial germination tests and cold moist stratification treatment. The initial seed water content was 6.62 ± 0.11 % (mean ± SE, n = 4).
Cold moist stratification and germination of seeds
Cold moist stratifications of seeds were performed in moist (18.5 ± 0.4 %) perlite (perlite/seeds = 3/1, v/v) in sealed polyethylene plastic bags in the dark. After stratification at 5 °C for 0 (untreated control), 15, 30, 45, 60 and 75 days, respectively, four replicates of 50 un-germinated seeds (a few seeds germinated during stratification; data not shown) from each stratification treatment were further incubated at 10 °C (the optimum germination temperature, Deng et al. 2010) under dark (negative photoblastic seeds, Deng et al. 2010) for 30 days, respectively. Radicle protrusion (2 mm) was the criterion for germination and the cumulative seed germination percentage was then determined from the daily counts. Seeds that germinated during the cold moist stratification were excluded from the percentage germination calculations.
Determination of seed respiration rate
Seed respiration rate is an indicator of energy metabolism during dormancy release. Intact seeds and bisected seeds (cut lengthways) from cold moist stratification and untreated control were used to determine the respiration rate (nmol O2/ml/min/seed) on the Oxytherm (Hansatech Instruments, Pentney, King’s Lynn, Norfolk, UK) according to the Instrumental Manual. For the determination of the respiration rate, one milliliter of reaction solution followed by four replicates of ten intact seeds or 20 bisected seed sections (derived from ten intact seeds) were added into the reactor of the Oxytherm. The reaction solution contained 0.4 M Mannitol, 10 mM 3-(N-Morpholino) propanesulfonic acid (MOPS) (pH 7.5), 10 mM KH2PO4, 10 mM KCl, 5 mM MgCl2 and 0.1 % (w/v) bovine serum albumin (BSA).
Protein extraction
Un-germinated seeds (100) that had been given 0, 30 and 60 day stratification under 5 °C were pulverized with liquid nitrogen, and soluble proteins for electrophoresis were extracted based on a modified method of Wang et al. (2007), as follows. One gram of frozen seed powder was homogenized in 2 ml ice-cold extraction buffer containing 100 mM Tris (pH 8.0), 100 mM EDTA, 50 mM borax, 50 mM ascorbic acid, 1 % (w/v) PVPP, 1 % (v/v) Triton X-100, 2 % (v/v) β-mercaptoethanol and 30 % (w/v) sucrose. The homogenate was firstly vortexed for 10 min at 4 °C, centrifuged at 16,000×g (relative centrifugal force, RCF) for 20 min at 4 °C, and the supernatant was then centrifuged at 32,000×g for 20 min at 4 °C. Two volumes of Tris-saturated phenol (pH 8.0) were added into the supernatant and the mixture was then vortexed for 10 min at 4 °C. After centrifugation at 16,000×g for 20 min at 4 °C, an equal volume of extraction buffer was added into the supernatant, the mixture was vortexed for 10 min at 4 °C and was again centrifuged at 16,000×g for 20 min at 4 °C. The supernatant was transferred to a new centrifuge tube, the protein was precipitated by five volumes of (NH4)2SO4 saturated-methanol and were incubated at −20 °C for >6 h. After centrifugation at 16,000×g for 20 min at 4 °C, the protein pellet was re-suspended and rinsed with ice-cold methanol once and in ice-cold acetone four times, the washed pellet was air dried at −20 °C and then stored at −80 °C.
Two-dimensional electrophoresis (2-DE)
Three replicate samples were prepared and 2-DE were performed to ensure the reliability of the assay. The dried protein was dissolved with the lysis buffer [8 M urea, 4 % (w/v) CHAPS, 65 mM DTT, 0.2 % (w/v) pharmalyte (pH 5–8), 0.001 % (w/v) bromphenol blue]. After two centrifugation periods (20 °C, 30 min, 48,000×g) twice, the total protein in the supernatant was measured (Bradford 1976). Samples containing 800 μg total proteins were loaded onto pH 5–8 linear gradient IPG strips (17 cm) (Bio-Rad, Hercules, California, USA) and rehydrated for 12–16 h at 20 °C. The strips were then subjected to iso electric focusing (IEF) in a Protean IEF Cell (Bio-Rad, Hercules, California, USA) system according to the manufacturer’s instruction (2-DE Manual, Bio-Rad, USA). After the IEF processing, these strips were transferred to perform the SDS-PAGE. The strips were equilibrated with a 1 % (w/v) DTT, and subsequently 4 % (w/v) iodoacetamide equilibration buffer (50 mM Tris pH 8.8, 6 M urea, 30 % glycerol, 2 % SDS, 0.002 % bromophenol blue) for 14 min. The separation of proteins in the second dimension was performed with SDS polyacrylamide (13.5 % separating and 4 % stacking) gels.
Protein staining and gel analyses
The gels were stained with 0.1 % Coomassie Brilliant Blue R-250, 45 % methanol and 10 % acetic acid and then destained with 25 % methanol and 7.5 % acetic acid. The analytical gels were scanned at 300 dpi with a UMAX Power Look 2100XL scanner (Maxium Technologies, Taipei, China) and visualized with an ImageMaster 2D Platinum 5.0 (Amersham Biosciences, Little Chalfont, UK) according to the user’s manual. The molecular weight of each protein in gels was determined with standard protein markers (BioDev, Beijing, China).
Protein identification by matrix assisted laser desorption/ionization time-of-flight/time-of -flight mass spectrometry (MALDI-TOF/TOF MS)
For the MALDI-TOF/TOF MS analysis, the differentially expressed protein spots (abundance variation ≥1.5-fold) were manually excised with clean tips from the gels and digested with trypsin according to the procedures of Shevchenko et al. (1996). Proteins were identified by searching the protein NCBInr databases using the MASCOT (http://www.matrixscience.com) search engine software (Matrix Science, Boston, Massachusetts, USA). To denote a protein as an unambiguously identified one, the Mowse scoring algorithms were used (Pappin et al. 1993). Only proteins whose score exceeded the statistical significance threshold (probability, which can be ambiguous if smaller than 0.05) were considered further.
Determination of abscisic acid and gibberellic acid
After stratification at 5 °C for 0, 15, 30, 45 and 75 days, un-germinated seeds were ground into powder with liquid nitrogen and stored at −20 °C overnight to extract of GA3 and ABA. 1.5 ml 80 % methanol was added into a 2 ml of centrifugal tube containing 0.5000–1.000 g of seed powder and then vortexted using an ultrasonic cleaner (Branson, SB2200, USA). After thawing, the extract was centrifuged at 15,200 g for 10 min. The supernatant was transferred into a new centrifugal tube and the deposit was centrifuged again twice with 1.5 ml 80 % methanol in a 2 h interval period. The supernatants from these three centrifugations were combined as one sample and dried using a vacuum freeze concentrator system (Jouan, RCT-60, France).
For purification, the freeze dried extract was dissolved with 200 μl 0.1 M Na3PO4 buffer (pH 7.8), and extracted with 400 μl petroleum ether and then the water phase was applied to a C18 column (Waters, USA). C18 columns were washed with 800 μl distilled water, and phytohormones were eluted with 1.4 ml 50 % methanol and subsequently freeze dried under low pressure. The phytohormone fraction was re-dissolved with 50 % methanol to 80 μl and measured by an Acquity SQD Ultra Performance Liquid Chromatography/Mass Spectrometry (UPLC/MS, Waters, USA).
For the UPLC/MS measurement, 5 μl purified sample was injected into a Waters Acquity UPLC BEH C18 chromatographic column (100 mm × 2.1 mm, 17 μm) with isocratic elution at a 0.25 ml/min flow rate at 35 °C, using 10 % (v/v) acetonitrile as the mobile phase. The conditions of mass spectrum analysis were: determination mode, single ion recording (SIR); ionization mode, electrospray ionization (ESI) negative; Capillary voltage, 2800 V; Source Temperature, 120 °C; Desolvation Temperature, 350 °C; Cone Gas, 50 L/Hr; Desolvation Gas, 600 L/Hr; for GA3, 40 V cone hole voltage with 345.3 charge/mass ratio (m/z); for ABA, 30 V cone hole voltage with 263.2 m/z. Each sample was measured at a minimum in biological triplicate, and the concentration of GA3 and ABA were calculated on a dry weight (DW) basis (ng/g DW).
Data analysis
All data were analyzed with the SPSS system 12.0 (SPSS 2003) to test for the effects of cold moist stratification on seed germination and relative abundance of protein spots during cold moist stratification. The changes in seed respiration rate and endogenous ABA and GA3 during dormancy release were analyzed using a one-way ANOVA followed by the Student–Newman–Keuls multiple comparisons test at P = 0.05. The relationship between seed respiration rate during dormancy release, and the relationship between percentage germination during dormancy release were examined further using curve fitting routines in GraghPad Prism 5.01 software (GraghPad Prism 2007) at P = 0.05. To stabilize the variances, all data were arcsine-transformed prior to statistical analysis.
Results
Dormancy release and germination of seeds by cold moist stratification
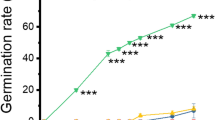
Dormancy of C. coggygria seeds was effectively released after they had been endured 5 °C pre-cold moist stratification for 15–75 days (Fig. 1, P < 0.05). Unstratified fresh mature seeds did not germinate. Percentage germination of stratified seeds was maximised following 30, 45, 60 and 75 days of treatment, with no significant difference (P > 0.05) between these treatments (Fig. 1). Percentage germination of seeds stratified for 15 days was significantly (P < 0.05) lower, at only about half of that achieved by seeds that received the other treatments. Using nonlinear regression analysis, the time taken for 50 % of final germination to occur at 10 °C was 12.1 ± 0.2, 7.9 ± 0.3, 8.4 ± 0.5, 7.1 ± 0.1 and 3.7 ± 0.2 days for cold stratification periods of 15, 30, 45, 60 and 75 days, respectively (Fig. 1). For these reasons, seeds that received 0 (control), 30 and 60 days stratification were used for the protein expression experiments since these treatments best represented the range in treatment responses.
Change of respiration rate of seeds during cold moist stratification
Respiration rate of C. coggygria seeds was increased with the increasing of time-period under cold moist stratification (Fig. 2). Respiration rates were similar for seeds given 15 or 30 days stratification, but values were significantly higher for seeds given 45 and 60 days stratification. In addition, the changes in respiration rates were similar in the bisected and intact seeds that had been released from dormancy (Fig. 2).
Proteome maps and mass spectrometry (MS) results
Significant differences in protein expression during dormancy release were found for seeds stratified for 30 or 60 days compared with no stratification (0 days) (Fig. 3). A total of 421 protein spots on the 2D-gels were observed using the ImageMaster 5.0 Platinum. The changes of 28 spots in relative abundance were significant (≥1.5 folds) (Table 1). Among these 28 differential protein spots, eight spots were up-accumulated, and other 20 spots were down-accumulated (Table 1). In addition, both the numbers and types of protein spots differed for seeds given 30 and 60 days stratification compared with no stratification (Fig. 3). Four of the 28 differently expressed protein spots were successfully identified, including adenosine triphosphatase (ATPase) β subunit, heat-shock cognate protein 70 (HSC70), aspartic proteinase 1 and actin (Table 2; Fig. 4).
Changes of ABA and GA3 content in seeds during cold moist stratification
The concentrations of ABA and GA3 declined as the period of stratification increased from 0 to 75 days, but most differences were not significant over the 15–75 day period of stratification (Fig. 5). In addition, the ABA/GA3 ratio was similar during the whole 75 days of cold moist stratification (P > 0.05, Fig. 5).
Discussion
Cold moist stratification at 5 °C effectively released the dormancy of C. coggygria seeds, and seed germination was increased with the increasing duration of cold moist stratification at 10 °C (optimum temperature) in dark (Fig. 1). Similar trends in dormancy release in response to cold stratification were reported previously (Guner and Tilki 2009; Deng et al. 2010). In this study, the final percentage germination of C. coggygria seeds was very low, the maximum percentage germination was only 56.7 ± 6.6 % (a 45-day stratification followed by a 30-day incubation at 10 °C in dark). The low percentage germination probably derived from the low seed maturation percentage, because C. coggygria possesses indeterminate inflorescences. When a indefinite inflorescence becomes mature, seed viability declines acropetally in indeterminate inflorescences and some seeds near the apical end of the inflorescence are not viable. This phenomenon is widespread in plants with indeterminate inflorescence.
The biosynthesis of GA is generally related to the promotion of seed germination but not to the release of seed dormancy (Bewley and Black 1994). Our results showed a significant decrease in both GA3 and ABA in C. coggygria seeds (Fig. 5), though the content of GA3 did not represent the total content of GA. In contrast, a significant increase in both GA1 and GA4 in A. thaliana seeds was observed during cold stratification (Yamauchi et al. 2004). However, the increase of GA content in A. thaliana seeds during stratification was not related to the regulation of seed dormancy (Derkx et al. 1994).
Seeds stratified at 5 °C for 0 (control, i.e. untreated fresh-mature dry seeds), 30 and 60 days were used to investigate protein expression during dormancy release and four proteins of ATPase β subunit, HSC70, aspartic proteinase 1 and actin were successfully identified (Table 2; Fig. 4). The release of seed dormancy is closely related to energy metabolism (Krawiarz and Szczotka 2000, 2005). Proteins associated with energy metabolism, one of the main protein categories, often changed during seed dormancy release of A. thaliana (Chibani et al. 2006) and Fagus sylvatica (Pawłowski 2007). The present study showed that ATPase β subunit (spot 31) was down-accumulated during dormancy release of C. coggygria seeds (Table 1; Figs. 3, 4a). ATPase was also down-accumulated during dormancy release in Fagus sylvatica, Acer platanoides and Acer pseudoplatanus seeds (Pawłowski 2007, 2009, 2010). ATPase is comprised of two complexes F0 and F1 (Wilkens et al. 2005). In F1 complex, α and β subunits form the catalytic core, which catalyzes ATP synthesis and hydrolysis (Amzel et al. 2003). In addition, the respiration rate of C. coggygria seeds significantly increased as the duration of cold stratification was extended (P < 0.05, Fig. 2). The down-accumulation of ATPase β subunit might have decreased ATP synthesis, thus perhaps having a feedback inhibitory effect of ATP function and hence enhancing oxidative respiration rates.
The expression of a group of functionally related proteins is both heat-inducible and constitutive when cells are exposed to elevated temperatures or other environmental stresses (Sorensen et al. 2003; Mizrahi et al. 2010). Such proteins are called “heat shock proteins” (HSPs) or heat-shock cognate proteins (HSCs). These two protein families have a high homology (up to 95 %) with similar biochemical characteristics, and considered as main stress proteins and the most conservative ones in the evolution of eukaryotes (Lindquist and Craig 1988; Mizrahi et al. 2010). Overexpression of HSP70s alters enzymatic activities, down-regulates signal transduction and protects cells from harmful damage as well as cohort proteins to other proteins in human, prokaryote and animal cells (Kiang and Tsokos 1998). In the present study, HSC70 (spot 91) declined to zero after 30 days stratification (Fig. 4b), indicating that seed dormancy is a stress response mechanism that prevents radicle emergence. With seed dormancy being released, the stress that impedes seed germination is removed, and expression of stress proteins (e.g. HSC70) that protected embryos from stress injuries started to decline and eventually reached zero. Similarly, Lee et al. (2006) also showed that dehydrin, another stress protein, was significantly down-accumulated during dormancy release of Prunus campanulata seeds following stratification.
Aspartic proteinases have been purified from both mono- and dico-tyledonous seeds. Their functions included: (1) hydrolysis as nitrogen resources (An et al. 2002); (2) stress reaction (Guevara et al. 2001; Xia et al. 2004); (3) plant senescence and programmed cell death (Bhalerao et al. 2003; Lindholm et al. 2000) and (4) plant reproduction (Vieira et al. 2001) respectively. Aspartic proteinase 1 (spot 227) was down-accumulated following 60 days of 5 °C pre-cold moist stratification (Fig. 4c), which might suggest its involvement in protein hydrolysis (An et al. 2002). The down-accumulation of aspartic proteinase 1 with dormancy release might accompany embryo growth that requires de novo synthesis of proteins during the initial stage of dormancy release, whereas the hydrolysis of storage proteins that occurs at this time provides amino acids for the de novo synthesis of proteins. Similarly, the down-accumulation of proteasome α subunit and α-gliadin (a storage protein) played important roles in the protein synthesis during cold stratification of F. sylvatica seeds (Pawłowski 2007).
It has been well known that chilling, i.e. cold stratification, releases seed dormancy in most tree species (Khan 1982). Actin, one of the main ingredients in cytoskeleton, existing in all plant cells, was involved in signal transduction of physical stimuli such as touching and chilling (Staiger and Schliwa 1987; McDowell et al. 1996; de Ruijter and Emons 1999). In this study, the change of expression and abundance of actin at this time may suggest that chilling signalled or initiated the dormancy release programme during stratification. A chilling stimulus would not be needed once it has initiated the programme of dormancy release. Accordingly, the role of actin involved in the signal transduction of chilling stimuli would also not be needed any more, since actin (spot 125) was nearly down-accumulated to 0 after a 30-day cold moist stratification (Fig. 4d).
The biosynthesis of GA is associated with the promotion of germination, but not the release of seed dormancy, and there was a strong antagonism between GA and ABA (Bewley and Black 1994). Yamauchi et al. (2004) found that the active GA1 and GA4 in A. thaliana seeds were increased during the cold stratification. However, the increase of GA content during stratification was not relevant to the regulation of seed dormancy in A. thaliana seeds (Derkx et al. 1994). We found that the concentrations of both GA3 and ABA in C. coggygria seeds were significantly decreased during cold moist stratification, but not the ABA/GA3 ratio (Fig. 5). Obviously, the change of GA3 could not represent the change of total GA content and hence the ABA/GA ratio. The decrease of GA3 concentration during cold moist stratification may implicate an unknown role of GA3 in seed dormancy release.
In conclusion, our study showed that the dormancy release of C. coggygria seeds resulted from a number of physiological and biochemical processes, including the synthesis of phytohormone (ABA and GA3) and proteins including those responsible for energy metabolism (ATPase β subunit), stress response (HSC70), hydrolysis and synthesis (aspartic proteinase 1) and signal transduction (actin). A better understanding of the seed dormancy release and germination mechanism has practical benefits to seedling production for artificial regeneration.
References
Ali-Rachedi S, Bouinot D, Wagner MH, Bonnet M, Sotta B, Grappin P, Jullien M (2004) Changes in endogenous abscisic acid levels during dormancy release and maintenance of mature seeds: studies with the Cape Verde Islands ecotype, the dormant model of Arabidopsis thaliana. Planta 219:479–488
Amzel LM, Bianchet MA, Leyva JA (2003) Understanding ATP synthesis: structure and mechanism of the F1-ATPase. Mol Membr Biol 20:27–33
An CI, Fukusaki E, Kobayashi A (2002) Aspartic proteinases are expressed in pitchers of the carnivorous plant Nepenthes alata Blanco. Planta 214:661–667
Baskin CC, Baskin JM (1998) Seeds: ecology, biogeography and evolution of dormancy and germination. Academic Press, San Diego
Baskin CC, Baskin JM (2004) A classification system for seed dormancy. Seed Sci Res 14:1–16
Bewley JD, Black M (1994) Seeds: physiology of development and germination. Plenum Press, New York
Bhalerao R, Keskitalo J, Sterky F, Erlandsson R, Bjorkbacka H, Birve SJ, Karlsson J, Gardestrom P, Gustafsson P, Lundeberg J, Jansson S (2003) Gene expression in autumn leaves. Plant Physiol 131:430–442
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Budiman MA, Mao L, Wood TC, Wing RA (2000) A deep-coverage tomato BAC library and prospects toward development of an STC framework for genome sequencing. Genome Res 10(1):129–136
Cadman CSC, Toorop PE, Hilhorst HWM, Finch-Savage WE (2006) Gene expression profiles of Arabidopsis Cvi seed during cycling through dormant and non-dormant states indicate a common underlying dormancy control mechanism. Plant J 46:805–822
Chibani K, Ali-Rachedi S, Job C, Job D, Jullien M, Grappin P (2006) Proteomic analysis of seed dormancy in Arabidopsis. Plant Physiol 142:1493–1510
de Ruijter NCA, Emons AMC (1999) Actin-binding proteins in plant cells. Plant Biol 1:26–35
Deng ZJ, Cheng HY, Song SQ (2010) Effects of temperature, scarification, dry storage, stratification, phytohormone and light on dormancy-breaking and germination of Cotinus coggygria var. cinerea (Anacardiaceae) seeds. Seed Sci Technol 38:572–584
Derkx MPM, Vermeer E, Karssen CM (1994) Gibberellins in seeds of Arabidopsis thaliana: biological activities, identification and effects of light and chilling on endogenous levels. J Plant Growth Regul 15:223–234
Gubler F, Millar AA, Jacobsen JV (2005) Dormancy release, ABA and pre-harvest sprouting. Curr Opin Plant Biol 8:183–187
Guevara MG, Daleo GR, Oliva CR (2001) Purification and characterization of an aspartic protease from potato leaves. Physiol Planta 112:321–326
Guner S, Tilki F (2009) Dormancy breaking in Cotinus coggygria Scop. seeds of three provenances. Sci Res Essays 4:73–77
Hilhorst HWM (2007) Definitions and hypotheses of seed dormancy. In: Bradford K, Nonogaki H (eds) Seed development, dormancy and germination. Annual plant reviews 27. Blackwell Publishing, Oxford, pp 50–71
Karssen CM, Laçka E (1986) A revision of the hormone balance theory of seed dormancy: studies on gibberellin and/or abscisic acid-deficient mutants of Arabidopsis thaliana. In: Bopp M (ed) Plant growth substances. Springer, Berlin, pp 315–323
Kermode AR (2005) Role of abscisic acid in seed dormancy. J Plant Growth Regul 24:319–344
Khan AA (1982) The physiology and biochemistry of seed development, dormancy and germination. Elsevier Biomedical Press, Amsterdam, p 280
Kiang JG, Tsokos GC (1998) Heat shock protein 70 kDa: molecular biology, biochemistry, and physiology. Pharmacol Ther 80:183–201
Krawiarz K, Szczotka Z (2000) Activity of ATPases during dormancy breaking in Norway maple (Acer platanoides L.) seeds. Acta Soc Bot Poloniae 69:119–121
Krawiarz K, Szczotka Z (2005) Adenine nucleotides and energy charge during dormancy breaking in embryo axes of Acer platanoides and Fagus sylvatica seeds. Acta Physiol Plant 27:455–461
Le Page-Degivry MT, Bianco J, Barthe P, Garello G (1996) Change in hormone sensitivity in relation to the onset and breaking of sunflower embryo dormancy. In: Lang GA (ed) Plant dormancy: physiology, biochemistry and molecular biology. CAB International, Wallingford, pp 221–231
Lee C, Chien C, Lin C, Chiu Y, Yang Y (2006) Protein changes between dormant and dormancy-broken seeds of Prunus campanulata Maxim. Proteomics 6:4147–4154
Li XB, Fan XP, Wang XL, Cai L, Yang WC (2005) The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell 17(3):859–875
Lindholm P, Kuittinen T, Sorri O, Guo DY, Merits A, Tormakangas K, Runeberg-Roos P (2000) Glycosylation of phytepsin and expression of dad1, dad2 and ost1 during onset of cell death in germinating barley scutella. Mech Dev 93:169–173
Lindquist S, Craig EA (1988) The heat-shock proteins. Ann Rev Genet 22:631–677
Liu CJ (2000) Cotinus coggygria Scop. var. cinerea Engl. In: The National Service Center for State-Owned Forest Farms and Forest Seed and Seedling Affairs of the Forestry Ministry (ed) Seeds of woody plants in China. China Forestry Publishing House, Beijing, p 870
McDowell JM, Huang S, McKinney EC, An Y-Q, Meagher RB (1996) Structure and evolution of the actin gene family in Arabidopsis thaliana. Genetics 142:587–602
Mizrahi T, Heller J, Goldenberg S, Arad Z (2010) Heat shock proteins and resistance to desiccation in congeneric land snails. Cell Stress Chaperon 15:351–363
Pappin DJ, Hojrup P, Bleasby AJ (1993) Rapid identification of proteins by peptide-mass fingerprinting. Curr Biol 3:327–332
Pawłowski TA (2007) Proteomics of European beech (Fagus sylvatica L.) seed dormancy breaking: influence of abscisic and gibberellic acids. Proteomics 7:2246–2257
Pawłowski TA (2009) Proteome analysis of Norway maple (Acer platanoides L.) seeds dormancy breaking and germination: influence of abscisic and gibberellic acids. BMC Plant Biol 9:48–58
Pawłowski TA (2010) Proteomic approach to analyze dormancy breaking of tree seeds. Plant Mol Biol 73:15–25
Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins from silver stained polyacrylamide gels. Anal Chem 68:850–858
Sorensen JG, Kristensen TN, Loeschcke V (2003) The evolutionary and ecological role of heat shock proteins. Ecol Lett 6:1025–1037
SPSS (2003) SPSS 12.0. SPSS Inc., Chicago
Staiger CJ, Schliwa M (1987) Actin localization and function in higher plants. Protoplasma 141:1–12
Steinbach HS, Benech-Arnold R, Sanchez RA (1997) Hormonal regulation of dormancy in developing sorghum seeds. Plant Physiol 113:149–154
Terauchi K, Asakura T, Nishizawa NK, Matsumoto I, Abe K (2004) Characterization of the genes for two soybean aspartic proteinases and analysis of their different tissue-dependent expression. Planta 218(6):947–957
Vieira M, Pissarra J, Verissimo P, Castanheira P, Costa Y, Pires E, Faro C (2001) Molecular cloning and characterization of cDNA encoding cardosin B, an aspartic proteinase accumulating extracellularly in the transmitting tissue of Cynara cardunculus L. Plant Mol Biol 45:529–539
Wang X, Li X, Deng X, Han H, Shi W, Li Y (2007) A protein extraction method compatible with proteomic analysis for the euhalophyte Salicornia europaea. Electrophoresis 28:3976–3987
White CN, Rivin CJ (2000) Gibberellins and seed development in maize. II. Gibberellin synthesis inhibition enhances abscisic acid signaling in cultured embryos. Plant Physiol 122:1089–1097
White CN, Proebsting WM, Hedden P, Rivin CJ (2000) Gibberellins and seed development in maize. I. Evidence that gibberellin/abscisic acid balance governs germination versus maturation pathways. Plant Physiol 122:1081–1088
Wilkens S, Zheng Y, Zhang Z (2005) A structural model of the vacuolar ATPase from transmission electron microscopy. Micron 36:109–126
Wu ZY, Raven PH, Hong DY (2008) Flora of China, vol 11. Science Press, Beijing, pp 343–345 (in Chinese)
Xia Y, Suzuki H, Borevitz J, Blount J, Guo Z, Patel K, Dixon RA, Lamb C (2004) An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO J 23:980–988
Yamauchi Y, Ogawa M, Kuwahara A, Hanada A, Kamiya Y, Yamaguchi S (2004) Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 16:367–378
Yasunari O, Kazuhiro I, Toshio K, Akira E, Mitsumasa H, Takashi S, Toru T, Shigeko U, Minoru M, Naoki M, Shigeo T, Kazuho I, Takashi G, Rika M, Koji T, Koichiro T (2000) Chinese spring wheat (Triticum aestivum L.) chloroplast genome: complete sequence and contig clones. Plant Mol Biol Rep 18:243–253
Acknowledgments
We are grateful to Drs. Shihua Shen, Xue Zhao and Kuixian Ji from the Institute of Botany, Chinese Academy of Sciences, Beijing, China and Dr. Langtao Xiao, Ms. Jianhua Tong and Qiong Peng from the Hunan Provincial Key Laboratory of Phytohormone and Growth Development, Hunan Agricultural University, Changsha, Hunan, China for providing assistances; and the National Natural Science Foundation of China (81303169), the Doctoral Scientific Research Foundation of Hubei University for Nationalities (MY2013B014), Guangxi Natural Science Foundation (2013jjBA40121) and the Open Fund of Key Laboratory of Biologic Resources Protection and Utilization of Hubei Province (PKLHB1304) for financial support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Deng, Z.J., Hu, X.F., Ai, X.R. et al. Dormancy release of Cotinus coggygria seeds under a pre-cold moist stratification: an endogenous abscisic acid/gibberellic acid and comparative proteomic analysis. New Forests 47, 105–118 (2016). https://doi.org/10.1007/s11056-015-9496-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11056-015-9496-2