Abstract
The operation of partitioning an image into a collection of connected sets of pixels is known as image segmentation. Expectation Maximization (EM) segmentation requires appropriate initialization of tissue class mean and variance, as it is get stuck on particular possible area of intensity of the probability nature. Decisive initialization is a necessary exploratory process for the forthcoming convergence of the algorithm to best regional maximum of possibility task. Indiscriminate initialization is not dossier directed, deep from optimal, outcomes are not reproducible, do not take advantage of deep rooted patterns in the data or may be loaded on outliers. This paper evaluates the performance of EM segmentation with random initialization, histogram guided initialization and initialization with k-means with respect to computational complexity and root mean squared error of tissue class mean and variance, updated by EM, with manually estimated tissue class mean and variance as ground truth, on paramount plane, T1 contrast and Magnetic resonance (MR) images of Glioblastoma Multiframe (GBM) Edema complex. The random initialization and histogram guided initialization was experimented for k-means, from the clustered output of which; initial tissue class mean and variance for EM are derived. RMS error remains the same for EM initialized and histogram guided K-means. EM initialized with K-means which has histogram guided initialization converges fast than the random initialization K-means, but the computational time is more for the former initialization than the latter. The experimental evaluation of EM initialization schemes and Fuzzy Cluster Means (FCM) were performed in MATLAB. The efficacy of Fuzzy Cluster Means clustering was analyzed qualitatively on tumour-edema complex. FCM could identify only 3 classes including background in the MR specimens. FCM consider edema and certain parts of WM as a single tissue class. Similarly, FCM clubs GM, CSF and necrotic focus into tissue class and produced empty clusters.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Expectation maximization [2, 5, 22] is a probabilistic segmentation scheme with wide acceptance on medical images. However, there is no broadly approved mechanism for criterion load-in of data in EM segmentation. Training Gaussian mixture by EM can be troublesome as the actual number of blend factors is normally exotic and the data can get captured in one of the many provisional area of the prospect activity [24]. The EM algorithm craves opportune compute, as it is especially recognized to pull data on limited maximal of the liability target [6, 7, 18, 25]. Valuable boot up is an crucial preparatory measure for the subsequent convergence of the algorithm to the best local maximum of the likelihood function [15]. This initialization is commonly chosen randomly, and as a consequence results are not reproducible, do not take advantage of inherent patterns in the data or may be initialized on outliers [14].
Random initialization is not data driven and is far from optimal and does not eliminate the problem of converging to local maxima [7] In addition, the segmentation results returned by the EM algorithm will be different any time the algorithm is executed, say the reproducibility of segmentation results are poor. To circumvent the problems caused by the random procedure, the result of the K-Means algorithm may be thought to set the initial cluster centers. The advantage of using this strategy is minimal since this does not assure that the K-Means algorithm will not be trapped itself in local maxima [1, 10].
The performance of four different initialization schemes of EM segmentation is analyzed with respect to the RMS error of the updated tissue class mean and variance by EM, and computational complexity, on axial plane T1 contrast test MR images. Analyzed initialization schemes of EM includes, random initialization, histogram guided initialization and initialization from K-means. Both random initialized K-means and K-means with histogram guided initialization were analyzed, providing initial tissue class mean and variance for EM. The ability of FCM and K-means in clustering the tissue classes present in the test MR images was qualitatively evaluated.
2 Reviews on initialization methods for EM
A review has been made on initialization schemes for EM segmentation. The initialization schemes include hierarchical clustering, rough set, histogram guided tree structure scheme [3], cross entropy based [25] K-means and the primitive random initialization literature [13] employed Expectation Maximization (EM) algorithm, with model parameters obtained from clustering technique known as hierarchical towards automatic segmentation of Diffusion-Weighted Images (DWI).
Literature [27] describes automatic dissection of perfusion compartments derived from Dynamic Susceptibility Contrast (DSC) MR images using a hybrid concept of multivariate Gaussians (MoMG) and the expectation-maximization (EM) algorithm. A semi-supervised top-down initialization method is used in [6], utilizing a priori knowledge about the number of tissues. Dual step procedure is adapted to initialize the data which are performed by means of down sampled version of brain volume. Gaussian modeling is suitable for regions split up.
In [16] an integration of a minimal spanning tree (MST) based graph-theoretic technique and expectation maximization (EM) algorithm with rough set initialization is described for non-convex clustering. Literature [18] also exploits rough-set-theoretic logical rules to obtain an initial approximation of Gaussian mixture model parameters. Ilea et al. [7] deals about the selection of extracting the dominant colors from the histogram to initialize EM algorithm. The methods include heuristic initialization by extracting the dominant colors after color quantization. Histogram for each color channel is constructed such that, n s refers to the number of pixels contained in the bin s. Histograms are partitioned linearly into R sections where R > M, number of structures or objects in the image.
In [26] initial statistics for MAP-EM framework was obtained from a histogram guided region labeling. Further, Vector Quantization (VQ) strategy was incorporated to refine the region labeling. In some literature review [3] proposed a Tree Structure (TS) algorithm whose scheme is similar to the TS vector quantization (TSVQ) method. Initialization chart in literature [25] is based on the Cross- Entropy method.
In the initialization method described in [11] individual voxel is initially drafted to one of k class labels by comparing its density to empirically defined k thresholds.
Multivariate Gaussian mixture models Expectation Segmentation J. Blomer and K. Bujina [12] presented a method based on K-means and Gonzalez algorithm. Literature [21] replaced the manual initialization of EM in slicer3 with K-means initialization. Most of the initialization schemes [3, 7, 14, 16, 25] and [12] available in literature are not tested or validated on medical images.
3 Preprocessing section
Axial plane T1 contrast enhanced (Series: AXT1, SE, FS&C Spin Echo Sequence (SE)) MR images were chosen for the preliminary interpretation of the proposed system. The stipulation of MR equipment manufacturer is: GE medical systems, Model Name: Sigma HDxt, Acquisition Type: 2D & 1.5 T girl strength.
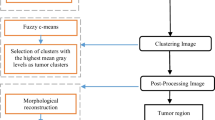
The prospective pre-processing was experimented on MATLAB Image Processing Tool Box. Demonstrates the hierarchy of steps involved is background elimination, bilateral filtering [23], Contrast Limited Adaptive histogram equalization [28] (CLAHE) and skull stripping (Figs. 1, 2, 3, and 4).
3.1 Background elimination
It was to bring out by augment the prime MR image with an amplification mask which was organized by a gradient based threshold is pursued by a series of semantic transactions. These include dilation, hope filling, border clearing and erosion. Sobel mask is used for computing the gradient for the prime image. The threshold of gradient based is [22],
From the above equations f^(x,y) is the threshold of edge image which are generated from gradient based [19]. Gr refers to an adaptive gradient threshold and similarly the gradient magnitude is denoted as G(x,y). The respective gradient are predicted along the x & y directions. The process of erosion and dilation was executed for the traced edges, horizontal & vertical structuring elements or steel objects with a diamond steel object having five neighbours or with a length one. [2] The optimum window size of about 11*11 with the variance of the spatial weight factor which is equal to about ten and variance of the radiometric weight factor kernel of the about 1.3 by mans of trial and error method. In this strategy, the weighted average of neighbouring intensities is used to replace the each pixel intensity. The adaptive threshold was computed using Otzu’s method [24].
3.2 Filtering
Bilateral filtering is an approach to gentle comparable region of images while perpetuate the edges. The mathematical concept [2] of bilateral filter is;
where Y is the corrupted signal.
In the given image the weighted average of pixels as follows.
The final weight is the product of these weight factors.
In the contrast limited adaptive histogram equalization [5], the clip limit and tile size was 0.12 and 8*8, respectively. The distribution opted is reliable distribution. Likewise it was determined provisionally, as the bilateral filter restriction. MR image afterwards recovery and contrast enhancement was intensity threshold and akin peripheral in emerging binary image were characterized. The adaptive threshold was computed using Otzu’s method.
Assume L be the maximum probable grey level in histogram [17] equalized MR image and image enclose grey levels {0,1,….L}. The normalized grey level histogram and the class variance
3.3 Contrast limited adaptive histogram equalization (CLAHE)
CLAHE has a positive noise elimination competence and the degree of contrast enhancement is satisfactory to backing characterization of tissue classes can yield accurate outcomes.
3.4 Skull stripping
It is based on an adaptive intensity threshold. The brain regions are effectively extracted from the skull and scalp using the technique called skull stripping. This technique will enhance the segmentation when the number of tissue class in the resultant image comes down.
4 Mathematical methodology for EM segmentation, initialization schemes and validation
In histogram guided initialization, let be the vector of mean intensity of different tissue classes present in the pre-processed MR image is given by
The range of pixel intensities of the pre-processed MR image is divided into intensity bins with k + 1 intensity points between the maximum and minimum intensity.
From the above equation we define the k as the number of tissue classes. IH refers to the maximum intensity in the pre-processed MR image. IL is the minimum intensity in the pre-processed image. Ii known as the intensities present in the bin.
The mean of pixel intensities present in a bin represent the mean intensity of tissue class corresponding to that bin. ‘Ii’ is the intensities present in the bin and nIi is the histogram of these intensities.
Similarly, variance of the jth tissue class is the variance of the intensities present in the jth intensity bin. The number of tissue classes ‘k’ are six i.e. GM, M, CSF, necrosis enhancing edema and background in specimen MR image 2, 3, 5 & 6 used to evaluate the initialization schemes of EM [8, 9].
In k-means clustering let the initialized cluster centre at the 1st iteration be μ(1) = {μ1(1),μ2(2)....μk(1)}.The variance of the tissue class is given by the equation
4.1 Algorithm
-
Early break the mean shift measure for all data point from x and stock its convergence point as z.
-
Associate all together from the z’s which are closer than 0.5 from every individual unit to form the clusters.
-
Accredit or match every individual point to its own cluster.
-
Eradicate the small regions obtain from the step 3
-
Load an estimate of θ l : l = 1. . L
-
Repeat
-
Step 1: (E step)
-
Obtain an estimate of the labels based on the current parameter estimates
-
Step 2: (M step)
-
Update the parameter estimates based on the current labelling
Until Convergence
-
The tissue class mean, variance and tissue fraction are estimated following
Where αi is the tissue fraction contributed by the jth tissue class which is he mixing parameter of Gaussian mixture in EM. The fractional contribution of a tissue class is the cumulative probability density of intensities present in the corresponding bin. The tissue class mean, variance and tissue fraction derived from histogram can be directly used to initialize the EM algorithm or the tissue class mean, estimated from histogram guided method can be used to initialize the K-means algorithm, which in turn initializes the EM. Individual tissue class mean, variance and tissue fraction, to initialize EM, are estimated from the classes, clustered by K-means. The initial tissue class mean of K-means may be randomly chosen or it can be derived from the histogram guided method as demonstrated. K-means clustering comprises initialization of cluster centre, initial clustering, cluster centre updating and final clustering.
The squared error (erms) of tissue class mean and variance is
The updated value of tissue class mean and variance from EM is compared with a manually estimated ground truth, to estimate the RMS error, thereby to evaluate the accuracy of these estimates. The RMS error, for different EM initialization schemes expresses the efficiency of the initialization. The ground truth tissue class mean and variance are estimated by manually cropping a portion of the tissue class from its middle and estimating the mean and variance of intensities present in that cropped region, in Mat lab. The computational complexity and accuracy of random, histogram guided, histogram guided K-Means initialization, and random initialized K-means initialization schemes for EM segmentation are evaluated in terms of computational time and Root Mean Squared error (erms) of tissue class mean and variance, on pre-processed six specimen MR images.
4.2 Fuzzy cluster means
FCM could be employed to initialize EM if it is able to identify the entire tissue classes in the specimen MR image, as K-means do or in a still better way. FCM [5] is occupying on minimizing an detached function, with esteem to fuzzy membership ‘U’, and agreed of cluster centroids [20]. In (1) X = {x1,x2,….j… N} is a p × N data matrix, where, p means the dimension of each xj “feature” vector, and represents the number of feature vectors pixel numbers in the image. UijЄU(p,N,C) is attribute to the membership function of vector from xj to the ith cluster, which amuse uij [0,1]
5 Result & discussion
The numerical values of tissue class mean & variance estimated by the initialization schemes, random histogram guided, random initialized k-means and k-means with histogram guided initialization are updated by EM segmentation. Each of the K Gaussians will have parameters θ j = (μ j ,Σ j ), where μ j is known as the mean of the jth Gaussian. Σ j is refers as the covariance matrix of the jth Gaussian. Covariance matrices are initialed to be the identity matrix. The means can be initialized by finding the average feature vectors in each of K windows in the image; this is data-driven initialization.
Eventually the efficacy of the above mentioned initialization schemes are qualitatively appreciated with the aid of images with tissue classes segmented by EM with these initialization and FCM (Tables 1 and 2).
From Table 3 inferred as the RMS errors of tissue class mean and variance estimated from EM with different initialization schemes, in comparison with manual ground truth, where Table 2 is for test MR images are furnished. Eventually the efficacy of the above mentioned an initialization scheme is qualitatively appreciated with the aid of images with the tissue classes segmented by EM with these initialization and FCM.
Random initialization, histogram guided initialization, initialization with histogram guided as well as random initialized K-means and Fuzzy C means are validated about specimen MR images in terms of concerning computational complexity and accuracy. The proposed, histogram guided K means initialization exhibited minimum RMS error, with manually estimated tissue class mean and variance as ground truth. Histogram guided K means initialization shows an average RMS error on six MR specimens, of 23.2386 which is 37.8446 and 36.2993 in histogram guided initialization and random initialization, respectively. The proposed initialization with histogram guided K-means is able to perform accurate segmentation with comparatively minimum computational time. From table RMS error remains the same for EM initialized with random initialized and histogram guided K-means. EM initialized with K-means which has histogram guided initialization converges fast than random initialized K-means. But the total computation time is more for the former initialization than the latter. Even though termed as random initialization, this also is partially image driven as the tissue class means are maximum and minimum intensities of the pre-processed MR image and intensities, spaced at equal intervals between maximum and minimum intensity points.
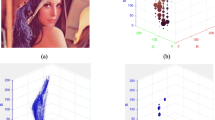
From the above figure it is vivid that the accuracy in clustering WM gradually increases such that the number of pixels identified to be WM increases as initialization progress from random to histogram guided K-means. Similarly, in Fig. 5b maximum number of CSF pixels are clustered by EM having histogram guided K-means initialization. The random initialized EM abruptly fails to cluster CSF pixels. The accuracy of clustering CSF and WM pixels exhibited by EM with histogram guided initialization is intermediate to random initialized EM and EM with histogram guided K-means initialization. The qualitative evaluation of the clustered morphological structures (Fig. 5d, e, and f) reveal that the performance of EM initialized from random initialized K-means and K-means having histogram guided initialization is similar.
The morphological structures present in these specimens are WM, GM, CSF, necrotic focus and edema. From the Fig. 6a–b it is apparent that FCM is able to identify only three classes, including the background, in both the MR specimens. FCM consider edema and certain parts of WM. In addition to that, FCM clubs GM, CSF and necrotic focus into a single tissue class. In other words, FCM produces barren clusters. This happens to certain regions of necrotic focus, CSF and GM. Histogram guided K means initialization shows an average RMS error on six MR specimens, of 23.2386 which is 37.8446 and 36.2993 in histogram guided initialization and random initialization, respectively. The proposed initialization with histogram guided K-means is able to perform accurate segmentation with comparatively minimum computational time [4]. RMS error remains the same for EM initialized with random initialized and histogram guided K-means. EM initialized with K-means which has histogram guided initialization converges faster than the random initialized K-means. But the total computation time is more for the former initialization than the latter. Even though termed as random initialization, this also is partially image driven, as the tissue class means are maximum and minimum intensities of the pre-processed MR image and intensities, spaced at equal intervals between maximum and minimum intensity points, calculate on the statistic of tissue classes, present in the latest specimen MR image.
6 Conclusion
The determination of image segmentation is to separate an image into worthwhile regions with respect to a distinct operation. The segmentation is established on assessments appropriated from the image and might be grey level, colour, texture, depth or motion. The proposed segmentation techniques have been used in real time clinical application. A ascendable lateral algorithm for active flock improves the act of fuzzy c-mean algorithm (FCM) for brain tumor detection was proposed. The accuracy and the computing time of the proposed algorithms were very impressive. The reckoning cost and the accuracy was improved by concluding a satisfactory set of initial band centers rather than that of accidental initial cluster center in FCM algorithm. Achieved segmentation accuracy in FCM was from 32% to 89% of detected tumor pixels based on ground truth. The performance of EM segmentation with random initialization, histogram guided initialization and initialization with K-means was analyzed with respect to computational complexity and RMS error of tissue class mean and variance, updated by EM, with manually estimated tissue class mean and variance as ground truth, on axial plane, T1 contrast, and MR images of GBM-edema complex. The random initialization and histogram guided initialization was experimented for K-means, from the clustered output of which; initial tissue class mean and variance for EM are derived. The efficacy of FCM clustering also was analyzed qualitatively on tumor-edema complex. FCM could identify only three classes, including the background, in the MR specimens. FCM consider edema and certain parts of WM as a sole tissue dashing. Furthermore, FCM clubs GM, CSF and necrotic focus into a single tissue class and produced desolated clusters. Alike assuming that EM with K-means initialization analyzes all the tissue classes, assertive parts of WM are bashed with vasogenic edema considering their analogous intensity features. This happens to certain regions of necrotic focus, CSF and GM. These segmentations techniques are found to be very successful and reliable. In terms of execution time, tremendous quickness parallel FCM algorithm is found directed towards best than the other methods.
References
Dempster AP, Laird NM, Rubin DB (1977) Maximum likelihood estimation from incomplete data via the EM algorithm. J R Stat Soc 39(1):1–38
Fatakdawala H, Xu J, Basavanhally A, Bhanot G, Ganesan S, Feldman M, Tomaszewski JE, Madabhushi A (2010) Expectation– Maximization-Driven Geodesic Active Contour with Overlap Resolution (EMaGACOR): application to lymphocyte segmentation on breast cancer histopathology. IEEE Trans Biomed Eng 57(7):1676–1689. https://doi.org/10.1109/TBME.2010.2041232
Fwu JK, Djuric PM (1997) EM algorithm for image segmentation initialized by a tree structure scheme. IEEE Trans Image Process 6(2):349–352
Gonzalez R, Woods R (1992) Digital image processing. Addison-Wesley Publishing Company, Boston, p 191
Gooya A, Pohl KM, Bilello M, Cirillo L, Biros G, Melhem ER, Davatzikos C (2012) GLISTR: Glioma Image Segmentation and Registration. IEEE Trans Med Imaging 31(10):1941–1954. https://doi.org/10.1109/TMI.2012.2210558
Greensp H, Ruf A, Goldberger J (2006) Constrained Gaussian mixture model framework for automatic segmentation of MR brain images. IEEE Trans Med Imaging 25(9):1233–1245
Ilea DE, Whelan PF (2006) Color image segmentation using a self- initializing EM algorithm. Proc. 6th international conference on visualization, imaging and image processing, 28–30 August 2006, Palma De Mallorca, ISBN 0–88986–598-1 Source: OAI
Karthikeyan A, Kumar PS (2017) Cluster comput. https://doi.org/10.1007/s10586-017-0979-0
Karthikeyan A, Senthil Kumar P (2017) Randomly prioritized buffer-less routing architecture for 3D network on chip. Comput Electr Eng 59:39–50. https://doi.org/10.1016/j.compeleceng.2017.03.006 ISSN 0045-7906
Khan S, Ahmad A (2004) Cluster center initialization algorithm for K-means clustering. Pattern Recogn Lett 25(11):1293–1302
Liang Z, Wang S (2009) An EM approach to MAP solution of segmenting tissue mixtures: a numerical analysis. IEEE Trans Med Imaging 28(2):297–310
Lomer JB, Bujna K (2013) Simple methods for initializing the EM algorithm for Gaussian mixture models. Computing Research Repository, http://arxiv.org/abs/1312.5946
Lu C-F, Wang P-S, Chou Y-C, Li H-C, Soong B-W, Wu Y-T (2008) Segmentation of diffusion-weighted brain images using expectation maximization algorithm initialized by hierarchical clustering. Engineering in Medicine and Biology Society, EMBS 2008. 30th Annual International Conference of the IEEE, pp. 5502–5505, 20–25 Aug. 2008, https://doi.org/10.1109/IEMBS.2008.4650460
Lynch M, Ilea D, Robinson K, Ghita O, Whelan PF (2007) Automatic seed initialization for the expectation-maximization algorithm and its application in 3D medical imaging. J Med Eng Technol 31(5):332–340
Melnykova V, Melnykovb I (2012) Initializing the EM algorithm in Gaussian mixture models with an unknown number of components. Comput Stat Data Anal 56(6):1381–1395
Mitra P, Pal SK, Siddiqi MA (2003) Non-convex clustering using expectation maximization algorithm with rough set initialization. Pattern Recogn Lett 24(6):863–873
Otsu N (1979) A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern 9(1):62–66
Pal SK, Mitra P (2002) Multispectral image segmentation using the rough- set-initialized EM algorithm. IEEE Trans Geosci Remote Sens 40(11):2495–2501
Ray S, Turi RH (1999) Determination of number of clusters in-means clustering and application in colour image segmentation. Proc 4th International Conference on Advances in Pattern Recognition and Digital Techniques (ICAPRDT'99), Calcutta, pp. 137–143
Shen S, Sandham W, Granat M, Sterr A (2005) MRI fuzzy segmentation of brain tissue using neighborhood attraction with neural-network optimization. IEEE Trans Inf Technol Biomed 9(3):459–467. https://doi.org/10.1109/TITB.2005.847500
Srinivasan P, Shenton ME, Bouix S, EM segmentation: automatic tissue class intensity initialization using K-means. http://hdl.handle.net/10380/3300
Tian GJ, Xia Y, Zhang Y, Feng D (2011) Hybrid genetic and variational expectation-maximization algorithm for gaussian-mixture-model- based brain MR image segmentation. IEEE Trans Inf Technol Biomed 15(3):373–380. https://doi.org/10.1109/TITB.2011.2106135
Tomasi C, Manduchi R (1998) Bilateral filtering for gray and color images. Proceedings of the 1998 I.E. international conference on computer vision, Bombay
Vlassis N, Likas A (2002) A greedy EM algorithm for Gaussian mixture learning. Neural Process Lett 1(1):77
Volkovich Z, Avros R., Golani M (2011) On initialization of the expectation maximization clustering algorithm. Transaction on Evolutionary Algorithm and Clustering Online Publication, June 2011, www.pcoglobal.com/gjto.htm, CA-O34/GJTO, ISSN: 2229–8711
Wang S, Lu H, Liang Z (2009) Theoretical solution to MAP-EM partial volume segmentation of medical images. Int J Imaging Syst Technol 19(2):111–119
Wu YT, Chou YC, Lu CF, Huang SR, Guo WY (2009) Tissue classification from brain perfusion MR images using expectation- maximization algorithm initialized by hierarchical clustering on whitened data. 11th international conference on biomedical engineering, IFMBE proceedings vol. 23, pp 714–717
Zuiderveld K (1994) Contrast limited adaptive histogram equalization. Graphic Gems IV. San Diego: Academic Press Professional, pp 474–485
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Prabhu, V., Kuppusamy, P.G., Karthikeyan, A. et al. Evaluation and analysis of data driven in expectation maximization segmentation through various initialization techniques in medical images. Multimed Tools Appl 77, 10375–10390 (2018). https://doi.org/10.1007/s11042-018-5792-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-018-5792-0