Abstract
Brain tumors are a leading cause of death in humans of various ages, making early detection and treatment crucial for improving patient outcomes. It is important to accurately determine the precise location, size, and dimensions of the tumor for successful radiotherapy. One reliable method for diagnosing brain tumors is Magnetic Resonance Imaging (MRI), as it can detect small, non-invasive lesions in the brain with great clarity and contrast. However, manual segmentation of tumors on MRI images is time-consuming, despite its accuracy. To address this challenge, computerized techniques can provide more precise and extensive results in less time. In this article, we propose a three-part method for segmenting tumors on MRI images. In the pre-processing stage, the contrast of the image is improved by matching the histogram, removing noise, and sharpening the image. In the next step, the tumor-related cluster is identified using fuzzy clustering. In the post-processing stage, the tumor areas are delineated using morphological reconstruction and active contour techniques. The proposed approach is evaluated on the training portions of two datasets: BraTS 2012 and BraTS 2013. This approach has shown robustness against noise, and intensity non-uniformity in experiments. Additionally, it is quick and more precise than other state-of-the-art segmentation methods, with an average running time of 2.33 s. Additionally, the average segmentation Sensitivity, Dice, Precision, Accuracy, Jaccard, and Hausdorff distance are 92.10%, 0.92, 92.05%, and 99.06%, 0.86, 3.60, respectively. The proposed method demonstrates satisfactory results for Glioma brain tumor segmentation due to fuzzy c-means clustering, morphological reconstruction, and active contour accurate segmentation results and short time. Since most medical images suffer from these issues, this method has the potential to be more effective in the segmentation of complex medical images.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Brain tumor segmentation from brain MRI is critical work in medical image processing. Image segmentation is a challenging operation that involves dividing a digital image into various sections The purpose of image segmentation is to ease the analysis process and determine the position of objects and boundaries in images [1, 2]. Early recognition and treatment of brain tumors are one of the most critical factors in the recovery process and immensely increase patient survival rate [3].
MRI images are more acceptable among doctors due to their sensitivity in contrast and the possibility of revealing very small, non-invasive lesions. Without causing radiation exposure to other regions of the brain and causing damage to the patient's skull artifacts, MR imaging is helpful in the identification and treatment of brain cancers [4]. Therefore, there is a demand for an effective approach to segmenting medical images that have several favored characteristics, such as minimal interaction with the user, precise outcomes, rapid calculation, and reliable segmentation outcomes [5].
The segmentation method is based on dividing the processed image according to intensity variation, such as edges and corners. The second method is based on dividing an image into sections based on a set of specified criteria that are similar. As a result, numerous segmentation techniques can be widely applied, including histogram-based techniques, edge-based techniques, artificial neural network-based segmentation methods, physical model-based approaches, techniques for dividing, expanding, and merging areas, as well as clustering techniques such as Fuzzy c-means clustering, K-means clustering, Partition Clustering, and Mean Shift [6].
A clustering method determines which pixels in an image integrate the best. There are two clustering methods: either by partitioning or by grouping pixels. Partitioning starts with the entire image and splits it into increasingly smaller clusters. In contrast, the grouping type starts by identifying each item as a separate cluster and then merging the single clusters to form bigger ones [7]. Fuzzy c-means is an important and popular unsupervised partitioning algorithm that is used in this study.
Morphology is one of the methods of image analysis, the output of this method describes the contents of the image. Morphology is based on a set of mathematical tools that extract a specific feature from the image furthermore removing unnecessary details and is therefore widely used in histology [8]. This method displays the edges of elements more prominently than other points and its purpose is to change or correct the elements inside a binary image. This operation is usually performed one step before the final processing operation. The final operation means the operation in which information is extracted from the image. For example, the perimeter or area of the image components is calculated. The mathematical morphological approach is the most effective in extracting tumor areas more precisely in the shortest amount of time [9]. A morphological reconstruction is proposed in order to improve the accuracy and reduce the computation time.
The proposed method utilizes image segmentation techniques based on clustering to identify the brain tumor and compute the tumor region. We introduce a new method for segmenting abnormal MRI images, named FCM + MR + AC. In the pre-processing stage, it improves the contrast of the images using histogram matching, and after removing the noise with the median filter, it sharpens the image with unsharp masking. In the next step, it uses the Fuzzy c-means algorithm to segment the brain image. After the clustering stage, in the post-processing stage, it extracts the tumor automatically by combining morphological reconstruction with the active contour. This integrated approach leverages the advantages of both techniques to achieve accurate boundary detection, precise measurements, noise reduction, and time efficiency. It gives an accurate result compared to the Fuzzy c-means algorithm. The original Fuzzy c-means algorithm works well for images without noise. By removing noise from input data in pre-processing, it can improve FCM convergence and create more accurate clusters. As a result, our method gains from combining these algorithms to lower the number of iterations, which influences execution time and provides an accurate result in tumor detection.
The remainder of the paper is organized as follows: Section 2 discusses the related work, Section 3 presents the proposed technique, Section 4 presents experimental research, Section 5 provides the comparative analysis, while Section 6 includes the conclusion and future work.
2 Related work
This section contains relevant research on brain tumor detection using MRI of the brain as well as analysis of previous algorithms used for brain tumor segmentation using MRI of the brain. The technique of grouping similar regions or segments of an image is known as image segmentation. Image segmentation is commonly used to find objects and boundaries in images. More precisely, brain digital image segmentation is the process of assigning a label to every pixel in an image that distinguishes a malignant component from a non-cancerous part. Maksoud et al. [10] proposed a methodology for brain tumor segmentation based on a hybrid-type approach, using the K-means clustering approach in combination with the Fuzzy c-means algorithm. The K-Means algorithm takes less time to compute than the Fuzzy c-means technique but the latter can predict tumor cells faster and more accurately. Melegy et al. [11] used the fuzzy technique to segment brain tumors. This method was developed using the FCM methodology. The suggested PIGFCM algorithm divides the brain MRI volume into basic tissues. The main tissues include the following parts: cerebrospinal fluid (CSF), white matter (WM), gray matter (GM), and background. Previous knowledge-based information improved the segmentation accuracy of brain tissues and accelerated the algorithm to the correct result. This approach dramatically improved segmentation accuracy, noise resiliency, and reaction time. Makropoulos et al. [12], suggested an algorithm for precise segmentation based on the intensity of the developing brains of infants that simulates the intensities throughout the brain. This method was exact and reliable throughout a wide variety of fetal ages. Hamamci et al. [13], proposed a seeded tumor segmentation approach based on cellular automata. The cellular automata technique has been presented to find the smallest path in graph theory. Validations were conducted using both clinical and artificial datasets. The tumor-cut method was used to further divide tumor tissue into necrotic and augmenting sections. The suggested technique highlighted the reduction in the method's sensitivity to initialization, robustness to diverse and heterogeneous tumor categories, and computational performance. Wilson and Dhas [14] used K-means and Fuzzy c-means to recognize the iron in brain SWI. The SWI for brain iron is compared using the K-means and FCM approaches. Experiments performed on Fuzzy c-means showed that the iron areas are more obvious than the result of the K-means method on the image. The main problem with this strategy was that the two procedures are not combined to gain the benefits of both. Sheela and Suganthi [15] introduced Greedy Snake Model and Fuzzy c-means optimization in their research in order to provide an effective automated brain tumor segmentation method. This method first explored the approximate region of interest (ROI) using two-level morphological reconstruction such as dilation and erosion to remove the non-tumor section. In the Greedy Snake approach, a mask was produced and degraded to enhance segmentation precision. The greedy snake model calculates new tumor borders by utilizing the mask border as the snake's primary contour. These limitations were more precise in areas with sharp edges and less accurate in areas with ramp edges. The Fuzzy c-means technique is used to improve the incorrect borders and generate the correct segmentation result. Fuzzy C means clustering, created by Patil et al. [16], firstly uses contrast enhancement in order to enhance the quality of the brain MRI image, and morphological operations are provided for removing the skull. Secondly, the result is entered into the Fuzzy c-means clustering, then a feature extraction approach combining GLRLM and GLCM is presented. The SVM classifier separates tumor and normal brain images based on retrieved features. The suggested approach detects tumor and non-tumor images. C. Narmatha et al. [17] proposed a fuzzy brainstorm optimization approach toward classifying MRI images. This approach combined both fuzzy and brainstorm algorithms. Brainstorm optimization focused on cluster centers and gave them the most priority. This approach might slip into the local optimum, similar to any other swarm approach. The fuzzy executed numerous iterations to offer an ideal network topology, and the brainstorm optimization delivered satisfactory results. In this research, they used BraTS 2018 dataset. Zahra Shahvaran et al. [18] used k-means clustering to detect brain tumors, then the active contour model based on morphological areas was employed for tumor detection, using a primary contour determined according to the border of the observed brain tumor areas. Morphological operators are used sequentially to evolve the contour. Local image intensities are modeled using a Gaussian distribution. BraTS 2013 was used to evaluate this approach. This method had better performance for tumor detection compared to other contour-based techniques.
Sajid et al. [19] proposed a fully automated deep learning method for brain tumor segmentation. They created a hybrid model that combined the effectiveness of a two-stream parallel network with a three-path network. and efficient. This model was designed to take into consideration both contextual and local data in order to address the problem of unbalanced data. To achieve this, a two-phase training process was used. Ma et al. [20] proposed a new method for automated brain tumor segmentation of MR images. In this approach, a multiscale patch-driven active contour model is combined with concatenated and connected random forests.
Lankton et al. [21] suggested a reformulation of the Chan-Vese model and the Yezzi model [22]. Instead of relying on global image information, this approach focused on localized methods for more effective contour localization. A Local Binary Fitting (LBF) model was developed by Li et al. [23] that utilized a kernel function to incorporate local intensity image details. This method involved implicit active contours driven by the energy derived from local binary fitting. A Local Gaussian Distribution Fitting (LGDF) was developed by Wang et al. [24] in order to separate the regions based on mean intensity and variance, the approach used a Gaussian distribution to define the local neighborhood. LGDF was employed in region-based Active Contour Models (ACMs) and was widely used in many fields, including medicine. Ilunga-Mbuyamba et al. [25] presented a localized active contour model with background intensity compensation for automatic MR brain tumor segmentation. The method effectively handled variations in background intensity and foreground objects in medical images. Compared to the traditional localized mean separation active contour model, this approach improved accuracy and reduced computation time.
In this work, we propose a novel approach that combines the power of morphological reconstruction with an active contour algorithm to achieve enhanced object analysis in post-processing. This integrated approach leverages the advantages of both techniques to achieve enhanced object representation, accurate boundary detection, precise measurements, noise reduction, adaptability, and time efficiency. Additionally, Fuzzy c-means clustering has been used to enhance the accuracy of brain tumor segmentation. Median filtering is also applied to remove noise from input data, which helps FCM provide results in a short time. We demonstrate the effectiveness of our approach on various datasets of brain tumor images and show that it outperforms traditional post-processing methods in terms of accuracy, time efficiency, and reliability.
3 Proposed algorithm
The K-means technique is used by several medical image segmentation systems to detect a brain mass tumor [52]. On large datasets, the K-means algorithm runs quickly, however, it suffers from insufficient tumor detection, mainly if the tumor is malignant. In contrast, other methods use the Fuzzy c-means algorithm since it maintains more details from the main image in favor of finding aggressive tumor cells more reliably than the K-means method [53].
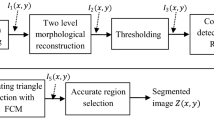
The Fuzzy c-means algorithm is useful in our suggested medical segmentation system. Algorithm 1 shows the pseudo-code for identifying tumors in the brain from MR images. Furthermore, As shown in Fig. 1, our method consists of three stages: 1) pre-processing, 2) clustering, and 3) post-processing.
3.1 Pre-processing stage
This stage is executed by performing several preliminary pre-processing steps on the image before any particular processing. Image pre-processing decreases noise and improves the brain's MR image for further processing. Since brain images are more vulnerable; they should preferably have minimum noise and the highest possible quality. Therefore, this process consists of several steps:
3.1.1 Histogram matching
In image processing, histogram matching is a technique used to change an image so that its histogram matches a pre-determined histogram [51]. To do this, an image with appropriate contrast is selected as a reference, and the histogram of the input image is then matched to it. In the context of detecting tumors in medical images, the reference image used must have enough contrast to clearly identify the tumor area in the input image.
3.1.2 Median filtering
The most often used filtering technique is median filtering, which filters distortion and artifacts without sacrificing key properties such as edge information. Nonlinear median filters are used to effectively remove noise while maintaining edges. The median filter approach shown in Eq. 1 is based on replacing each pixel's gray-level with the median of the pixels in the adjacent neighborhood.
where k is a neighborhood chosen by the user and centered on the image's position [x,y]. The "window" refers to the order of neighbors, and it moves across the entire image pixel by pixel. The median is calculated by first numerically ordering all of the pixel values in the window, then, the desired pixel is replaced with the median pixel.
3.1.3 Unsharp masking
A way to create an image with well-defined edges is through the use of non-sharp filters. This method involves applying a smoothing filter to the main image and then subtracting the smoothed image from the original image. The result is an image with sharp edges, which is then added back to the original image. The specific smoothing filter affects the final result. For example, the result of using the Gaussian filter is better than the box filter, but the result of both amplifies the noise. Another leading challenge is what coefficient of high-frequency information (this image is obtained by reducing the smoothed version of the original image known as a mask) should be added to the original image. This amount is different depending on the images. Another issue is the effect of kernel size in filters such as box and median, which adds more noise to the image by increasing the size. An additional method of sharpening the edges is the direct approach of finding high-frequency information and adding it to the image. Applying the Laplacian operator to the image results in obtaining high-frequency information, and subtracting (adding) it to the image leads to the sharpening of the edges.
3.2 Clustering
One of the most well-known algorithms, FCM, develops an optimal cluster pattern through iteration by minimizing an objective function based on cluster center positions and membership. It enables the division of available sets of points of power (n) by a given number of fuzzy groups. A distinguishing aspect of the technique is the calculation of a fuzzy membership matrix, W = {wik}, made up of the membership grade of the pattern \({x}_{k}\) to each cluster (i). Therefore, points at the cluster's edge with lower membership degrees may be included in the cluster to a smaller extent than points in the cluster's center. Considering a set of clusters (c), FCM separates the info X = {x1, x2, …, xn} into C-fuzzy clusters including centroids that are consistent with V = (v1, v2, …, vc) which minimizes the objective function's value with Eq. 2.
where the parameter m shows the fuzzy exponent and m \(\in\) (1, ∞) and \(\left\|{\boldsymbol x}_{\mathbf k}-{\boldsymbol v}_i\right\|\) is the Euclidean norm between \({\mathbf{x}}_{k}\) and \({\mathbf{v}}_{i}\), \({\mathbf{v}}_{i}\) is the center of the ith cluster, each element of \({w}_{i,k}\), defines the degree to which element, \({\mathbf{x}}_{k}\), belongs to a cluster, \({\mathbf{v}}_{i}\).
Every time the FCM clustering iterates, matrix W is created using Eq. 3, and Eq. 4 computes the related cluster centers. The square error in Eq. 1 is then calculated. The algorithm terminates when either the error or the tolerance value is reached. The performance index's membership grades are controlled by the number m. The partition grows fuzzier as m increases and if m → the degree of membership for each object in each cluster is the same.
Considering that the number of clusters must be determined using the number of white matter, gray matter, cerebrospinal fluid, and tumor areas, the number of areas has been considered four. After calculating the degree of belonging of each pixel, it is related to the cluster that has the highest degree of membership. To find the area related to the tumor, considering that this area has the highest brightness, the average gray levels of each cluster are calculated, and the cluster with the highest value is considered the tumor cluster. The rest of the clusters are attributed to other parts of the brain according to the volume of the samples inside them.
3.3 Morphological reconstruction
Morphological operation is a broad category of image processing procedures that process images depending on their size and shape. A morphological operation adjusts each pixel in an image depending on the values of neighboring pixels. By selecting the local size and shape, a morphology operation sensitive to specific shapes in the input image can be created. The most basic morphological operations are dilation and erosion, which aid in performing various operations in image processing [54]. The morphological operation of the binary image A by the structuring element B is defined in Table 1.
An opening operation is described as an erosion followed by dilation, and a closing operation is an opening performed in reverse using the same structuring element. Morphological reconstruction aims not only to reduce false positives but also to increase true positives. For this purpose, we first remove the small non-tumor areas in the image related to the tumor cluster with the opening operator and then use the closing operator to eliminate the effect of the opening operator on the tumor area. Morphological reconstruction operations require two images which are marker and mask images. The mask image is selected as the initial image. The marker image is obtained by performing morphological opening and closing operations on the candidate regions acquired in the previous step and subtracting them from the initial image. The difference between the reconstructed image and the initial image is considered the tumor regions.
3.4 Fill regions and use the binary image in active contour
In recent research, active contour models (ACM) have been used for image segmentation. Active contour models are based on the surface evolution theory and geometric flows. Snakes are known as active control and are initially made available in 1987 by Kass et al. [26]. Active contour models are used for edge detection, shape modelling, motion tracking, medical image analysis, and especially the segmentation of objects [27, 28].
In the active contour method, the contours of an image are changed based on an energy function derived from the image data to minimize energy and accurately place them on the object's boundary. The energy function is related to a curve, the goal is to find the object's boundary by minimizing the energy function. The active contour models work by evolving the curve to identify objects within the image. The contours change shape according to the features present in the image [29].
In general, active contour models can be divided into two groups: a) edge-based models and b) region-based models. Each of these strategies has advantages and limitations. Region-based active contour models use statistical data of the image in curve evolution but edge-based active contour models use image edge and gradient data. Region-based active contour models have more advantages than edge-based models. One of the most famous region-based models is the ChanVese model [30], which has been used in this study.
4 Experimental results
Implementation of all steps is done using MATLAB 2018. We run our experiments on a PC (Intel Core i7) with 16 GB of RAM and an NVIDIA/(1 GB VRAM) VGA card.
4.1 Data sets
The BraTS 2012 and 2013 dataset is a collection of multimodal brain tumor images from synthetic and real patients. The training and testing sets are different for the BraTS 2012 and 2013 datasets. For the BraTS 2012 dataset, the training set consists of 30 patients with HG and LG and includes the ground truth segmentations. The testing set consists of 15 real patients with HG and LG and does not include the ground truth segmentations. For the BraTS 2013 dataset, the training set consists of 10 real patients and includes the ground truth segmentations. The testing set consists of 10 real patients with HG and LG and does not include the ground truth segmentations. The testing set is used for ranking the algorithms on the BraTS website. The dataset also provides manual segmentations of the tumor regions for each image. The segmentations have four labels: necrosis (1), edema (2), non-enhancing tumor (3), and enhancing tumor (4). The segmentations are used as ground truths for evaluating the performance of brain tumor segmentation algorithms. Figure 2 shows some sample images from the dataset with different modalities and tumor types.
The image patches show tumor structures in different MRI modalities: FLAIR (A), T2 (B), and T1c (C). The segmentations are combined to generate the final labels for the dataset (D): edema (yellow), non-enhancing solid core (red), necrotic/cystic core (green), and enhancing core(blue) [31]
4.2 Evaluation
A valid evaluation method provides quantitative and comparative information necessary for performance evaluation. The criteria of Sensitivity, Dice score, Accuracy, and Precision have been used in the proposed algorithm. These factors are mainly relying on the value of TP, TN, FP, and FN.
Where TP, TN, FP, and FN can be described as follows:
-
(i)
TP is a tumor that is real and has been properly identified.
-
(ii)
TN is a tumor that does not exist and cannot be found.
-
(iii)
FP is a tumor that is not present but is detected.
-
(iv)
FN is a tumor that is present but not found.
$$\mathrm{Dice}=\frac{2\mathrm{TP}}{2\mathrm{TP}+\mathrm{FP}+\mathrm{FN}}$$(5)$$\mathrm{Sensitivity }= \frac{\mathrm{TP}}{\mathrm{TP}+\mathrm{FN}}$$(6)$$\mathrm{Accuracy}= \frac{\mathrm{TP }+\mathrm{ TN}}{\mathrm{ TP }+\mathrm{ FP }+\mathrm{ TN }+\mathrm{ FN}}$$(7)$$\mathrm{Precision}= \frac{\mathrm{TP}}{\mathrm{TP}+\mathrm{FP}}$$(8)$$\mathrm{Hausdroff\, Distance}\left(\mathrm{A},\mathrm{ B}\right)=\mathrm{max}\left\{\mathrm{h}\left(\mathrm{a},\mathrm{ B}\right)\right\}=\mathrm{max}\{\mathrm{min}\{\mathrm{d}(\mathrm{a},\mathrm{ b})\}\}$$(9)$$\mathrm{Jaccard}\left(\mathrm{A},\mathrm{ B}\right)=\frac{\mathrm{TP}}{\mathrm{ TP }+\mathrm{ FP }+\mathrm{ FN}}$$(10)
The Dice score is used to calculate the overlap between manually created ground truth masks and automatically segmented tumors in medical imaging. It is expressed as Eq. (5). Its main application is in the evaluation of segmentation models. Sensitivity is another metric used to predict real positive cases that are predicted as positive. It is also known as recall and can be expressed as Eq. (6). Accuracy is a statistical measure that indicates how well the predicted value matches the actual value. It is used in the method of analyzing the system’s performance. It is expressed as Eq. (7). In addition, the most common method of evaluation is the accuracy of the tumor diagnosis. A high level of accuracy indicates that the classification of MRI images is accurate and therefore, the proposed algorithm is effective. Precision is a measure of how well a model can identify true positive pixels that belong to a cluster compared to all the pixels that the model predicted as positive which is shown in Eq. (8). The results of each technique are recorded in Tables 2 and 3 according to Precision, Sensitivity, Accuracy, and Dice that are mentioned before and represented. The Hausdorff distance can be calculated using Eq. (9), where A and B are two sets of points, h(a, B) is the minimum distance between a and B, and d(a, b) is the Euclidean distance between points a and b. In the context of brain tumor segmentation, the Hausdorff distance metric measures how well the boundary voxels of the segmented regions match between the ground truth and the results of the algorithms used for brain tumor segmentation. Furthermore, we used the Jaccard coefficient. It is a measure of similarity between two sets of data and can be expressed as Eq. (10).
4.3 Results and discussion
The output results of the figure are discussed in this section. The proposed tumor segmentation method is validated by performance parameters such as accuracy, specificity, sensitivity, and dice. In this technique, Image preprocessing reduces noise and increases the image's clarity. FCM clustering is used to initially identify the tumor areas. Also, morphology operators based on reconstruction are used to properly refine the appeared frills in the images. Finally, by applying active contour, the boundaries of the tumor became more precise. The results in Table 2 demonstrate the reliability and accuracy of our method. We discuss the proposed method in the following.
In the pre-processing step, the images used in the study are 16 bits, and no reduction in gray levelshave been made to prevent information loss. Figure 3a shows the image considered as a reference in the histogram matching step. Figure 3c shows the result of histogram matching on the input image. The tumor region is completely uniform and includes all the tumor regions that are shown in the ground truth. In the next step, Fig. 3d and e illustrate the noise removal and contrast improvement utilized for the image. MRI is often affected by noise and artifacts that need to be removed before the image can be processed to determine whether or not it has a tumor. The median filter is the preferred tool for filtering noise in MRI brain images [32]. Unlike other filters, this one doesn't blur or smooth out the image's edges, maintaining the image's sharpness.
In the clustering step, the FCM clustering algorithm assigns membership to each data depending on its distance from the cluster's center. If there is less distance between the cluster's center and the data point, the data point is more likely to be a member of that cluster. The number of clusters must be determined using the number of white matter, gray matter, cerebrospinal fluid, and tumor areas; therefore, the number of clusters has been considered 4. For better understanding, in Fig. 4d, the pixels whose degree of membership is less than 0.6, which may belong to several categories, are shown in yellow. These pixels represent the regions where the segmentation is uncertain, and where different tumor structures may overlap or coexist with other brain tissues, such as white matter, gray matter, and cerebrospinal fluid. In Fig. 5, the clusters are determined according to the average gray levels and the number of pixels in each category. The order of the clusters based on the average of pixels gray level follows this sequence: tumor, white matter, gray matter, and cerebrospinal fluid, which means that the brightest cluster is considered a tumor. The tumor cluster has a much higher gray level than the other clusters, indicating its high intensity and contrast in the MRI images. Figure 5e also shows that the tumor cluster has a lower pixel count than the other clusters, indicating its small size and shape.
According to Fig. 6, the tumor cluster includes non-tumor areas in addition to the tumor area. To partially remove the above-mentioned areas, firstly, the opening operator is applied to the image. Then, the closing operator is applied to the image to cause the remaining areas to completely cover the tumor.
Figure 7 illustrates the steps of morphological reconstruction and the difference between the reconstructed image and the original image is considered a tumor area. After morphological reconstruction, the issue is that the border of the tumor areas is not accurate enough. As shown in Fig. 7d, the boundaries of the tumor are still not exact. To improve the tumor boundaries, after filling the empty areas, the boundaries of the tumor are preceded by active contour. As the output of this step is shown in Fig. 7e, the boundaries obtained by the active contour method have been improved. In the following, the output of the proposed approach is shown in detail in Fig. 8.
The proposed technique was used to produce the experimental results for the segmented regions for the three classes of WM, GM, and CSF, and for the extracted tumor region are given in Fig. 8. The figure shows the different steps and outputs of the proposed technique applied to a brain MR image. Figure 8a shows the input image, and Fig. 8b shows the ground truth segmentation. Figure 8c shows the result of removing noise from the input image using a median filter. Figure 8d shows the result of enhancing the edges and details of the image using unsharp masking. Figure 8e shows the result of computing the degree of membership for each pixel using a Fuzzy c-means. Figure 8f shows the result of clustering the pixels into four groups using the Fuzzy c-means algorithm. Figure 8g, h, and i show the result of extracting the WM, GM, and CSF regions from the clustered image, respectively. Figure 8j shows the result of extracting the tumor region from the clustered image using a morphological reconstruction and active contour. The figure demonstrates that the proposed technique can effectively segment and extract brain tissues and tumors from a noisy and low-contrast image. In addition, the other images from BraTS 2013 that are shown in Figs. 9, 10, and 11 are added.
4.4 Computation time
As shown in Table 2, it is important to notice that the FCM + MR + AC technique has the lowest computation time compared to previous methods for performing brain tumor segmentation. This method achieves more promising results and reduces the running time by using a combination of fuzzy c-means, morphological reconstruction, median filtering, and active contour. The proposed method exhibits a considerable reduction in computation time as compared to the LACM-BIC solution, with a speed advantage of approximately six times.
The median filter for pre-processing is a tool of choice for filtering noise in MRI brain images. By removing noise from the input data, median filtering can help FCM converge faster and produce more accurate clusters. The reason behind this significant difference can be attributed to the iterative nature of the Fuzzy c-means solution which leads to computationally expensive processing whereas by using fewer clusters and a quicker termination condition, it can speed up the image processing for brain tumor segmentation. Conversely, the proposed method employs a non-iterative and quick-to-compute morphological step, resulting in reduced computational overhead. These findings also hold true for brain tumor images of varying types. Using region-based active contours, also known as Chan-Vese models, can reduce computation time by exploiting global information about the image.
4.5 Experimental results
Table 3 shows the results for complete tumor detection on the BraTS 2012 dataset. 30 images from the training dataset are utilized to test the suggested approach. The proposed method is compared with SVM and ERT methods [39]. Results present the evaluation measures for SVM, ERT, and FCM + MR + AC, respectively. It is clear that FCM + MR + AC results in a slightly improved performance, compared to that of SVM and ERT, with an overall classification Precision of 89.09%, Sensitivity of 88.09%, and Dice 0.88% for ERT, Accuracy of 83.79, the Sensitivity of 82.72 and Dice 0.83% for SVM and 92.05%, 92.10% and 0.92 for FCM + MR + AC, respectively. Furthermore, we use the Jaccard coefficient and Hausdorff distance that averages are 0.86 and 3.60 respectively. The dice score of ERT, SVM classifiers, and FCM + MR + AC for the BraTS dataset is presented in Table 4. It can be seen that result for FCM + MR + AC is more accurate than that of SVM-based and ERT-based. In addition, the proposed algorithm is evaluated on BraTS 2013 and achieves excellent results in terms of commonly used metrics such as Sensitivity, Accuracy, Dice score, Jaccard, and Hausdorff distance with values of 93.15%, 99.56%, 0.93, 0.87, and 3.21, respectively.
4.6 Comparative analysis
Regarding all the evaluations in Tables 5 and 6, it can be seen that the performance of the FCM + MR + AC method is superior to other algorithms, and a similar value is obtained only in Reza's method [40].
In Fig. 12, it can be seen that all evaluation parameters of our method are much higher than that of SVM-based and ERT-based. Figures 13 and 14 show the bar graph comparing different Dice methods.
We emphasize the significance of the pre-processing stage in our study, which is MRI histogram matching. This is especially crucial when using the BraTS dataset, which contains data from multiple scanners and multicenter [39]. With the purpose of brain tumor segmentation and the regionalization of brain tumor segments in this study, FCM fuzzy clustering has been used to identify and extract the tumor region. Morphology reconstruction has also been used to reduce the areas that are wrongly placed in the tumor cluster, which resulted in a positive effect. In the last step, the use of active contour has resulted in obtaining better boundaries for the tumor area.
5 Conclusion and future work
This study proposes a novel algorithm for the automatic detection and segmentation of brain tumors from MRI images. The algorithm combines Fuzzy c-means clustering and morphological reconstruction to achieve fast and accurate tumor segmentation. The Fuzzy c-means clustering is used to partition the image into clusters based on the intensity values, and the morphological reconstruction is used to refine the tumor cluster by removing the non-tumor regions. The algorithm is applied to the BraTS 2012 and BraTS 2013 data sets, which contains multimodal MRI images of brain tumors with different grades and types. The proposed algorithm is effective and robust, as demonstrated by its excellent performance in terms of commonly used metrics such as Sensitivity, Accuracy, Dice score, Jaccard, and Hausdorff distance for BraTS 2012, 92.10%, 99.06%, 0.92, 0.86, and 3.60 and BraTS 2013, 93.15%, 99.56%, 0.93, 0.87, and 3.21. The diagnosis can be improved by using the proposed algorithm which provides valuable information and treatment of brain tumors, which are among the most serious and challenging diseases.
In future work, we plan to use U-net [50], a state-of-the-art deep learning network for brain tumor segmentation that can learn complex features from MRI images and perform end-to-end segmentation with high accuracy and robustness. We will compare the performance of U-net with our proposed method and investigate how to improve it further with data augmentation, transfer learning, or self-supervised learning techniques.
Data availability
The datasets analysed during the current study are available in the BraTS repository, https://www.smir.ch/BRATS/Start2012, https://www.smir.ch/BRATS/Start2013
References
Janani V, Meena P (2013) Image segmentation for tumor detection using fuzzy inference system. Int J Comput Sci Mobile Comput (IJCSMC) 2(5):244–248
Hu K, Gan Q, Zhang Y, Deng S, Xiao F, Huang W, ... Gao X (2019) Brain tumor segmentation using multi-cascaded convolutional neural networks and conditional random field. IEEE Access 7:92615–92629
Kabade RS, Gaikwad MS (2013) Segmentation of brain tumour and its area calculation in brain MR images using K-mean clustering and fuzzy C-mean algorithm. Int J Comput Sci Eng Technol 4(5):524–531
Bauer S, Wiest R, Nolte LP, Reyes M (2013) A survey of MRI-based medical image analysis for brain tumor studies. Phys Med Biol 58(13):R97
Aslam HA, Ramashri T, Ahsan MIA (2013) A new approach to image segmentation for brain tumor detection using pillar K-means algorithm. Int J Adv Res Comput Commun Eng 2(3):1429–1436
Naik D, Shah P (2014) A review on image segmentation clustering algorithms. Int J Comput Sci Inform Technol 5(3):3289–3293
Kaur J, Agrawal S, Vig R (2012) Integration of clustering, optimization and partial differential equation method for improved image segmentation. Int J Image, Graphics Sig Process 4(11):26
Haralick RM, Sternberg SR, Zhuang X (1987) Image analysis using mathematical morphology. IEEE Trans Pattern Anal Mach Intell 4:532–550
Yazdani S, Yusof R, Karimian A, Riazi AH, Bennamoun M (2015) A unified framework for brain segmentation in mr images. Comput Mathematic Methods Med 2015:829893. https://doi.org/10.1155/2015/829893
Abdel-Maksoud E, Elmogy M, Al-Awadi R (2015) Brain tumor segmentation based on a hybrid clustering technique. Egypt Inf J 16(1):71–81
El-Melegy MT, Mokhtar HM (2014) Tumor segmentation in brain MRI using a fuzzy approach with class center priors. EURASIP J Image Video Process 2014(1):1–14
Makropoulos A, Gousias IS, Ledig C, Aljabar P, Serag A, Hajnal JV, ... Rueckert D (2014) Automatic whole brain MRI segmentation of the developing neonatal brain. IEEE Trans Med Imag 33(9):1818–1831
Hamamci A, Kucuk N, Karaman K, Engin K, Unal G (2011) Tumor-cut: segmentation of brain tumors on contrast enhanced MR images for radiosurgery applications. IEEE Trans Med Imaging 31(3):790–804
Wilson B, Dhas JPM (2014) An experimental analysis of Fuzzy C-means and K-means segmentation algorithm for iron detection in brain SWI using Matlab. Int J Comput Appl 104(15):36
Sheela CJJ, Suganthi G (2022) Automatic brain tumor segmentation from MRI using greedy snake model and fuzzy C-means optimization. J King Saud Univ-Comput Inf Sci 34(3):557–566
Patil PG, Karande KJ, Surwase SV (2022) Detection of brain tumor using optimized fuzzy C-means and SVM classifier. In: AIP Conference Proceedings, vol. 2494, no. 1). AIP Publishing
Narmatha C, Eljack SM, Tuka AARM, Manimurugan S, Mustafa M (2020) A hybrid fuzzy brain-storm optimization algorithm for the classification of brain tumor MRI images. J Ambient Intell Humanized Comput. https://doi.org/10.1007/s12652-020-02470-5
Shahvaran Z, Kazemi K, Fouladivanda M, Helfroush MS, Godefroy O, Aarabi A (2021) Morphological active contour model for automatic brain tumor extraction from multimodal magnetic resonance images. J Neurosci Methods 362:109296
Sajid S, Hussain S, Sarwar A (2019) Brain tumor detection and segmentation in MR images using deep learning. Arab J Sci Eng 44:9249–9261
Ma C, Luo G, Wang K (2018) Concatenated and connected random forests with multiscale patch driven active contour model for automated brain tumor segmentation of MR images. IEEE Trans Med Imaging 37(8):1943–1954
Lankton S, Tannenbaum A (2008) Localizing region-based active contours. IEEE Trans Image Process 17(11):2029–2039
Yezzi A, Jr, Tsai A, Willsky A (2002) A fully global approach to image segmentation via coupled curve evolution equations. J Visual CommunImage Represent 13(1–2):195–216
Li C, Kao C-Y, Gore JC, Ding Z (2007) Implicit active contours driven by local binary fitting energy. In: 2007 IEEE Conference on computer vision and pattern recognition. IEEE, pp 1–7. https://doi.org/10.1109/CVPR.2007.383014
Wang L, He L, Mishra A, Li C (2009) Active contours driven by local Gaussian distribution fitting energy. Signal Process 89(12):2435–2447
Ilunga-Mbuyamba E, Avina-Cervantes JG, Garcia-Perez A, de Jesus Romero-Troncoso R, Aguirre-Ramos H, Cruz-Aceves I, Chalopin C (2017) Localized active contour model with background intensity compensation applied on automatic MR brain tumor segmentation. Neurocomputing 220:84–97
Kass M, Witkin A, Terzopoulos D (1988) Snakes: Active contour models. Int J Comput Vision 1(4):321–331
McInerney T, Terzopoulos D (1995) A dynamic finite element surface model for segmentation and tracking in multidimensional medical images with application to cardiac 4D image analysis. Comput Med Imaging Graph 19(1):69–83
Xu C, Pham DL, Rettmann ME, Yu DN, Prince JL (1999) Reconstruction of the human cerebral cortex from magnetic resonance images. IEEE Trans Med Imaging 18(6):467–480
Cheng J, Liu Y, Jia R, Guo W (2007) A new active contour model for medical image analysis-wavelet vector flow. IAENG Int J Appl Math 36(2):2–6
Zhang K, Song H, Zhang L (2010) Active contours driven by local image fitting energy. Pattern Recogn 43(4):1199–1206
Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, ... Van Leemput K (2014) The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans Med Imag 34(10):1993–2024
Dandıl E, Çakıroğlu M, Ekşi Z (2015) Computer-aided diagnosis of malign and benign brain tumors on MR images. In: Bogdanova A, Gjorgjevikj D (eds) ICT innovations 2014: World of data (Advances in intelligent systems and computing, vol 311). Springer. https://doi.org/10.1007/978-3-319-09879-1_16
Li C, Kao C-Y, Gore JC, Ding Z (2007) Implicit active contours driven by local binary fitting energy. In: 2007 IEEE Conference on computer vision and pattern recognition. IEEE, pp 1–7. https://doi.org/10.1109/CVPR.2007.383014
Hu K, Gan Q, Zhang Y, Deng S, Xiao F, Huang W, ...,Gao X (2019) Brain tumor segmentation using multi-cascaded convolutional neural networks and conditional random field. IEEE Access 7:92615–92629
Chen G, Li Q, Shi F, Rekik I, Pan Z (2020) RFDCR: Automated brain lesion segmentation using cascaded random forests with dense conditional random fields. Neuroimage 211:116620
Zhang D, Huang G, Zhang Q, Han J, Han J, Yu Y (2021) Cross-modality deep feature learning for brain tumor segmentation. Pattern Recogn 110:107562
Zhang D, Huang G, Zhang Q, Han J, Han J, Wang Y, Yu Y (2020) Exploring task structure for brain tumor segmentation from multi-modality MR images. IEEE Trans Image Process 29:9032–9043
Zhou C, Ding C, Wang X, Lu Z, Tao D (2020) One-pass multi-task networks with cross-task guided attention for brain tumor segmentation. IEEE Trans Image Process 29:4516–4529
Soltaninejad M, Yang G, Lambrou T, Allinson N, Jones TL, Barrick TR, ... Ye X (2017) Automated brain tumour detection and segmentation using superpixel-based extremely randomized trees in FLAIR MRI. Int J Comput Assist Radiol Surg 12:183–203
Reza S, Iftekharuddin KM (2013) Multi-class abnormal brain tissue segmentation using texture features. In: Proceedings of NCI-MICCAI BRATS, pp 38–42
Baid U, Talbar S, Talbar S (2016) Comparative study of K-means, Gaussian mixture model, fuzzy C-means algorithms for brain tumor segmentation. In: Proceedings of the international conference on communication and signal processing 2016 (ICCASP 2016) . Atlantis Press, pp 583–588. https://doi.org/10.2991/iccasp-16.2017.85
Tustison N, Wintermark M, Durst C, Avants B (2013) ANTs and arboles. In: Proceedings of NCI-MICCAI BRATS, pp 47–50
Pereira S, Pinto A, Alves V, Silva CA (2016) Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging 35(5):1240–1251
Kwon D, Shinohara RT, Akbari H, Davatzikos C (2014) Combining generative models for multifocal glioma segmentation and registration. Med Image Comput Comput-Assist Interven 17(Pt 1):763–770. https://doi.org/10.1007/978-3-319-10404-1_95
Havaei M, Davy A, Warde-Farley D, Biard A, Courville A, Bengio Y, ... Larochelle H (2017) Brain tumor segmentation with deep neural networks. Med Image Anal 35:8–31
Urban G, Bendszus M, Hamprecht F, Kleesiek J (2014) Multi-modal brain tumor segmentation using deep convolutional neural networks. In: MICCAI BraTS (Brain Tumor Segmentation) Challenge, pp 31–35. https://doi.org/10.1007/978-3-319-12057-3_4
Havaei M, Larochelle H, Poulin P, Jodoin PM (2016) Within-brain classification for brain tumor segmentation. Int J Comput Assist Radiol Surg 11:777–788
Dvořák P, Menze B (2016) Local structure prediction with convolutional neural networks for multimodal brain tumor segmentation. In: Menze B et al (eds) Medical computer vision: Algorithms for big data (Lecture Notes in Computer Science, vol 9601). Springer, pp 77–84. https://doi.org/10.1007/978-3-319-42016-5_6
Zikic D, Ioannou Y, Brown M, Criminisi A (2014) Segmentation of brain tumor tissues with convolutional neural networks. Proc MICCAI-BRATS 36(2014):36-39
Ronneberger O, Fischer P, Brox T (2015) U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N, Hornegger J, Wells W, Frangi A (eds) Medical image computing and computer-assisted intervention – MICCAI 2015, vol 9351). Lecture Notes in Computer Science. Springer, pp 234–241. https://doi.org/10.1007/978-3-319-24574-4_28
Gonzalez RC, Woods RE (2008) Digital image processing, 3rd edn. Prentice Hall, Upper Saddle River
Mohan P, AL V DB, Kavitha BC (2013) Intelligent based brain tumor detection using ACO. Int J Innovat Res Comput Commun Eng 1(9):2143–2150
Anandgaonkar G, Sable G (2014) Brain tumor detection and identification from T1 post contrast MR images using cluster based segmentation. Int J Sci Res 3(4):814–817
Yousuf M, Khan KB, Azam MA, Aqeel M (2020) Brain tumor localization and segmentation based on Pixel-Based thresholding with morphological operation. Intelligent technologies and applications (Communications in Computer and Information Science, vol. 1198). Springer Singapore, pp 562–572. https://doi.org/10.1007/978-981-15-5232-8_48
Ranjbarzadeh R, Bagherian Kasgari A, Jafarzadeh Ghoushchi S, Anari S, Naseri M, Bendechache M (2021) Brain tumor segmentation based on deep learning and an attention mechanism using MRI multi-modalities brain images. Sci Rep 11(1):1–17
Davy A et al (2014) Brain tumor segmentation with deep neural networks. In: MICCAI Multimodal Brain Tumor Segmentation Challenge (BraTS), pp 1–5
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors whose names are listed immediately below certify that they have NO affiliations with or involvement in any organization or entity with any financial interest (such as honoraria; educational grants; participation in speakers' bureaus; membership, employment, consultancies, stock ownership, or other equity interest; and expert testimony or patent-licensing arrangements), or non-financial interest (such as personal or professional relationships, affiliations, knowledge or beliefs) in the subject matter or materials discussed in this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shekari, M., Rostamian, M. Brain tumor segmentation from MRI using FCM clustering, morphological reconstruction, and active contour. Multimed Tools Appl 83, 42973–42998 (2024). https://doi.org/10.1007/s11042-023-17233-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-17233-5