Abstract
An automated plant biometric system is now an important step in preserving nature’s biodiversity. This paper presents a novel Relative Sub-image Sparse Coefficient (RSSC) algorithm for mobile devices (MDs) representing plant leaves into a mathematically compact vector for its classification. The RSSC feature vector includes local Statistical Entropy Texture (SET) information inter-related to all the sub-images within a leaf. RSSC space is merged with Gray Level Co-occurrence Matrix (GLCM) feature to refine the outputs using best-Nearest Neighbor (best-NN), designed for MDs. The experiments were performed on three different types of leaf datasets: (i) Flavia, (ii) ICL and (iii) Diseased leaf datasets. The results proves our method more accurate and better compared to other existing plant identification systems. The proposed approach is also tolerant under shape distortion caused while capturing. The mobile machine learning system for leaf image informatics is deployed on Android devices which helps botanists, agriculturists and medical biologists to recognize ubiquitously the herbs and plant species anywhere-anytime.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
The growth of mobiles and ubiquitous technologies are serving in many application domains allowing information accessibility anywhere-anytime. The users of smartphones / phablets / tablets are increasing day-by-day and more than the population, as predicted by Cisco [6]. In mobile agriculture informatics, m-agriculture includes agricultural statistics, monitoring, commerce and botanical research which can aid farmers and botanists in their daily routines. As we know, the manual experimenting and monitoring the biodiversity is a tedious job involving human experts and is a time consuming practice - for both experts and non-experts. Therefore, an automated plant biometric system heightens such traditional identification process.
A human vision system (HVS) recognizes the multitude of an object in a real and digital world even with diversified changes in the viewpoints. Whereas, the artificial or computer vision (CV) fails to analyze them as accurately and timely as HVS. Thus, leaving several challenges for CV researchers including leveraging human intelligence, object recognition, invariant to orientation and occlusion.
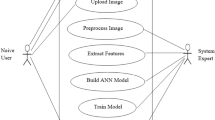
With increasing global warming, it’s important to recognize and preserve the rare and the new hybrid plant species for better nature understanding by just looking at their leaves, fruits, flowers, stems, and roots. However, leaf information is sufficiently rich and is available throughout the year for species identification based on its shape, color, and/or texture [9]. In the past few years, various machine learning algorithms were proposed and used in field of species identification [9] such as LeafSnap [19], LeafView [24] and ApLeaf [43]. In general, an automated plant leaf image informatics involves: (i) leaf acquisition, (ii) pre-processing, (iii) feature extraction and (iv) leaf learning/matching, as listed in Fig. 1. In this paper, we are mainly focusing on novel feature extraction from plant leaves.
As we know, a colored leaf image \(I_{leaf}^{3}\), with resolution (x×y), is a 3D matrix having 0 to 255 intensity values; representing three different colors: red, green and blue. Similarly, a sub-image is a 3D matrix selected using some pattern or relation from \(I^{3}_{leaf}\), as shown in Fig. 2. These sub-images may be called as relative sub-images if they share some common information. In an ongoing research AgroMobile [30] and Agriculture-as-a-Service (AaaS) [34], proposes a cloud based mobile assistant for farmers (or for any non-experts) to monitor their crops in real-time by just using their MDs as their third eye. AaaS framework may also be used by the experts like botanists in their research to find the new species and explore the huge biodiversity of 4,20,000 species (reported by Govaerts [14]) within few seconds.
In this paper, three basic contributions are highlighted. Firstly, a novel local statistical entropy-based texture (SET) leaf matrix for mobile plant leaf informatics is proposed. Secondly, the plant leaves are uniquely represented using the proposed derivational relative sub-image sparse coefficient (RSSC) from SET matrix for classification of plant species, efficiently and robustly. It makes feature space more rich and compact, suitable for MDs and has high accuracy rate compared to state-of-the-art (see Section 5). Lastly, we have also designed an Android based mobile client-server architecture for plant leaf analysis accessing information related to it, anytime-anywhere.
Rest of the paper is organized as follows: Section 2 presents a brief literature background on the existing systems, Section 3 proposes a new RSSC sparse transform for plant image informatics, Section 4 shows the architecture for Mobile Vision (MV) system to retrieve plant species, and in Section 5 the experimental results and discussions are covered. Lastly, the paper concludes with few directions on future scope in Section 6.
2 Background and existing systems
In past, several machine learning methods have been proposed for identification of plant species using their fruits, flowers or leaves. Majority of these methods are plant leaf features based such as leaf shape, leaf margin, leaf vein and/or leaf texture [9]. These classification are briefly described in Table 1.
Warren [38], in 1997, proposed an automated system to recognize plant genus Chrysanthemum with 30 species. In this, the three basic mathematical descriptors such as shape, color and size of leaf, flower and petal of a species were used for identification. White [24, 39, 40] used morphological descriptor for shape information extraction from a complete leaf image. They designed a MV Tablet-PC-based an electronic field guide to identify plant leaves. Since LeafSnap [19] is designed for iOS, Zhao et al. [43], in 2015, proposed ApLeaf – an Android based species identification.
On the other side, Mobile Computing (MC) can aid anytime-anywhere accessing agricultural related information such as production, monitoring, banking policy, m-agriculture commerce, and so on [30, 34]. As a third eye to farmers, Mobile Cloud Computing (MC2) automatically monitors the crop field by remote experts. MC2 also manages the energy consumption of MD by supporting an off-line accessibility of plant and pathological information. Kim et al. [18] proposed a self-growing agriculture knowledge using cloud computing (CC) services assisting farmers to make smart decisions.
In plant identification, Bai et al. [4] used graph transductive learning approach on Swedish leaf dataset and achieved an accuracy of 93.8 % in 2010. Whereas, in 2011-12, ImageCLEFs plant identification task [12, 13] provided a testbed for plant leaf based automated plant species identification. Further, in 2013, Mouine et al. [25] proposed a multi-scale triangular shape representation method for plant species recognition. This triangle side lengths (TSL) gives an accuracy of 95.73 % on Swedish leaf dataset. While in the latest literature, Wang et al. [37] proposed MARCH description for mobile retrieval of leaf images.
There must be a generic low-cost automated plant leaf informatics system in every ones pocket which is easy to operate by anyone-anywhere-anytime. Hence, this paper aims to increase the reliability and accuracy of classification by reducing the complexity such that it is suitable for MDs. This proposed MV system, aims to various groups working with and for agricultural sectors such as botanists, agronomists, gardeners, foresters, land managers and farmers.
3 Proposed methodology
To automate the process of plant leaf identification in real-time, accurately and efficiently, we proposed SET texture based RSSC algorithm. The system is designed as a combination of several modules, viz image acquisition followed by extraction of RSSC features with a detailed discussion on the algorithm properties, matching and computational complexity in further sections. The complete flow graph of the approach is shown in Fig. 3 for an overview.
3.1 Image acquisition and pre-processing
In pre-processing three basic steps are involved: (i) transformation of device dependent RGB color space to device independent L*a*b* color space, (ii) noise removal and (iii) extraction of region of interest (ROI) with minimal computational power and memory consumption.
While capturing leaf image MD uses a device dependent RGB sensors and therefore for same leaf samples the color may vary from one device to another device. Hence, a device independent color space transform such as RGB to CIE L*a*b* color space is used [32], (1). In all cases, the color space volume is same.
Then an average filter is applied to remove the unwanted non-uniform pixel distribution of size (5×5) by using (2).
Now, in order to have translation and scale invariant leaf features, \(I_{leaf_{\eta }}^{3}\) is cropped and rescaled to a standard resolution (w×h), by using (3) and (4). It helps to distinctively identify and focus on ROI [32].
where, (x ′×y ′) is the translation image size. For example, as in Fig. 4a, the leaf sample of resolution (1600×1200) is cropped and scaled to (w×h)=(460×460) and Fig. 4b, to focus on ROI.
3.2 Relative sub-image sparse coefficient (RSSC)
RSSC features are extracted from a series of 3D sub-image matrices in such a way that each sub-image within the image has a relation with the other adjacent sub-images. The colored leaf, \(I_{leaf}^{3} (w,h)\) is divided into (s 1×s 2) grid matrix each of size ((w/s 1 + c)×(h/s 2 + c)), lets say it as (x ″×y ″). Here, s i denotes the grid size and c is the number of common overlapping pixels between the two sub-images. Note: c is a whole number and s i is a natural number, i.e. \(c \in \mathbb {R}\) and \(s_{i} \in \mathbb {N}\).
s i and c highly influence the computation cost because if they are increased the grid matrix tends to behave like \(I_{leaf}^{3}\) and therefore increases the computation cost. Here, in this paper, we have considered a square grid matrix, that is s 1 = s 2 = s to study the behavioral analysis of c in species classification.
On the other hand, the criteria for selecting s i depends on the ratio of w and h to s i which must be a natural number \(\mathbb {N}\). Depending upon s i , in total for a colored image, there will be (s 2×3) sub-images and in case of gray image (L*) its only s 2, (5).
To clearly visualize and represent the local leaf patterns in \(I_{leaf}^{3} (w,h)\), each and every sub-image, \(I_{leaf_{k_{l}}}^{2} (x^{\prime \prime },y^{\prime \prime })\), denoted as nodes in Fig. 5, are inter-related to adjacent nodes by minimum of c 2 pixels where k={L∗,a∗,b∗} and l={1,…s 2}. If c is non-zero, x ″ = y ″ and s=5 then the connected weighted graph is as shown in Fig. 5.
\(I_{leaf_{l}}^{3}\) is then further processed to extract the local statistical entropy texture (SET) information of every sub-image. Entropy is a key feature of a matrix used to uniquely identify it. Whereas SET is a quantitative measure of randomness of intensities within the sub-image locally. So it is the suitable parameter to measure irregular leaf patterns with minimum energy consumption and is computed using (6).
where, \(p_{k_{l}} (L)\) is the probability of l th sub-image in k th layer, defined by (7).
Here, \(ngray_{k_{l}} (L)\) is the number of pixels in \(I_{leaf_{k_{l}}}^{2}\) with maximum intensity L for total number n=(x ″×y ″) pixels. Note that, the sum of \(p_{k_{l}} (i)\) is always a unity, where 0<i<L and \(ngray_{k_{l}}\) is calculated using (8).
The SET, \(epy_{k_{l}}\) defines the information present in l th sub-image of k th layer by analyzing its gray level distribution. The higher the entropy, the lower the information, see Fig. 6b.
A series of these local SET characterizes the leaf and independently used to represent them. But in majority of times it happens that the other leaf species have few SET elements similar and so may misclassify. Therefore, to separate such SETs, a derivative relative sub-image sparse coefficients are computed. A simple relative approach given by (9), relates \(epy_{k_{i}}\) with \(epy_{k_{j}}\), such that the min-max ranges between −1 to +1.
where, α is the total number of RSSC features in a single color space, i,j∈l. Note that i is paired with every other j and i≠j. The only condition is that the denominators must not be zero, i.e. \(|~epy_{k_{i}}+epy_{k_{j}} ~|\neq 0\), in any situation. The total \(RSSC_{k_{\alpha }}\) features of a leaf for a single color layer is \(C_{2}^{s^{2}}\), a combination formula, which is again multiplied by 3 for colored space. The majority of adjacent sub-images may have similar pixel intensity and the entropy texture possibly be same so (9) results in many zeros. This vector resembles a sparse vector and thus, the coefficients are named as sparse coefficients.
RSSC presents a local block-by-block SET feature vector representation of an image. The SET graph comparison of colored layers with gray leaf is shown in Fig. 7. It is seen that even grayscale vector is sufficient to represent leaf in feature space. But along with this RSSC, to refine the classification, there must be a global texture information too and so GLCM is fused to complete the vector.
3.3 Gray level co-occurrence matrix (GLCM)
In this subsection, \(I_{leaf}^{3} (w,h)\) is processed to extract global GLCM texture features [15]. GLCM is a well-known second order derivative texture matrix [15]. It considers the reference pixel and the neighbor pixel relationship which is a user defined entity. GLCM offset is the reference window length used to find relationship between the pixels within that window. As we know GLCM is not rotation invariant and so in this paper a circular GLCM [5] is used.
The circular GLCM is applied on \(I_{leaf_{gray}}^{2} (w,h)\) to get a global leaf texture, \(GLCM_{leaf}^{2} (i,j)\) by using (10).
where, 0≤i,j≤(L−1), and (δ x,δ y) is the offset, i.e., {[0 δ],[−δ δ],[−δ 0],[−δ −δ]}, as shown in Fig. 8. In L*a*b* color space, the L* component shows the intensity of an image and independently it acts as a gray image. Therefore, \(I_{leaf_{gray}}^{2}\) is the L* component of L*a*b* space (11).
The four basic statistical properties of \(GLCM_{leaf}^{2}\) matrix: energy or uniformity E, contrast C o n, homogeneity H, and correlation C r n, are calculated using Equations, as shown in Table 2.
In Table 2, (μ i ,μ j ) and (σ i ,σ j ) are the means and the standard deviations of the marginal probability of \(I_{leaf_{gray}}^{2} (w,h)\), respectively. This is calculated for all four angles (0∘,45∘,90∘,135∘). Therefore, GLCM feature vector is a set of all 16 features computed at four different orientations, i.e., G L C M={E a ,C o n a ,H a ,C r n a } such that a={0∘,45∘,90∘,135∘}.
RSSC and GLCM are fused to form a single vector, F l e a f ={G L C M + R S S C}. Note that, the GLCM features are normalized to a range [−1+1] before F l e a f . Thus, the feature length of F l e a f is \(((4\times 4)+(C_{2}^{s^{2}} \times 3)) = (16 + (C_{2}^{s^{2}} \times 3)) = N\), which is a high dimensional feature vector and requires N dimension feature space to represent it. In such a high dimension space, feature vector faces the curse of dimensionality where it has a high chance to mis-classify a leaf with the other species. This vector, \(F_{leaf} \in \mathbb {R}^{N}\) (12) is a type of sparse vector and so it may be possible to reduce the feature space dimension by reducing the zeros by using Multi Linear Discriminant (MLD).
Here, m is the total number of samples from each C class, having N attributes with zeros and non-zeros.
3.4 Multi linear discriminant (MLD)
The MLD is the commonly used dimension reduction method in pattern recognition [10]. It is a generalized form of Fishers LD for multiple classes which is used to extract the linear combination of features such that it clearly separates the objects. Unlike principal component analysis (PCA), MLD explicitly models the difference between the classes to clearly separate them in feature space. Basically, PCA seek directions that are efficient for presentation of the classes while MLD seeks for efficient discriminations of classes. MLD maximizes the ratio of between-class variance \(\delta _{between}^{2}\) to within-class variance \(\delta _{within}^{2}\) so that the adequate class separability is obtained. The separation between the classes is
where, \(\delta _{between}^{\mathbf {C}}\) and \(\delta _{within}^{\mathbf {C}}\) are defined by (14) and (15), respectively.
Here, m ′ is the number of samples per class, \(\mu _{i}^{\prime }\) is the mean of i th leaf sample, μ ′ is mean of complete leaf data and \(\delta _{i}^{\mathbf {C}}\) is the class dependent scatter variance matrix, given by (16):
Since F l e a f is a sparse matrix, the computation cost required to compute MLD basis vector to represent leaf will be reduced. The Fishers basis vector is calculated using eigenvalue (λ) and eigenvector (ω), that is
The high dimensional RSSC feature vector can be now projected to a low dimensional Fishers hyperplain by using (18):
That is, the F M L D will be minimized to a vector with optimal hyperplane having minimum zeros, similar to (19).
The maximum length of Fishers hyperspace to separate plant species is (C−1). This is because the maximum rank of \(\delta _{between}^{\mathbf {C}}\) is (C−1), which can be increased by its recursive application.
Now after dimension reduction, F M L D is used for classification via a well-known k-Nearest Neighbor (k-NN) classifier. Since k-NN is a simple, powerful, accurate and efficient method; easily implemented on a low processing device like mobile phone. The value k is set to one an so named best-NN. The experimental results and discussion are presented in detail in Section 5 after discussing the proposed architecture for mobile device.
4 Proposed architecture for mobile device
Today, the smart ubiquitous device possesses all required sensors for the proposed system: mobile machine learning for plant leaf image informatics. Mobile phones with camera, processor and Internet connection (not always compulsory) are sufficient for proposed species identification. Android is today’s major market capturing operating system (OS) and is open source. Therefore, we targeted to deploy the proposed system on Android.
Due to MD constraints, the majority of high computing processes are offloaded to a high-end processing unit - the Server. To offload the tasks, unfortunately a regular communication connectivity is required and in countries like India where communication channels are very poor and overloaded, the system may not be so feasible. Thus, to have a real-time efficient system, the algorithms must directly execute on device without depending up on a remote Server. Therefore, in our implementation, both the problems are solved by introducing an intelligent agent to adaptively decide whether to compute-on device or to offload the job.
The complete architecture of the proposed mobile plant leaf informatics system is shown in Fig. 9. As discussed previously (Section 3.1), MD is used to capture leaf image and pre-process, extract features, then classify and augment results on the screen. Here, in this architecture, there are three scenarios: (1) execute everything on MD, if it is capable, (2) computer pre-processing and SET extraction on MD and rest is processed on the remote Server, and (3) perform pre-processing on MD and transmit rest to the Server for further processing, as labeled in Fig. 9. There is an Agent module in pre-processing phase, designed to record the current health status of MD and decide the transmission. The decision is made by estimating the processing capacity, battery life and communication cost of mobile device at that instant [32].If the bandwidth is very less and requires more energy and time to transmit compared to computing on MD, then only the RSSC feature vector is transmitted for classification on the server.
On the other hand, whenever MD is online, an update module automatically updates the online database storage. This helps in getting better performance time-to-time and explore new plant species even if there is no connectivity. The description of various modules used in Fig. 9 are given below and is briefed in Table 3.
-
1.
Pre-Processing: As the very first module of the proposed mobile client-server architecture, it captures and transforms image to device independent color space (as in Section 3.1). The L*a*b* color image is denoised and transformed to translation and scale invariant space (Section 3.1). All these tasks are carried on mobile itself.
Table 3 Module description of the proposed mobile architecture -
2.
Agent: Then the Agent sitting on mobile device estimates the computation cost and transmission cost, decides whether further high-level computations can be carried on on-board or offloaded to the server [32]. For the estimation, Agent computes following parameters: battery strength, network strength, bandwidth, image size, and computation power of MD. This task is also performed on MD.
-
3.
SET Matrix: This module of mobile client -server architecture computes SET matrix of pre-processed images for further RSSC features (Section 3.2). Depending upon the transmission cost it can be performed either on-board or on the server. If the bandwidth is small then mobile computes the SET values and transmits on the SET matrix else the pre-processed image is transmitted, as shown in Fig. 9.
-
4.
RSSC Feature: The SET matrix is derived to RSSC for classification as discussed in Section 3.2. This task can also be performed on both the sides: client or server depending upon Agent. Since RSSC is local vector it is fussed with the global GLCM leaf features to increase the efficiency.
-
5.
MLD & Classification: The RSSC vector is further reduced to MLD feature space and then classified using best-NN either on-board using online dataset or on server using offline dataset (Section 3.4).
-
6.
Result Display: The result module simply transforms the information into the readable form such as species name and augmented on mobile screens. It helps user in understand nature more continently anywhere-anytime. This module is performed on client device and so is a light-weighted thread.
-
7.
Update: The Update module updates the parameters of mobile-based plant leaf informatic system whenever it is connected to Internet so that the result is up to the users satisfaction. It also uploads and downloads the online dataset on local mobile machines from server. It fetches a connection with server and updates the dataset with the key instances. This task is performed on client side.
-
8.
Online & Offline Database: In this phase, the online database, that is, the subset of the original plant leaf database is stored on MD to perform species identification in offline mode. Whereas, the offline database is the actual complete leaf database that performs the complete match and validates the results before any decision is made. The offline database is updated by the research term day-to-day and is also updated with exceptions received from users.
5 Experimental results and discussion
In this segment of paper, the experimental results and the performances are shown on three different types of datasets: two are publically available leaf datasets and the third is a diseased leaf dataset. The first dataset is ICL (Intelligent Computing Laboratory) Leaf dataset [17], second is Flavia Leave dataset [11, 41] and the third is a diseased leaf dataset collected from Indian Institute of Technology (IIT) Roorkee, India and Forest Research Institute (FIR) Dehradun, India campuses [32, 33], as seen in Fig. 10. The ICL leaf dataset includes 220 different plant species (Fig. 10a) whereas Flavia leaf dataset contains around 1905 leaf samples for 32 different plant species (Fig. 10b). The diseased leaf dataset [33] is a small dataset with four different species having five different diseases and are around 297 leaf samples, Fig. 10c.
5.1 Implementation
The proposed system is implemented using Android SDK and OpenCV on Eclipse Windows environment. The system is deployed on an original Micromax mobile device of configuration: 1GHz processor, 512MB internal RAM, and Android 4.1.x or above. We used different evaluation approach to compare the proposed method with the existing systems; discussed in Section 5.2.
5.2 Parameters
In this paper, for experiment, the parameters selected are: w = h=460, a square matrix, to acquire at least a standard of VGA quality image, the size of grid is five (s=5), bigger blocks to reduce the computational cost and memory requirement. The third and the most important parameter is c=10, i.e. at least 100 pixels are common with their adjacent sub-images, (x ″=100 = y ″) and therefore, the length of vector will be 916. Whereas in case of gray image, it will be 316, which is reduced further by using MLD.
5.3 Experiments
With these parameters, F M L D is computed for a plant leaf to recognize it using best-NN. The accuracy of the proposed RSSC algorithm is better than other existing methods discussed in this paper and is fast and efficient. The state-of-the-art comparison of all different datasets are shown in Table 4. Here, the accuracy of algorithms like Riemannian Elastic Metric (REM), probability neural network (PNN), RSC, CT, Multi-scale Convexity Concavity (MCC), Triangle-Area Representation (TAR), Inner Distance (IDSC), Multiscale-ARCH-height (MARCH), FT, GT, Elliptic Fourier transform (EFT), curvature scale scope corner detection (CSSCD) method and Triangle Oriented Angle (TOA) are compared with RSSC.
From Table 4, the maximum accuracy achieved with Flavia dataset is 99.36 % and 92.95 % for ICL leaf dataset using our proposed RSSC approach. The accuracy rate tested with our diseased leaf dataset is 100 %. Even a single dimension feature space clearly separates the diseased leaves, Fig. 11a. This is due to the small size of dataset.
Further, experiments are carried out to find the minimum feature dimension of F M L D required to represent a leaf image properly, Fig. 11. This approach again optimizes the classification computation cost and increases the accuracy rate, avoiding the curse of dimensionality. Figure 11a, shows an accuracy rate of diseased leaf dataset with varying F M L D length followed by Flavia and ICL leaf datasets, Fig. 11b and c, respectively. The comparison is also made with other different local statistical textures: energy and sum, (20) and (21), respectively. It is seen that, in Fig. 11a, even single dimension is sufficient to separate the species. But in Flavia dataset, the vector of length 20 successfully represents the leaf for mobile vision identification and in case of ICL leaf dataset, its 31.
In shape recognition, Mouine et al. [25], proposed TOA and triangle side length and angle (TSLA) to represent the object shape uniquely, but it is a complex approach, see Table 6. Unlike TOA, RSSC is a simple and less complex to identify species/shape. Figure 12 shows the RSSC comparison with different color dimension local SET features for all three different datasets using different classifiers. The bar graph compares five different types of classifiers: Bayes - Nave Bayes, Functional - Sequential Minimal Optimization (SMO), Lazy - k-NN, Meta-classifier, and Tree - Random Forest and found that best-NN outperforms in all the cases. Whereas, Nave Bayes also results equally good compared to best-NN but due to MD limitations best-NN is chosen. It is clear that the local entropy texture of \(I_{leaf}^{3}\) results better using RSSC representation. The computation cost is even reduced in case of grayscale image with marginal accuracy compensation.
Another comparison graph, Fig. 13, shows an error rate change with varying Flavia training dataset size for different vector length (N). The graph shows a relation between grayscale image, i.e. L* component of L*a*b* color space with L*a*b* color image. The graph compares entropy texture with energy and sum texture at 15, 25, 50 and 80 percent of training sets, \(F_{MLD_{train}}\). Similarly, Fig. 14 shows the error rate change for ICL leaf dataset with feature length: 1, 5, 10, 20, 31 and 100.
The time complexity of computing F M L D , in both L*a*b* and grayscale, is compared in Table 5. It is clear that the cost involved in RSSC texture feature calculated of grayscale leaf is almost three times smaller than L*a*b* image and thus highly suitable for MD computing.
5.4 Performance on mobile
The research is also carried on optimizing the different parameters (w,h,s, and c) in terms of accuracy and complexity, see Table 6. The third column in Table 6 gives the execution time for SET-based RSSC feature extraction (in seconds) for Flavia leaf dataset. Here, the L* (grayscale) image is used for comparison. The snapshots of leaf image informatics is shown in Fig. 15 where user captures the plant leaf and partially processed on the device and rest of the computations are pushed to the computing server.
The detailed misclassification of each class of a dataset can easily be shown by using a confusion matrix, representing the confusing classes. A confusion matrix or the error (matching) matrix is the visualization of performance of any algorithm. A confusion matrix, \(\mathfrak {C}_{matrix}\) of Flavia dataset is shown in Table 7. In \(\mathfrak {C}_{matrix}\), the values in each column represents the prediction class and each row represents the actual class. Note, for the readability purpose, the scientific names of species in \(\mathfrak {C}_{matrix}\) are represented by their serial numbers as used in [41]. The accuracy rate A, in this paper, is a ratio of total number of predictions that were correct to the total number of observations and is defined by (22).
where, \(total_{cor_{prdt}}\) and \(total_{incor_{prdt}}\) are the count of correct and incorrect prediction in \(\mathfrak {C}_{matrix}\), respectively.
Wang and Gao [35], proposed and RST-mirror invariant hierarchical string cuts (HSC) algorithm for shape retrieval with a maximum accuracy of 89.40 %. The accuracy achieved by this approach is around 86.03 % for ICL dataset which is very less compared to our proposed algorithm, see Table 4.
5.5 Computational complexity
The final parameter to measure any algorithm is its computational complexity. Since the proposed model is executed on MD, the cost must be minimum. The computational complexity of the proposed system is a sum of two models: the cost involved in RSSC feature extraction and the cost involved in best-NN classification.
Suppose, the size of a normalized input leaf image is (w×h), divided into s 2 sub-images, each of (x ″×y ″) then the complexity required to extract RSSC feature is O(s 2 x ″ y ″) which can be rewritten as O(w h). If (w h)=N, then the complexity will be O(w h) = O(N). At the classification end, the time complexity required for best-NN is O(F M L D m C). If (m×C)=M, that is, if the dataset size is M and F M L D =20 then complexity reduces to O(M). Therefore, the total time complexity involved for our proposed system is (O(N) + O(M)) and if M is reduced to 25 % for MD the complexity is directly affected, without effecting accuracy much (Figs. 12 – 13). In Table 8, a complexity comparison of different algorithms used in leaf shape identification is compared.
The proposed leaf informatics system helps in identification of species and further provide information for plant leaf disease diagnosis [14, 33, 34]. Species identification reduces the search database for disease recognition and enhance the system performance.
6 Conclusion and future work
This paper presents a novel RSSC algorithm for mobile vision based plant leaf informatic system. The RSSC entropy texture feature describes uniquely the texture of a plant leaf captured via MD. F l e a f is a combination of four orientation independent GLCM features and local SET information. Best-NN classifier designed for MD is used for species classification. To validate the proposed system, the experiments were carried out on three different leaf datasets: ICL, Flavia and Diseased leaf datasets. The algorithm is compared with various other methods and techniques in state-of-the-art and come up with a decision that the proposed algorithm is more robust and efficient in terms of accuracy and computational complexity.
The proposed system is designed for isolated leaf and so leaf segmentation in a complex background in a low vision is still an open challenge for future. Secondly, for real-time feasibility and scope, mobile augmented reality in agriculture can be a new direction. And thirdly, the RSSC can be tested on other texture datasets to study its robustness.
References
Adamek T, O’Connor N E (2004) A multiscale representation method for nonrigid shapes with a single closed contour. IEEE Trans Circ Syst Video Technol 14(5):742–753
Alajlan N, El Rube I, Kamel M S, Freeman G (2007) Shape retrieval using triangle-area representation and dynamic space warping. Pattern Recogn 40 (7):1911–1920
Backes A R, Gonċalves W N, Martinez A S, Bruno O M (2010) Texture analysis and classification using deterministic tourist walk. Pattern Recogn 43(3):685–694
Bai X, Yang X, Latecki L J, Liu W, Tu Z (2010) Learning context-sensitive shape similarity by graph transduction. IEEE Trans Pattern Anal Mach Intell 32 (5):861–874
Barrera M, Andrade M, Kim H Y (2012) Texture-based fuzzy system for rotation-invariant classification of aerial orthoimage regions. jip 1(1):4
C V N I (2014) Cisco: global mobile data traffic forecast update, 2013-2018. White Paper, pp 1–40
Clark J Y (2009) Neural networks and cluster analysis for unsupervised classification of cultivated species of tilia (malvaceae). Botan J Linnean Soc 159(2):300–314
Cope J S, Remagnino P, Barman S, Wilkin P (2010) Plant texture classification using gabor co-occurrences. In: International Symposium on Visual Computing. Springer Berlin Heidelberg, pp 669–677
Cope J S, Corney D, Clark J Y, Remagnino P, Wilkin P (2012) Plant species identification using digital morphometrics: a review. Expert Syst Appl 39 (8):7562–7573
Deypir M, Boostani R, Zoughi T (2012) Ensemble based multi-linear discriminant analysis with boosting and nearest neighbor. Scientia Iranica 19(3):654–661
Flavia leave dataset. http://flavia.sourceforge.net/. Accessed: 2012-10-14
Goëau H, Bonnet P, Joly A, Bakic V, Barthélémy D, Boujemaa N, Molino J-F (2013) The imageclef 2013 plant identification task. In: CLEF
Goëau H, Bonnet P, Joly A, Boujemaa N, Barthélémy D, Molino J-F, Birnbaum P, Mouysset E, Picard M (2011) The imageclef 2011 plant images classi cation task. In: ImageCLEF 2011, pp 0–0
Govaerts R (2001) How many species of seed plants are there? Taxon 50 (4):1085–1090
Haralick R M, Shanmugam K, Dinstein I H (1973) Textural features for image classification. IEEE Trans Syst Man Cybern 6:610–621
Hearn D J (2009) Shape analysis for the automated identification of plants from images of leaves. Taxon 58(3):934–954
Intelligent computing laboratory. http://www.intelengine.cn/English/dataset. Accessed: 2012
Kim T, Bae N-J, Shin C-S, Park J W, Park D, Cho Y-Y (2013) An approach for a self-growing agricultural knowledge cloud in smart agriculture. In: Multimedia and ubiquitous engineering. Springer, pp 699– 706
Kumar N, Belhumeur P N, Biswas A, Jacobs D W, Kress W J, Lopez I C, Soares J V (2012) Leafsnap: a computer vision system for automatic plant species identification. In: Computer vision–ECCV 2012. Springer, pp 502–516
Laga H, Kurtek S, Srivastava A, Golzarian M, Miklavcic S J (2012) A riemannian elastic metric for shape-based plant leaf classification. In: 2012 International conference on digital image computing techniques and applications (DICTA). IEEE, pp 1–7
Lee C-L, Chen S-Y (2006) Classification of leaf images. Int J Imag Syst Technol 16(1):15–23
Li Y, Chi Z, Feng D D (2006) Leaf vein extraction using independent component analysis. In: IEEE International conference on systems, man and cybernetics, 2006. SMC’06, vol 5. IEEE, pp 3890– 3894
Ling H, Jacobs D W (2007) Shape classification using the inner-distance. IEEE Trans Pattern Anal Mach Intell 29(2):286–299
Marino S M W D M, Feiner S K Leafview: a user interface for automated botanical species identification and data collection
Mouine S, Yahiaoui I, Verroust-Blondet A (2013) A shape-based approach for leaf classification using multiscaletriangular representation. In: Proceedings of the 3rd ACM conference on international conference on multimedia retrieval. ACM, pp 127–134
Mullen R J, Monekosso D, Barman S, Remagnino P, Wilkin P (2008) Artificial ants to extract leaf outlines and primary venation patterns. In: Ant colony optimization and swarm intelligence. Springer, pp 251–258
Neto J C, Meyer G E, Jones D D, Samal A K (2006) Plant species identification using elliptic fourier leaf shape analysis. Comput Electron Agri 50(2):121–134
Prasad S, Kumar P, Tripathi R (2011) Plant leaf species identification using curvelet transform. In: 2011 2nd International conference on computer and communication technology (ICCCT). IEEE, pp 646– 652
Prasad S, Kudiri K M, Tripathi R (2011) Relative sub-image based features for leaf recognition using support vector machine. In: Proceedings of the 2011 international conference on communication, computing & security. ACM, pp 343–346
Prasad S, Peddoju S K, Ghosh D (2013) Agromobile: a cloud-based framework for agriculturists on mobile platform. Int J Adv Sci Technol 59:41–52
Prasad S, Peddoju S K, Ghosh D (2013) Mobile plant species classification: a low computational aproach. In: 2013 IEEE Second international conference on image information processing (ICIIP). IEEE, pp 405–409
Prasad S, Peddoju S K, Ghosh D (2014) Energy efficient mobile vision system for plant leaf disease identification. In: Wireless communications and networking conference (WCNC), 2014 IEEE. IEEE, pp 3314–3319
Prasad S, Peddoju S K, Ghosh D (2016) Multi-resolution mobile vision system for plant leaf disease diagnosis. Signal Image Video Process 10(2):379–388
Prasad S, Peddoju S K, Ghosh D (in press) Agriculture-as-a-service. IEEE Potentials, Accepted
Wang B, Gao Y (2014) Hierarchical string cuts: a translation, rotation, scale, and mirror invariant descriptor for fast shape retrieval. IEEE Trans Image Process 23 (9):4101–4111
Wang Z, Chi Z, Feng D (2003) Shape based leaf image retrieval. In: IEEE Proceedings vision, image and signal processing, vol 150. IET, pp 34–43
Wang B, Brown D, Gao Y, La Salle J (2015) March: multiscale-arch-height description for mobile retrieval of leaf images. Inf Sci 302:132–148
Warren D (1997) Automated leaf shape description for variety testing in chrysanthemums. In: Sixth international conference on image processing and its applications, vol 2. IET, pp 497–501
White S, Feiner S, Kopylec J (2006) Virtual vouchers: prototyping a mobile augmented reality user interface for botanical species identification. In: IEEE symposium on 3D user interfaces, 2006. 3DUI 2006. IEEE, pp 119–126
White S M, Marino D, Feiner S (2007) Designing a mobile user interface for automated species identification. In: Proceedings of the SIGCHI conference on human factors in computing systems. ACM, pp 291–294
Wu S G, Bao F S, Xu E Y, Wang Y-X, Chang Y-F, Xiang Q-L (2007) A leaf recognition algorithm for plant classification using probabilistic neural network. In: 2007 IEEE International symposium on signal processing and information technology. IEEE, pp 11–16
Yanikoglu B, Aptoula E, Tirkaz C (2014) Automatic plant identification from photographs. Mach Vis Appl 25(6):1369–1383
Zhao Z-Q, Ma L-H, Y-m Cheung X Wu, Tang Y, Chen C L P (2015) Apleaf: an efficient android-based plant leaf identification system. Neurocomputing 151:1112–1119
Zulkifli Z, Saad P, Mohtar I A (2011) Plant leaf identification using moment invariants & general regression neural network. In: 2011 11th International conference on hybrid intelligent systems (HIS). IEEE, pp 430–435
Acknowledgments
Authors would like to thank MHRD for financially supporting S. Prasad throughout his PhD work at IIT Roorkee.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Prasad, S., Peddoju, S.K. & Ghosh, D. An adaptive plant leaf mobile informatics using RSSC. Multimed Tools Appl 76, 21339–21363 (2017). https://doi.org/10.1007/s11042-016-4040-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-016-4040-8