Abstract
Aim
Plant metal tolerance proteins (MTPs) are plant membrane divalent cation transporters that specifically contribute to heavy metal stress resistance and mineral uptake. However, little is known about this family’s molecular behaviors and biological activities in soybean.
Methods and results
A total of 20 potential MTP candidate genes were identified and studied in the soybean genome for phylogenetic relationships, chromosomal distributions, gene structures, gene ontology, cis-elements, and previous gene expression. Furthermore, the expression of MTPs has been investigated under different heavy metals treatments. All identified soybean MTPs (GmaMTPs) contain a cation efflux domain or a ZT dimer and are further divided into three primary cation diffusion facilitator (CDF) groups: Mn-CDFs, Zn-CDFs, and Fe/Zn-CDFs. The developmental analysis reveals that segmental duplication contributes to the GmaMTP family’s expansion. Tissue-specific expression profiling revealed comparative expression profiling in similar groups, although gene expression differed between groups. GmaMTP genes displayed biased responses in either plant leaves or roots when treated with heavy metal. In the leaves and roots, nine and ten GmaMTPs responded to at least one metal ion treatment. Furthermore, in most heavy metal treatments, GmaMTP1.1, GmaMTP1.2, GmaMTP3.1, GmaMTP3.2, GmaMTP4.1, and GmaMTP4.3 exhibited significant expression responses.
Conclusion
Our findings provided insight into the evolution of MTPs in soybean. Overall, our findings shed light on the evolution of the MTP gene family in soybean and pave the path for further functional characterization of this gene family.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Metals function as cofactors, which is important in inactivating enzymes in plant cells to complete particular biological reactions [1]. The optimum concentration of numerous metal ions such as manganese (Mn2+), iron (Fe2+,3+), zinc (Zn2+), copper (Cu2+), nickel (Ni2+), vanadyl (VO2+), and cobalt (Co2+) is required for regulation of enzymatic activities in various biological processes. Still, the excess of these transition ions stimulates over-accumulation of reactive oxygen species (ROS), which seriously affects crop yield [2, 3]. For instance, two amidohydrolase compound families utilize Zn2+, Ni2+, and Fe2+ as cofactors during hydrolysis. Contrastively, minuscule amount of non-essential metals including mercury (Hg2+), silver (Ag+), cadmium (Cd2+), chromium (Cr+2,Cr+3,Cr+6), selenium (Se−2,Se+4,Se+6), lead (Pb2+) and arsenic (As−3) is harmful to plant cells. [4, 5]. Plant roots assimilate metal ions from soil dissolved in water and store them in different tissues [6].
To deal with the harmful impacts of heavy metals stress, plants’ biochemical and metabolic processes go through exact changes at a physiological and molecular level [7, 8]. Metal transporters are important for plant homeostasis in specific metal transport proteins, natural resistance-associated macrophage proteins, cation diffusion facilitators (CDFs), and heavy metal ATPase, which are initiated to prevent cell harm by metal stress, consistently [9, 10]. Physiological processes, for example, metal absorption, transport, accumulation, chelation, and efflux, are plant elements to keep up with metal homeostasis [11]. Some specific membrane-bound protein families serve as metal ions transport channels. Cation diffusion facilitators (CDFs) proteins serve as divalent cation transporters responsible for metal ions efflux from the cytoplasm in subcellular organelles or cell export [12]. In light of substrate diffusion, there are three classes of CDF families; Zn-CDF, Fe/Zn-CDF and Mn-CDF, which are responsible for Zn2+, Fe2+ and Mn2+ moves alongside Cu2+, Co2+, Cd2+ and Ni2+ [11].
These CDFs transporters are also known as metal-tolerance proteins in plants, as explained in the accompanying seven gene subfamilies 1, 5, 6, 7, 8, 9, and 12 [12]. There are 12 MTP proteins in Arabidopsis thaliana [13], 11 in Solanum Lycopersicum [8], 11 in Vitis vinifera [14], 22 in Populus trichocarpa [15], 20 in Triticum aestivum [16], 12 in Medicago truncatula [17], 13 in Camellia sinensis [18], 26 in Nicotiana tabacum, 13 in Nicotiana sylvestris, and 12 in Nicotiana tomentosiformis [7], 18 in Brassica rapa [19], 12 in Citrus sinensis [20] and 7 in Pyrus bretschneideri [21]. Initially, a group of 8 MTPs was distinguished in Stylosanthes hamata, which increased resistance against Mn2+ toxicity on overexpression in Arabidopsis [22].
Numerous Zn-CDFs have been identified in Arabidopsis [13], such as AtMTP1 and AtMTP3 performs a vital role in Zn2+and Co2+ disposition into vacuole [23], and AtMTP5 and AtMTP12 in the accumulation of Zn2+ into Golgi bodies [24]. Correspondingly, the elements of the Mn-CDF group, such as AtMTP8, play a significant function in Mn2+ homeostasis via their disposition as well as deposition of Fe2+ and Mn2+ in seeds of Arabidopsis during seed development and germination [22, 25, 26]. OsMTP8.1, OsMTP8.2, HvMTP8.1, and HvMTP8.2 are localized in tonoplast and serve as Mn2+ deposition in Golgi apparatus in rice and barley [27,28,29]. Furthermore, cucumber CsMTP8 confers Mn2+ tolerance when overexpressed in Arabidopsis and yeast [30].
Soybean is a leguminous crop rich in seed oil, proteins, vitamins, and isoflavones. Genetically, soybean is a legacy paleopolyploid and a pending model for elucidating the sequels of genome duplication in higher eukaryotes [31]. As of late, endeavors have been led to investigate model plants and common commercial species genomes [32]. Also, the accomplishment of great draft genomes in the soybean MTP gene family at the genome-wide level gave an event to play out an orderly examination. In this article, we recognized GmaMTP genes in soybean and investigated their structure, 3D protein structure, gene ontology, and cis-regulating elements. Expression outlines of GmaMTP genes in various soybean tissues under the five divalent heavy metals stresses were examined.
Our findings will provide a deep understanding of the MTP gene family involved in heavy metal stress response in a plant cell, as well as the founders and biological functions of GnaMTP proteins, which will open up new avenues of research in the area of molecular mechanisms of homeostasis and heavy metal transport, and will ultimately help to precisely engineer soybeans plants for heavy metal stress.
Materials and methods
Identification of soybean GmaMTP genes
The candidate soybean GmaMTP gene sequences were retrieved via blast analysis of open reading frames (ORFs) of AtMTPs, MtMTPs, and PtMTPs in an Integrating Genetics and Genomics to Advance Soybean Research (soybase.org/) database by adjusting the default parameters. Subsequently, BioEide 7.0 software was employed to construct a local database. Furthermore, candidate GmaMTP genes were analyzed for HMM profiling of two conserved MTP domains, such as PF16916 and PF01545, by searching the Pfam website (sanger.ac.uk/Software/Pfam). Every gene was assigned a specific name following the standard Mendel database for plant gene families listed in Commission on Plant Gene Nomenclature (CPGN) (mbclserver.rutgers.edu/CPGN/), International Society of Plant Molecular Biology (ISPMB) [33].
Whole-genome blast analysis of putative MTP protein sequences of soybean was performed on the NCBI database (blast.ncbi.nlm.nih.gov/blast.cgi) and phytozome database (phytozome.jgi.doe.gov/). Finally, all retrieved protein sequences were confirmed by exploring CCD (www.ncbi.nlm.nih.gov/cdd/) and SMART (smart.embl-heidelberg.de/) programs. All retrieved GmaMTP protein sequences were anatomized at E-value < 10−5 to identify the MTP domain by employing SMART (smart.embl-heidelberg.de/) tools [34]. All detailed inheritable information of the putative GmaMTP gene family, including chromosomal location and CDS, were acquired from the phytozome database (phytozome.jgi.doe.gov/). Likewise, MTP family proteins were analyzed for their molecular weight, number of atoms, number of amino acids, isoelectric point, and instability index using EXPASY PROTOPARAM (expasy.org/tools/protparam.html) [35]. Eventually, calculations of theoretical isoelectric point (pI) and molecular weight (MW) were attained using ExPASy ProtParam Tools (web.expasy.org/portparam) [18].
Phylogenetic analysis
In addition to protein sequences of GmaMTPs of Glycine max, AtMTP protein sequences of Arabidopsis thaliana (arabidopsis.org), CsaMTP sequences of Cucumis sativus (cucurbitgenomics.org/), PtrMTPs protein sequences of Populus trichocarpa (plantgdb.org/PtGDB/), OsMTPs protein sequences of Oryza sativa (rapdb.dna.affrc.go.jp/) and TaMTPs protein sequences of Triticum aestivum (wheatgenome.org/) were also recaptured and anatomized for genetic mapping. The ClustalX2.0 software with default parameters was used for multiple sequence alignments of all retrieved MTP protein sequences [36]. All alignments were uploaded in MEGA6.0 software with a Neighbor-Joining method to construct a phylogenetic tree [37]. Finally, bootstrap analysis was performed at iterations with a pair-wise gap deletion mode [38, 39].
Chromosomal localization and gene synteny analysis
Soybean genetic database (phytozome.jgi.doe.gov/) was searched to retrieve data about chromosomal localization of GmaMTP genes, and a high-fidelity genetic map was constructed by assuming MapChart (wur.nl/en/show/Mapchart.htm) software. The genes of single species placed in the same clade were declared asco-paralogs which were further explored for possible duplication events. The Phytozome (phytozome.jgi.doe.gov/) database was searched to explore segmental duplications among GmaMTP genes. The presence of paralogs was supposed to result from tandem duplication due to splicing two genes into five or further within a 100 kb stretch [40]. Also, co-paralogs located within duplicated chromosomal regions were supposed to be segmentally duplicated [41]. Smith-Waterman algorithm (ebi.ac.uk/Tools/psa/) was employed to calculate the local alignments of two protein sequences. The synteny analysis of the GmaMTP family members was performed using circos (circos.ca/) tools to localize different alleles distributed among chromosomes [42].
Gene structures, motif analyses, and cis-regulatory elements prediction
Each GmaMTP gene sequence was analyzed to investigate for number, size, and location of introns and exons in both gDNA and CDS sequences by deploying the Genes Structure Display Server program (GSDS) (http://gsds.gao-lab.org/) [43]. Furthermore, conserved gene family motifs were also identified by deploying a Multiple EM for motif elicitation (MEME) (meme.nbcr.net/meme3/meme.html) tool with the following parameters; a maximum of 20 motifs harboring 6–200 amino acids per motif [44]. To investigate regulatory mechanisms of GmaMTP genes, 2 kb upstream 3’UTR promoter region of each gene was downloaded from plant genomic resource (phytozome.jgi.doe.gov/). The cis-elements of GmaMTPs promoter were identified using the PLACE (dna.affrc.go.jp/PLACE/?action=newplace) and PlantCARE programs (bioinformatics.psb.ugent.be/webtools/plantcare/html/) [45, 46].
Protein modeling, protein–protein interaction, and Gene ontology (GO) analysis
The Phyre2 (http://www.sbg.bio.ic.ac.uk/~phyre2/html/page.cgi?id=index) website was searched to perform protein modeling, prediction of protein–protein interactions and detailed analysis of GmaMTP polypeptides [47]. The amino acid sequences of each GmaMTP family member were used to construct a protein–protein interaction network in the STRING database (string-db.org/). Finally, the following two softwares, Blast2GO v3.0.11 (www.blast2go.com) and OmicsBox were employed to predict GO enrichment of all identified GmaMTP protein sequences[48].
RNA-seq-based gene expression analysis
RNA-seq data of different organs of soybean plants were retrieved by integrating genetics and genomics to advance soybean research (soybase.org/soyseq/). The expression level of each GmaMTP gene was calculated in young leaves, one cm long pod, pod shell after 10 and 14 days of fertilization, seeds 10, 14, 21, 25, 28, 35, and 42 days after fertilization, roots, flowers, and nodule of soybean under normal conditions by deploying cufflinks v.2.2.1. Finally, absolute FPKM values were divided by their mean, transformed into a ratio of log2 and MeV 4.5 was employed to cluster expression data into a heat map (heatmapper.ca/) [49].
Growth conditions and heavy metal treatments
The seeds of the soybean variety Zhonghuang-39 were sown during the Autumn of 2020 and placed in the experimental greenhouse of Yibin University (China). In order to perform surface sterilization, soybean seeds were first washed with 10% hypochlorous acid, followed by three washing items with distilled deionized water (ddH2O). Subsequently, soybean seeds were spread on water-saturated filter papers and incubated in the dark for germination. After 10 days of germination, four uniform seedlings were shifted from filter paper to fertilized pit moss soil-filled plastic pots and placed under the following conditions; 16 h light (27ºC) and 8 hours dark (18ºC) with a relative humidity of 70%. For metal ions treatments, thirty-day-old soybean plantlets were dipped in 1/2 Hoagland solution (pH 6.0) supplemented with following different concentrations of heavy metals; 0.1 mM CdCl2, 0.1 mM CoCl2, 0.5 mM FeSO4,1 mM MnSO4, 0.5 mM ZnSO4 and plane 1/2 Hoagland (CK)solution as a control [15]. After 24 h, the leaves and roots of soybean plantlets were detached and immediately preserved in liquid nitrogen for further experiments. The whole experiment was performed in a completely randomized design comprised of 6 treatments with three scientific repeats.
RNA extraction and qRT-PCR analysis
To extract RNA, each metal ion solution treated soybean leaf and root tissue was separately homogenized in liquid nitrogen in triplicate manners. Subsequently, the Trizol® reagent method (Invitrogen, USA) was used to extract RNA; integrity confirmation was performed by running 2 μl in 1% agarose gel prepared in RNase-free 0.5 × TAE buffer and reverse transcribed to cDNA by using SuperMix Kit (Transgen, Beijing) [50, 51]. The primers of all selected GmaMTP genes and β-actin as an internal control were designed using Primer Premier 5.0 software (Table S1). The real-time PCR was performed with the following ingredients in 20 μL reaction volume; 10 μl of 2 × SYBR premix Taq mixture, 1 μl of cDNA,0.5μL of each primer, and 8 μl of ddH2O. The PCR conditions were adjusted as follows: initial denaturation at 95 °C for 10 min, extension at 95 °C for 15 s, total cycles 40, final extension at 60 °C for 1 min, and finally, storage at 4 °C for infinity [52]. The relative expression level of each GmaMTP gene was calculated using Livak 2−ΔΔCT values in triplicate manners for each sample [53].
Statistical analysis
The results of three biological replicates of expression analysis of each gene are presented in mean ± standard deviation (SD) at p < 0.05. The analysis of variance (ANOVA) among different values was compared at p < 0.05 using a student t-test.
Results
Identification of GmaMTP genes in soybean
After excluding reads with incomplete functional domains, only 20 candidate GmaMTP genes were obtained after blast analysis of 30 soybean GmaMTP gene sequences. Each gene’s characteristics, such as gene locus, molecular weight, number of amino acids, grand average of hydropathicity, and isoelectric points, have been listed (Table 1). Except for the following seven chromosomes; 1,4,5,6,13,17, and 20, the remaining 13 chromosomes of soybean contain GmaMTP gene loci. The molecular weight of GmaMTP protein molecules differs from 38,221.62 to 53,254.09 Da (Table 1). We observed a variable number of inter and intra-protein ionic residues, such as the highest anionic residues observed in GmaMTP1.2 and the lowest in GmaMTP5.2. Essentially, the most elevated cationic residues were seen in GmaMTP10.2, while the lowest was in GmaMTP3.1 and GmaMTP3.2.).
Phylogenetic analysis of GmaMTP genes
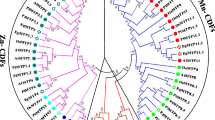
An evolutionary relationship-based phylogenetic tree showed that all GmaMTP family genes and their orthologs AtMTP, PtrMTP, OsMTP, and CsMTP were divided into the following seven groups; 1, 5, 6, 7, 8, 9, and 12 (Fig. 1).The biggest number of MTP genes are exist in Group 9 including GmaMTP9, GmaMTP10.1, GmaMTP10.2 and GmaMTP11.1 along with AtMTP9, AtMTP10 and AtMTP11; followed by Group 1 which is comprised of GmaMTP1.1, GmaMTP1.2 and GmaMTP3.1 along with AtMTP1, AtMTP2,AtMTP3 and AtMTP4; followed by Group 8 which is comprised of GmaMTP4.1 and GmaMTP4.2 along with OsMTP4; followed by Group 5, 6 and 7 which are comprised of GmaMTP5.1, GmaMTP5.2, GmaMTP2.1, GmaMTP2.2, GmaMTP7.1and GmaMTP7.2 along with AtMTP5, AtMTP6 and AtMTP7. Notably, no GmaMTP secured a position in Group 12. Ionic bunching uncovered that 6 GmaMTPs were bunched in the Zn-CDFs group, 4 GmaMTPs were clustered in Fe/Zn-CDFs group, and 10 GmaMTPs were crowded in the Mn-CDFs group (Fig. 1).
Phylogenetic tree of 80 MTP proteins: 12 Glycine max (marked by a red circle),12 Arabidopsis (blue circle), 8 Wheat (purple circle), 10 Rice (brown circle), 9 Cucumber ( green circle), and 21 Black Poplar (black circle). ClustalX1.83 was used for protein alignments and the phylogenetic tree’s construction Neighbor-Joining (NJ) level with MEGA5.0 software at 1,000 replications bootstrap
Gene synteny analysis of GmaMTPs
Assessment of gene family increase and novel functions with the help of circos in the Plant Genome Duplication Database (PGDD) revealed segmental and tandem gene pair duplications (Fig. 2). We observed 70–100% identical collinearity after the ablation of segmental replication in numerous gene sets (Table S2). Segmental duplication resulted in a considerable level of homologies in GmaMTP gene pairs such as GmaMTP1.1/GmaMTP1.2, GmaMTP1.2/GmaMTP3.2, and GmaMTP1.2/GmaMTP4.2 (Fig. 2). Similarly, two tandem replicated gene clusters were seen on chromosomes 3 and 15.
Gene structures and Construction of Conserved Motifs
Entire GmaMTP family genes were partitioned into six subfamilies; A, B, C, D, E, and F (Fig. 3a). Subfamily A has seven members, followed by subfamily F with four and then B with three. Finally, subfamilies C, D, and E each had only two genes. (Fig. 3a). All GmaMTP gene sequences harbor a variable number of introns except GmaMTP1.1, GmaMTP1.2, GmaMTP3.1, and GmaMTP3.2. Intron and exon analysis of all MTP genes revealed that each retrieved sequence of the GmaMTP gene family is a correct and true member of the six subfamilies (Fig. 3c).
Phylogenetic relationship, gene structure and conserved motif analysis of GmaMTP genes; a The neighbor-joining phylogenetic tree was constructed with MEGA7 using GmaMTP amino acid sequences with 1000 times replicate. b The motif composition of GmaMTP proteins using ten conserved motifs is represented by the unique colour mentioned in the box on the top lift. c The exon–intron structure of Glycine max MTP proteins where dark green boxes presented the exons and the black lines represented the introns. The blue boxes represented the untranslated regions (UTRs), with detailed size scales at the bottom
All GmaMTP subfamilies harbor various introns except subfamily F, which does not harbor any intron. All GmaMTP genes have different introns and exons. Nonetheless, the similarity index in sub-families is very high, indicating a close evolutionary link among all retrieved GmaMTP genes. Conserved protein motifs in GmaMTP polypeptide are composed of various amino acids. The largest motif was 6, observed in 14 MTP members (Fig. 3b and Table 2). Noticeably, motifs’ number, type, and order were more similar in the intrasubfamily than in the intersubfamily.
Cis-regulatory element in the upstream sequence of the MTP Family
Cis-elements associated with various stress reactions (ARE, WUN-motif, LTR, and MBS) were also discovered to support the majority of GmaMTPs (Fig. 4). Furthermore, cis-elements involved in plant advancement (MBS1, CAT-box, ERE, O2-site, and EBRE) were discovered in the promoter district of nearly all GmaMTPs. In the meantime, the cis-elements of the entire GmaMTPs family were separated into various classifications given capacity expectations. The outcome showed that the most significant classification of cis-elements was hormone-related, trailed by stress-related and advancement-related cis-elements. In any case, numerous themes have not yet been practically described and whether these themes present remarkable utilitarian roles to GmaMTPs remains to be researched.
Cis-regulatory elements in the promoter region of GmaMTP genes. Cis-regulatory elements were identified in the 2000 bp promoter sequence upstream of the start codon of GmaMTP genes using PlantCARE and PLACE. These elements are related to different functional diversity, represented by different colors
Protein modeling, protein–protein interactions
Phyre 2 web-based interface (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index) was utilized for protein displaying utilizing all MTP amino acid sequences (Figure S1 and Table S4). Each of the twelve anticipated models for MTP proteins was 100 percent in light of c6xpdB, c3j1zP, c2qfiB, and d2qfia2 layouts. On the contrary, the STRING examination of protein–protein interplay showed the physical (immediate) and the practical (circuitous) affiliations (Figure S2 and Table S4).
The outcome showed various associations inside the studied proteins, where the all-out number of hubs was 20, with an intermediate of 11. 4. The STRING information base investigation showed 114 edges and ten expected local network clusters, which were CL:48,378, 48,377, 48,387, 48,345, 48,298, 48,258, 48,504, 49,055, 48,255, and 48,260. Group 48,255 was the greatest, including 17 MTP proteins (Table S5). In addition, our protein investigation showed the presence of two ordinary spaces inside the checked member from the MTPs family, which were PF01545 (all proteins) and PF16916 (12 proteins) (Table S6).
GO enrichment analysis
The cellular component, molecular function, and biological processes were anticipated by GO improvement examination (Fig. 5 and Table S5). In subcellular localization investigation, the anticipated conveyance scores of MTP proteins were as follows; 20/71% in all layers 4/14% in the plasma membrane, vacuole, and Golgi apparatus. The GmaMTP1.1 and GmaMTP1.2 proteins were identified in 14 of the 17 sub-cellular compartments, highlighting their importance in metal stress resistance. The aggregate scores of MTP protein molecules during the biological process were as follows: trans-membrane transport of Zn + was 4/31%, Mn + ions were 6/46%, and trans-membrane transport of protons was 3/23%. GmaMTP1.1, GmaMTP1.2, GmaMTP3.1, and GmaMTP3.2 performs a significant part in transmembrane transport of Zn+, while GmaMTP4.1, GmaMTP4.2, GmaMTP4.3, GmaMTP9, GmaMTP11.1,and GmaMTP11.2 plays an essential part in transmembrane transport of Mn+. The investigation of molecular function and biological processes uncovered huge roles in GmaMTP1.1, GmaMTP1.2, GmaMTP3.1, and GmaMTP3.2, which assume a critical part in the transmembrane transport of Zn+. It affirmed the fundamental pieces of GmaMTP4.1, GmaMTP4.2, GmaMTP4.3, GmaMTP9, GmaMTP11.1, and GmaMTP11.2 in transmembrane transport of Mn+.
Gene Ontology analysis of Glycine max MTP genes. Gene ontology showed the distribution of every GmaMTP gene in the plant, where a blue colour column mentioned the cellular component. In contrast, the MTP family participation’s biological processes were mentioned in the red color column, and the molecular function was mentioned in the green colour
Gene expression analysis by RNA-seq data
The tissue expression models of GmaMTPs were researched using transcriptome data in various soybean tissues (Fig. 6. A). As displayed in Fig. 6b and Table S6, all 20 GmaMTP genes were identified in the 14 tissues tested (log2(FPKM + 1) > 0), except for GmaMTP4.2 (expressed only in root tissue), GmaMTP4.1 (expressed in young leaf, bloom, one cm unit, case shell, seed 21 DAF, root, and knob), and GmaMTP10.2 (only expressed in flower, seed 42DAF, root, and nodule). Among these,13 genes (GmaMTP1.1, GmaMTP1.2, GmaMTP2.1, GmaMTP2.2, GmaMTP3.1, GmaMTP4.3, GmaMTP5.1, GmaMTP5.2, GmaMTP7.1, GmaMTP7.2, GmaMTP9, Gma TP11.1 and Gma TP11.2) expressed integral expression (log2(FPKM + 1) > 1 in all tissues), and GmaMTP11.1 had the most elevated expression levels contrasted and other GmaMTPs in completely identified tissues, besides in-unit shell 14 DAF, seed 14 DAF, seed 25 DAF, seed 42 DAF, root and knob, though GmaMTP4.2 displayed the least expression levels in all tissues (0 < log2(FPKM + 1) < 1). Besides, a few genes displayed tissue-explicit expression. For example, four genes (GmaMTP7.1, GmaMTP9, GmaMTP10.2, and GmaMTP11.1) in the root, one gene (GmaMTP7.1) in the nodule, three genes (GmaMTP1.2, Gma MP7.1, and Gma TP11.1) in young leaf and two genes (GmaMTP7.1 and GmaMTP11.1) in flower showed the highest transcript abundances.
Expression analysis of GmaMTPs in response to heavy metals treatment
Entire GmaMTP genes showed discrepant gene expression levels when treated with various heavy metals examined in the root and leaf tissues (Fig. 7). GmaMTP1.1, GmaMTP1.2, GmaMTP3.1, GmaMTP4.1, GmaMTP4.3, GmaMTP10.4, and GmaMTP11.1 were upregulated in roots, while GmaMTP5.1 and GmaMTP7.2 were downregulated. Similarly, Co2+ increased the expression of GmaMTP1.1, GmaMTP2.1, GmaMTP2.2, GmaMTP3.2, GmaMTP5.2, GmaMTP9, GmaMTP10.1, GmaMTP10.2, GmaMTP10.3, GmaMTP10.4, and GmaMTP11.2. Under Fe2+ treatment, GmaMTP2.2, GmaMTP4.1, GmaMTP4.3, and GmaMTP10.3 were upregulated, whereas GmaMTP5.1 and GmaMTP7.2 were downregulated. Under Mn2+ treatment, GmaMTP1.1, GmaMTP1.2, GmaMTP2.1, GmaMTP3.2, GmaMTP4.1, GmaMTP4.2, GmaMTP4.3, GmaMTP5.2, GmaMTP9, GmaMTP10.1, GmaMTP10.4, GmaMTP11.1 and GmaMTP11.2 were upregulated, while GmaMTP2.2 was downregulated Under Zn2+ treatment, GmaMTP10.4 and GmaMTP11.1 were upregulated, whereas GmaMTP2.1 and GmaMTP7.1 were downregulated.
The qRT-PCR expression of the Glycine max MTP genes from root and leaf samples. The reactions were normalized using the β-actin reference gene. The standard deviations have been represented by the error bars from three independent technical replicates. The mean expression levels of three replicates were analyzed with the five heavy metals treatments (Cd2+, Co2+, Fe2+, Mn2+, and Zn2+) using t-tests (p < 0.05), while the CK represents control samples. Different letters (a, b, and c) indicate significant differences between the roots, and leaves under normal conditions. Asterisks indicate significant differences between the treatment samples and the corresponding control samples in the roots, and leaves. (n = 9, p < 0.05, Student’s t-test)
In leaf, Cd2+ treatment conclusion in enhancement expression of GmaMTP1.1, GmaMTP1.2, GmaMTP3.1, GmaMTP3.2 and GmaMTP4.1, yet, a huge end in expression of GmaMTP2.1 and GmaMTP5.1. Comparatively, Co2+ treatment fundamentally expanded the expression of GmaMTP1.1, GmaMTP1.2, GmaMTP3.1, GmaMTP3.2, GmaMTP4.1, GmaMTP4.3 and GmaMTP9 but decreased the expression of GmaMTP5.1. Fe2+ treatment decreased in expression of GmaMTP2.1, GmaMTP7.2 and GmaMTP10.1 but resulted in increased the expression of GmaMTP1.1, GmaMTP1.2, GmaMTP3.1, GmaMTP3.2, GmaMTP4.1, GmaMTP4.3, GmaMTP5.2, GmaMTP9 and GmaMTP10.3. Mn2+ treatment brought about expanded expression of GmaMTP1.1, GmaMTP1.2, GmaMTP3.1, GmaMTP3.2, GmaMTP4.1, GmaMTP4.3, GmaMTP9 and GmaMTP11.1, but decreased expression of GmaMTP2.1. Finally, Zn2+ treatment eventuated in improved expression of GmaMTP1.1, GmaMTP1.2, GmaMTP3.1, GmaMTP3.2, GmaMTP4.1, GmaMTP9, GmaMTP10.4 and GmaMTP11.1, but decreased expression of GmaMTP2.1 and GmaMTP5.2.
Discussion
Heavy metals majorly affect the environment and make them unsuitable for human utilization [54, 55]. Once delivered into the environment, they accumulate in plants and other living tissues utilizing the order of things and cause harmfulness even at lower concentrations [56]. MTP genes (membrane divalent cation transporters) are fundamental for transporting different heavy metals and upgrading plant tolerance against heavy metals stress [57]. They likewise play a normal part in plant mineral sustenance upkeep [7]. Additionally, these metal-binding proteins are currently being used as bio-natural markers for foreseeing heavy metal pollution in light of their expression levels [58]. They also function normally in plant mineral nourishment upkeep [7, 57]. The MTP family has formerly been examined in various plants, like Arabidopsis [13], tobacco [7], wheat [16], and Black poplar [15]. While this is the first genetic characterization research of the MTPs family in soybean. As a result, we identified 20 MTP genes in soybean, which were named based on the sequence similarities and orthologous relationships between them and AtMTPs. The MTP proteins’ evolutionary links between soybeans and other relevant plant species were first determined in A. thaliana, which contained 12 MTPs in past research (AtMTP1 to 12) [16]. These findings would be the basis for determining the practical characteristics, especially the GmaMTP protein substrate-specificities. The CDS length, protein size, MW, pI, GRAVY, sub-cellular localization, and TMD quantity of the GmaMTPs were dissected and subsequently predicted. Following the investigation of Vatansever, Filiz and Eroglu [16], NNtMTPs may act as vacuole-localized cation transporters, as some MTP proteins are anticipated to be found in the vacuole while others are found in the cellular membrane or nucleus. Unlike other plant MTP families, where the MTP12 has the largest molecular size [7], GmaMTP2.1 and GmaMTP2.2 were roughly one-third the mass of the other GmaMTPs. (Table 1). In our article, we explored the gene synteny and duplication examination for more information about the genome annotation and the development system of the MTP gene family in soybean (Fig. 2 and Table S2). The exit of at least two genes on a similar chromosome is frequently connected with tandem duplication, while segmental duplication regularly happens on various chromosomes [31].Our article explains two pair duplication sets while there were 28 segmental duplications. In-plant gene families, the gene duplication occasions in its sorts are trailed by difference, think about standard highlights, and are connected with auxiliary plant metabolic gene [59]. Consequently, our outcomes about the gene replications affirmed their fundamental parts in the MTP gene family development. Practically all subfamilies contained similar quantities of introns and theme sequences, predictable with the past examinations where a comparative gene construction was found inside similar subfamilies [60, 61]. For instance, all of the gene individuals from subfamily A contain five introns. Be that as it may, subfamily F individuals contain no introns. These results showed that during the soybean advancement of GmaMTPs, some intron gain and misfortune occurred. A few genes have no intron except for one exon, which causes the lower capacity of exons in gain/loss rate because of higher selection pressure in the exons sequences [62]. In this way, with this multitude of perceptions, it is plausible that the situation divergences in intron numbers consider shared occasions connected with the gene family advancement [49]. In a comprehensive consideration of the MTP proteins, we anticipated their 3D design, which was considered a supportive tool for inspecting their function [63]. The four temples in soybean MTP proteins demonstrate that these proteins, with heavy metals where these carrier proteins are in the plant, are characterized as metal-take-up proteins that shift fundamental and poisonous heavy metals to the cytoplasm and metal-take-up proteins. Simultaneously, the other is metal-efflux proteins that assist the cell with eliminating any excess heavy metals [64]. Then again, the protein–protein connection investigation gave us more information about the plant developmental processes and their roles in response to environmental stresses [65]. Moreover, the cis-regulatory elements (CREs) examination investigated the potential administrative variables influencing the expression of GmaMTPs. At long last, we recognized 3440 CREs involved in numerous biological processes. Past examinations showed that a portion of these recognized CREs were more related to pressure reactions [7].On the other hand, gene ontology is a fundamental analysis to predict putative functional contributions across living organisms [66]. Additionally, gene ontology classes and ideas have been utilized to characterize the connections and gene functions existing between these concepts [67]. Our gene ontology examination uncovered the critical role of the soybean GmaMTP genes with heavy metals (Fig. 5). Besides, the GO showed the molecular functions, where most of them participate in metal-related processes, including cation transmembrane transporter activity, transporter activity, transmembrane transporter activity, and ion transmembrane transporter activity.
The previous transcriptomic data helps detect the presence, structure, and amount of RNA in any biological sample under certain conditions [68]. In this manner, we explored the expression profile of all individuals from the MTP gene family from recently distributed RNA-sequencing information, which showed the declaration of all gene individuals in thoroughly chosen soybean tissues (Fig. 6 and Table S5). Digital data analysis showed that the MTP gene’s critical part could contribute to development and advancement. Considerable evidence about the fundamental part of soybean MTPs after tissue expression assessment has been obtained. For example, the exclusive expression of the three genes GmaMTP1.2, GmaMTP7.1, and GmaMTP11.1 were in the young leaf, though GmaMTP7.1 and GmaMTP11.1 were most abundant in flowers, showing that they may be engaged with early leaf and flower enhancement. Besides the crucial expected part of GmaMTP7.1in nodule development and maturation, its appearance has expanded.
Nonetheless, just GmaMTP4.2 was rarely expressed in totally analyzed tissues from all GmaMTPs. The archived down-regulation in some gene expressions is fundamental for keeping up with the gene duplicates and ancestral functions [69]. Subsequently, in our finding, the down-regulation of GmaMTP4.2 expression is relied upon as essential for keeping their biological processes and keeping up with them from misfortune during cell evaluation.
The dependability of the transcriptome information was additionally approved by qRT-PCR; nevertheless, the minor imbalance between the two examinations might be because of various development conditions and soybean assortments, which at last impacted the spatial expression. We analyzed the expressional behavior of MTP genes under five divalent metals (Mn2+, Cd2+, Co2+, Fe2+, and Zn2+). Various examinations in different plants showed the massive part of the MTP gene family to improve the plant tolerance against these metals [11, 15] as it was depicted as metal efflux transporters out of the cytoplasm, basically shipping Zn2+, yet additionally moves Ni+2, Co2+, Cd2+, Fe2+, and Mn2+ [57]. The transcription of the transcript amassing a record of MTPs in light of different heavy metals fluctuated and confounded. Nonetheless, the gene’s expressional reaction to various stresses usually is reflected in comparing gene functioning. In Arabidopsis, the tonoplast-limited Zn carrier AtMTP1 expressed bit modifications in expression with an overabundance of Zn openness at transcription and translation levels [70, 71]. Furthermore, despite the fact that the high expression of CsMTP1 encoded protein, gene expression in cucumber was consistent despite the high amount of Zn2+ [72].
As previously stated, AtMTP12 up-regulation is independent of Zn concentration. Still, it can carry Zn via joining AtMTP5 in a heterodimeric complex form [24], which is comparable to the findings of et al. [7] in tobacco. Furthermore, differing Mn2+ sources had little impact on Mn-CDF expression (AtMTP8, 9, 10, and 11). [22]. Close Same findings have been reported in the tobacco industry [7]. Except for GmaMTP5.1 and GmaMTP5.2, all Zn-CDF members showed a considerable increase in expression when exposed to high Zn2+ in our investigation. Additionally, in various soybean tissues, the up-regulation of GmaMTP7.2 of Zn/Fe-CDFs exceeds Zn2+, but it is down-regulated by Fe2+. Moreover, the deposition of Mn2+ significantly impacted all members of the Mn-CDF class, except for the tree genes GmaMTP10.1 and GmaMTP10.3. As a result, our research will be critical in determining the molecular functions of MTP in soybeans during diverse heavy metal stresses. Generally, a total of 20 potential GmaMTPs in the soybean genome were successfully identified and analyzed for a phylogenetic relationship, chromosomal distributions, gene structures, gene ontology, cis-elements, and previous gene expression. Besides, the expression assay of MTPs has been examined under five divalent heavy metals (Cd2, Co2, Mn2, Zn2, and Fe2) treatments.
Substantially, our discoveries will help to understand the functions of GmaMTP proteins in heavy metal resistance as well as the methodology of heavy metal transport regulated by GmaMTP proteins. These findings, taken collectively, would provide a practical and theoretical platform for future studies on the operational identification of GmaMTP genes. Consequently, depending on their expression levels, the most highly expressed MTPs (GmaMTP1.1, GmaMTP1.2, GmaMTP3.1, GmaMTP3.2, GmaMTP4.1, and GmaMTP4.3) can be used as bio-environmental markers for predicting the heavy metal accumulation.
Conclusions
In soybeans, genome-wide recognition identified twenty MTP genes, phylogenetically and extensively studied. The GmaMTPs were split into three substrate-specific clusters (Zn/Fe-CDFs, Zn-CDFs, and Mn-CDFs). Six groups seemed to have undergone expansion and gene loss after polyploidization via segmental duplication. The cation efflux region and/or ZT dimerization domain are projected to be present in all GmaMTPs, and each MTP within the same group has the same structural characteristics. Aside from predicting cis-elements, gene ontology provides valuable information regarding the critical functions of MTPs during plant growth and tolerance. The expression patterns of each GmaMTPs gene in retaliation to several heavy metals in diverse tissues revealed that these genes are essential for soybean growth and expansion. In addition, we discovered that GmaMTP1.1, GmaMTP1.2, GmaMTP3.1, GmaMTP3.2, GmaMTP4.1, and GmaMTP4.3 play a significant role in heavy metal stress tolerance in plants.
Data availability
Provided as supplementary material.
References
Thomine S, Vert G (2013) Iron transport in plants: better be safe than sorry. Curr Opin Plant Biol 16:322–327
Kolaj-Robin O, Russell D, Hayes KA, Pembroke JT, Soulimane T (2015) Cation diffusion facilitator family: structure and function. FEBS Lett 589:1283–1295
El-Sappah AH, Rather SA (2022) Genomics Approaches to Study Abiotic Stress Tolerance in Plants. Plant Abiotic Stress Physiology: Volume 2: Molecular Advancements 25
Clemens S (2001) Molecular mechanisms of plant metal tolerance and homeostasis. Planta 212:475–486
El-Sappah AHAM (2013) Utilization of micronucleus test and genetic markers for determining the pollution of Nile Tilapia (Oreochromis niloticus). Zagazig University
Ali I, Asim M, Khan TA (2013) Arsenite removal from water by electro-coagulation on zinc–zinc and copper–copper electrodes. Int J Environ Sci Technol 10:377–384
Liu J, Gao Y, Tang Y, Wang D, Chen X, Yao Y, Guo Y (2019) Genome-wide identification, comprehensive gene feature, evolution, and expression analysis of plant metal tolerance proteins in tobacco under heavy metal toxicity. Front Genet 10:345
El-Sappah AH, Elrys AS, Desoky E-SM, Zhao X, Bingwen W, El-Sappah HH, Zhu Y, Zhou W, Zhao X, Li J (2021) Comprehensive genome wide identification and expression analysis of MTP gene family in tomato (Solanum lycopersicum) under multiple heavy metal stress. Saudi J Biol Sci 28:6946–6956
Krzesłowska M (2011) The cell wall in plant cell response to trace metals: polysaccharide remodeling and its role in defense strategy. Acta Physiol Plant 33:35–51
El-Sappah AH, Seif MM, Abdel-Kader HH, Soaud SA, Elhamid MAA, Abdelghaffar AM, El-Sappah HH, Sarwar H, Yadav V, Maitra P, Zhao X, Yan K, Li J, Abbas M (2022) Genotoxicity and trace elements contents analysis in Nile Tilapia (Oreochromis niloticus) indicated the levels of aquatic contamination at three Egyptian areas. Front. Vet. Sci 9:818866
Montanini B, Blaudez D, Jeandroz S, Sanders D, Chalot M (2007) Phylogenetic and functional analysis of the Cation Diffusion Facilitator (CDF) family: improved signature and prediction of substrate specificity. BMC Genomics 8:1–16
Gustin JL, Zanis MJ, Salt DE (2011) Structure and evolution of the plant cation diffusion facilitator family of ion transporters. BMC Evol Biol 11:1–13
van der Zaal BJ, Neuteboom LW, Pinas JE, Chardonnens AN, Schat H, Verkleij JA, Hooykaas PJ (1999) Overexpression of a novel Arabidopsis gene related to putative zinc-transporter genes from animals can lead to enhanced zinc resistance and accumulation. Plant Physiol 119:1047–1056
Shirazi Z, Abedi A, Kordrostami M, Burritt DJ, Hossain MA (2019) Genome-wide identification and characterization of the metal tolerance protein (MTP) family in grape (Vitis vinifera L). 3 Biotech 9:1–17
Gao Y, Yang F, Liu J, Xie W, Zhang L, Chen Z, Peng Z, Ou Y, Yao Y (2020) Genome-wide identification of metal tolerance protein genes in Populus trichocarpa and their roles in response to various heavy metal stresses. Int J Mol Sci 21:1680
Vatansever R, Filiz E, Eroglu S (2017) Genome-wide exploration of metal tolerance protein (MTP) genes in common wheat (Triticum aestivum): insights into metal homeostasis and biofortification. Biometals 30:217–235
El-Sappah AH, Elbaiomy RG, Elrys AS, Wang Y, Zhu Y, Huang Q, Yan K, Xianming Z, Abbas M, El-Tarabily KA (2021) Genome-wide identification and expression analysis of metal tolerance protein gene family in Medicago truncatula under a broad range of heavy metal stress. Front Genet. https://doi.org/10.3389/fgene.2021.713224
Zhang X, Li Q, Xu W, Zhao H, Guo F, Wang P, Wang Y, Ni D, Wang M, Wei C (2020) Identification of MTP gene family in tea plant (Camellia sinensis L) and characterization of CsMTP8: 2 in manganese toxicity. Ecotoxicol Environ Saf 202:110904
Li X, Wu Y, Li B, He W, Yang Y, Yang Y (2018) Genome-wide identification and expression analysis of the cation diffusion facilitator gene family in turnip under diverse metal ion stresses. Front Genet 9:103
Fu X-Z, Tong Y-H, Zhou X, Ling L-L, Chun C-P, Cao L, Zeng M, Peng L-Z (2017) Genome-wide identification of sweet orange (Citrus sinensis) metal tolerance proteins and analysis of their expression patterns under zinc, manganese, copper, and cadmium toxicity. Gene 629:1–8
Hou L, Gu D, Li Y, Li J, Li J, Chen X, Zhang W (2019) Phylogenetic and expression analysis of Mn-CDF transporters in pear (Pyrus bretschneideri Rehd.). Plant Mol Biol Report 37:98–110
Delhaize E, Gruber BD, Pittman JK, White RG, Leung H, Miao Y, Jiang L, Ryan PR, Richardson AE (2007) A role for the AtMTP11 gene of Arabidopsis in manganese transport and tolerance. Plant J 51:198–210
Dhankhar R, Sainger PA, Sainger M (2012) Phytoextraction of zinc: physiological and molecular mechanism. Soil Sediment Contamin Int J 21:115–133
Fujiwara T, Kawachi M, Sato Y, Mori H, Kutsuna N, Hasezawa S, Maeshima M (2015) A high molecular mass zinc transporter MTP 12 forms a functional heteromeric complex with MTP 5 in the Golgi in Arabidopsis thaliana. FEBS J 282:1965–1979
Eroglu S, Meier B, von Wirén N, Peiter E (2016) The vacuolar manganese transporter MTP8 determines tolerance to iron deficiency-induced chlorosis in Arabidopsis. Plant Physiol 170:1030–1045
Chu H-H, Car S, Socha AL, Hindt MN, Punshon T, Guerinot ML (2017) The Arabidopsis MTP8 transporter determines the localization of manganese and iron in seeds. Sci Rep 7:1–10
Pedas P, Schiller Stokholm M, Hegelund JN, Ladegård AH, Schjoerring JK, Husted S (2014) Golgi localized barley MTP8 proteins facilitate Mn transport. PLoS ONE 9:e113759
Takemoto Y, Tsunemitsu Y, Fujii-Kashino M, Mitani-Ueno N, Yamaji N, Ma JF, Kato S-I, Iwasaki K, Ueno D (2017) The tonoplast-localized transporter MTP8. 2 contributes to manganese detoxification in the shoots and roots of Oryza sativa L. Plant Cell Physiol 58:1573–1582
Tsunemitsu Y, Yamaji N, Ma JF, Kato S-I, Iwasaki K, Ueno D (2018) Rice reduces Mn uptake in response to Mn stress. Plant Signal Behav 13:e1422466
Migocka M, Papierniak A, Maciaszczyk-Dziubińska E, Poździk P, Posyniak E, Garbiec A, Filleur S (2014) Retracted: cucumber metal transport protein MTP8 confers increased tolerance to manganese when expressed in yeast and Arabidopsis thaliana. Oxford University Press
Schlueter JA, Lin J-Y, Schlueter SD, Vasylenko-Sanders IF, Deshpande S, Yi J, O’Bleness M, Roe BA, Nelson RT, Scheffler BE (2007) Gene duplication and paleopolyploidy in soybean and the implications for whole genome sequencing. BMC Genom 8:1–16
Edwards K, Fernandez-Pozo N, Drake-Stowe K, Humphry M, Evans A, Bombarely A, Allen F, Hurst R, White B, Kernodle S (2017) A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genom 18:1–14
Price CA, Reardon EM (2001) Mendel, a database of nomenclature for sequenced plant genes. Nucleic Acids Res 29:118–119
Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz J, Ponting CP, Bork P (2004) SMART 4.0: towards genomic data integration. Nucleic Acids Res 32:D142–D144
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31:3784–3788
Li J, Zhang L, Yuan Y, Wang Q, Elbaiomy R, Zhou W, Wu H, Soaud S, Abbas M, Chen B, Zhao Dand El-Sappah AH (2021) In silico functional prediction and expression analysis of C2H2 zinc-finger family transcription factor revealed regulatory role of ZmZFP126 in maize growth. Front Genet. https://doi.org/10.3389/fgene.2021.770427
Li J, Zhang LRGE, Chen L, Wang Z, Jiao J, Zhu J, Zhou W, Chen B, Soaud SA, Abbas M, Lin N, El-Sappah AH (2022) Evolution analysis of FRIZZY PANICLE (FZP) orthologs explored the mutations in DNA coding sequences in the grass family (Poaceae). PeerJ 10:e12880
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Islam M, Qi S, Zhang S, Amin B, Yadav V, El-Sappah AH, Zhang F, Liang Y (2022) Genome-Wide Identification and Functions against Tomato Spotted Wilt Tospovirus of PR-10 in Solanum lycopersicum. Int J Mol Sci 23:1502
Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH (2008) Synteny and collinearity in plant genomes. Science 320:486–488
Wei F, Coe E, Nelson W, Bharti AK, Engler F, Butler E, Kim H, Goicoechea JL, Chen M, Lee S (2007) Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet 3:e123
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645
Hu B, Jin J, Guo A-Y, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31:1296–1297
Bailey TL, Williams N, Misleh C, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34:W369–W373
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27:297–300
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Kelley L, Mezulis S, Yates C, Wass M, Sternberg M (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protocols 10:845–858
Conesa A, Götz S (2008) Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genom 2008:1–12
Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, Wishart DS (2016) Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res 44:W147–W153
El-Sappah AH, Shawky A, Sayed-Ahmad MS, Youssef M (2017) Estimation of heat shock protein 70 (hsp 70) gene expression in nile tilapia (Oreochromis niloticus) using quantitative Real-Time PCR. Zagazig J Agric Res 44:1003–1015
Abbas M, Peszlen I, Shi R, Kim H, Katahira R, Kafle K, Xiang Z, Huang X, Min D, Mohamadamin M (2020) Involvement of CesA4, CesA7-A/B and CesA8-A/B in secondary wall formation in Populus trichocarpa wood. Tree Physiol 40:73–89
Liu J, Shi M, Wang J, Zhang B, Li Y, Wang J, El-Sappah A, Liang Y (2020) Comparative transcriptomic analysis of the development of sepal morphology in tomato (Solanum Lycopersicum L). Int J Mol Sci 21:5914
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25:402–408
El-Sappah A, Shawky A, Sayed-Ahmad M, Youssef M (2012) Nile tilapia as bio indicator to estimate the contamination of water using SDS-PAGE and RAPDPCR techniques. Egypt J Genet Cytol 41:209–227
Chidambaram R (2015) Isotherm modelling, kinetic study and optimization of batch parameters using response surface methodology for effective removal of Cr (VI) using fungal biomass. PLoS ONE 10:e0116884
Elrys AS, Merwad A-R, Abdo AI, Abdel-Fatah MK, Desoky E-SM (2018) Does the application of silicon and Moringa seed extract reduce heavy metals toxicity in potato tubers treated with phosphate fertilizers? Environ Sci Pollut Res 25:16776–16787
Ricachenevsky FK, Menguer PK, Sperotto RA, Williams LE, Fett JP (2013) Roles of plant metal tolerance proteins (MTP) in metal storage and potential use in biofortification strategies. Front Plant Sci 4:144
Samuel MS, Datta S, Khandge RS, Selvarajan E (2021) A state of the art review on characterization of heavy metal binding metallothioneins proteins and their widespread applications. Sci Total Environ 775:145829
Ober D (2005) Seeing double: gene duplication and diversification in plant secondary metabolism. Trends Plant Sci 10:444–449
Liu J, Pang X, Cheng Y, Yin Y, Zhang Q, Su W, Hu B, Guo Q, Ha S, Zhang J (2018) The Hsp70 gene family in Solanum tuberosum: genome-wide identification, phylogeny, and expression patterns. Sci Rep 8:1–11
Abbas M, Li Y, Elbaiomy RG, Yan K, Ragauskas AJ, Yadav V, Soaud SA, Islam MM, Saleem N, Noor Z, Zafar S, Hussain SS, Abbas M, Abbas S, Li J, El-Sappah AH (2022) Genome-wide analysis and expression profiling of SlHsp70 gene family in solanum lycopersicum revealed higher expression of SlHsp70-11 in roots under Cd(2+) stress. Front Biosci (Landmark edition) 27:186
Harrow J, Denoeud F, Frankish A, Reymond A, Chen C-K, Chrast J, Lagarde J, Gilbert JG, Storey R, Swarbreck D (2006) GENCODE: producing a reference annotation for ENCODE. Genome Biol 7:1–9
Büyükköroğlu G, Dora DD, Özdemir F, Hızel C (2018) Techniques for protein analysis. In Omics technologies and bio-engineering. Elsevier, pp 317-351.
Mani A, Sankaranarayanan K (2018) Heavy metal and mineral element-induced abiotic stress in rice plant. Rice Crop Curr Dev. https://doi.org/10.5772/intechopen.76080
Struk S, Jacobs A, Sánchez Martín-Fontecha E, Gevaert K, Cubas P, Goormachtig S (2019) Exploring the protein–protein interaction landscape in plants. Plant, Cell Environ 42:387–409
Consortium GO (2010) The gene ontology in 2010: extensions and refinements. Nucleic Acids Res 38:D331–D335
Purwantini E, Torto-Alalibo T, Lomax J, Setubal JC, Tyler BM, Mukhopadhyay B (2014) Genetic resources for methane production from biomass described with the GENE Ontology. Front Microbiol 5:634
Zambounis A, Ganopoulos I, Valasiadis D, Karapetsi L, Madesis P (2020) RNA sequencing-based transcriptome analysis of kiwifruit infected by Botrytis cinerea. Physiol Mol Plant Pathol 111:101514
Qian W, Liao B-Y, Chang AY-F, Zhang J (2010) Maintenance of duplicate genes and their functional redundancy by reduced expression. Trends Genet 26:425–430
Dräger DB, Desbrosses-Fonrouge AG, Krach C, Chardonnens AN, Meyer RC, Saumitou-Laprade P, Krämer U (2004) Two genes encoding Arabidopsis halleri MTP1 metal transport proteins co-segregate with zinc tolerance and account for high MTP1 transcript levels. Plant J 39:425–439
Kobae Y, Uemura T, Sato MH, Ohnishi M, Mimura T, Nakagawa T, Maeshima M (2004) Zinc transporter of Arabidopsis thaliana AtMTP1 is localized to vacuolar membranes and implicated in zinc homeostasis. Plant Cell Physiol 45:1749–1758
Migocka M, Kosieradzka A, Papierniak A, Maciaszczyk-Dziubinska E, Posyniak E, Garbiec A, Filleur S (2015) Retracted: two metal-tolerance proteins, MTP1 and MTP4, are involved in Zn homeostasis and Cd sequestration in cucumber cells. Oxford University Press
Acknowledgements
The authors are grateful to and acknowledge the Sichuan Province Government for providing such a well-equipped platform for conducting research, the management of Yibin University for their support and for providing us with a pleasant environment for research, and the Chinese Government and the Chinese public in general for their love of Science and Research. We thank Yibin University’s High-level Talent Project for its assistance during the experimental investigation.
Funding
We would like to thank the Sichuan Provincial Government, as well as Yibin University's High-level Talent Project (No.2018RC07), for their assistance throughout the experimental investigation.
Author information
Authors and Affiliations
Contributions
Conceptualization: AHES, MA, HQ and JL. Formal analysis: AHES, JL, SHW and HQ. Writing an original draft: AHES. Methodology: AHES, ASE, ZN and NS. Writing, review and editing; AHES, MA, SAR, SHW, MB and JL. Investigation: AHES, HQ and RRM.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest and nothing to disclose.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ahmed H. El-Sappah, Manzar Abbas, Shabir A. Rather and Huang Qiulan share the first authorship. Shabir A. Rather
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
El-Sappah, A.H., Abbas, M., Rather, S.A. et al. Genome-wide identification and expression analysis of metal tolerance protein (MTP) gene family in soybean (Glycine max) under heavy metal stress. Mol Biol Rep 50, 2975–2990 (2023). https://doi.org/10.1007/s11033-022-08100-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-08100-x