Abstract
Metal transport process in plants is a determinant of quality and quantity of the harvest. Although it is among the most important of staple crops, knowledge about genes that encode for membrane-bound metal transporters is scarce in wheat. Metal tolerance proteins (MTPs) are involved in trace metal homeostasis at the sub-cellular level, usually by providing metal efflux out of the cytosol. Here, by using various bioinformatics approaches, genes that encode for MTPs in the hexaploid wheat genome (Triticum aestivum, abbreviated as Ta) were identified and characterized. Based on the comparison with known rice MTPs, the wheat genome contained 20 MTP sequences; named as TaMTP1–8A, B and D. All TaMTPs contained a cation diffusion facilitator (CDF) family domain and most members harbored a zinc transporter dimerization domain. Based on motif, phylogeny and alignment analysis, A, B and D genomes of TaMTP3–7 sequences demonstrated higher homology compared to TaMTP1, 2 and 8. With reference to their rice orthologs, TaMTP1s and TaMTP8s belonged to Zn-CDFs, TaMTP2s to Fe/Zn-CDFs and TaMTP3–7s to Mn-CDFs. Upstream regions of TaMTP genes included diverse cis-regulatory motifs, indicating regulation by developmental stage, tissue type and stresses. A scan of the coding sequences of 20 TaMTPs against published miRNAs predicted a total of 14 potential miRNAs, mainly targeting the members of most diverged groups. Expression analysis showed that several TaMTPs were temporally and spatially regulated during the developmental time-course. In grains, MTPs were preferentially expressed in the aleurone layer, which is known as a reservoir for high concentrations of iron and zinc. The work identified and characterized metal tolerance proteins in common wheat and revealed a potential involvement of MTPs in providing a sink for trace element storage in wheat grains.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Metals are incorporated into proteins and activate enzymes, which catalyze essential biological reactions. Thus, a deficiency of metals results in serious negative impacts on plant biomass (Marschner 2012). Contrastingly, metals can cause toxicity when their concentration is in excess within the cytosol, (Thomine and Vert 2013). Therefore, their cellular concentrations must be tightly regulated to achieve optimum growth. In addition, as a parameter of food quality, metal concentrations in edible parts of the plants raise significant attention for human nutrition. In particular, insufficient intake of Fe and Zn from plant-based diets exacerbates deficiencies of those elements in humans, which are considered among the most widespread nutritional disorders in the world (White and Broadley 2009). To combat this, increasing the concentration of Fe and Zn in edible parts of the staple crops, also known as biofortification, has been considered to be one of the most sustainable approaches to the problem (White and Broadley 2009). A major factor determining the partition of nutrients in plant tissues is their relative sink strengths for the respective metal; where sink strength refers to the competitive ability of an organ to attract assimilates (Marcelis 1996). So, to transfer more of a metal to edible parts of the plant, sink strength can either be increased in the edible organs or decreased in the non-edible parts. For example, increasing Fe sink strength in transgenic rice seeds, by overexpressing ferritin, a protein that captures Fe in a nano-cage structure led to an increase in total seed Fe concentration (Qu et al. 2005). In a different study, a decrease of sink strength in the non-edible flag leaf of rice by disrupting proteins which sequestrate Fe into vacuoles resulted in an increase of total seed Fe concentration (Zhang et al. 2012). At the subcellular level, the sink strength of a particular metal in the cell is generated by the exclusion of this metal from the cytoplasm, which often depends on membrane-bound metal transporter proteins (Clemens et al. 2002).
The plant metal tolerance proteins (MTPs) are divalent-cation/H+ antiporters and generally effluxers of metals out of the cytoplasm (Gustin et al. 2011). MTPs usually act as homodimers and contain six transmembrane domains (TMDs) with cytosolic N- and C-terminals (Lu and Fu 2007; Lu et al. 2009; Kolaj-Robin et al. 2015). MTPs mainly transport Mn, Zn and Fe metals but they also have affinities for other divalent cations such as Ni or Cd (Peiter et al. 2007; Cubillas et al. 2013). MTPs are classified based on their metal specificities, as either of the Mn-, Zn- or Fe/Zn- CDF (cation diffusion facilitator family) type (Montanini et al. 2007). MTPs were further phylogenetically grouped and each phylogenetically distinct cluster was named according to the founding Arabidopsis thaliana member of the cluster (Gustin et al. 2011). In A. thaliana several MTPs have been characterized. AtMTP1 is ubiquitously expressed in all tissues and transport Zn into the vacuoles (Kobae et al. 2004). AtMTP3 and AtMTP8 are involved in metal tolerance during the Fe-deficiency response, the former transporting Zn and the latter Mn (Arrivault et al. 2006; Eroglu 2015; Eroglu et al. 2016). AtMTP11 confers Mn tolerance to the plants by transporting Mn into endosomal vesicles which are then secreted out of the cytoplasm (Delhaize et al. 2007; Peiter et al. 2007). AtMTP12 has recently been characterized as a Zn transporter and contrary to other MTPs which contain 6 TMDs, it possesses 14 TMDs and acts as a heterodimer with AtMTP5 (Fujiwara et al. 2015). Knowledge on MTPs is lower in species other than Arabidopsis (Ueno et al. 2015). Most of the characterized members of monocot MTPs belong to rice.
Although it is one of the most important staple crops in the world, knowledge on the metal transporters of wheat is scarce. Such information would aid in the development of wheat varieties that translocate more Fe and Zn to their seeds or that are more tolerant to metal stresses. This work aimed to identify and characterize wheat MTPs using various bioinformatics approaches.
Materials and methods
Mining of MTP genes in the wheat genome
Eight known rice MTP protein sequences,OsMTP1 (Q688R1.1), OsMTP2 (Q10LJ2.1), OsMTP3 (Q6Z7K5.1), OsMTP4 (Q10PP8.1), OsMTP5 (Q5NA18.1), OsMTP6 (Q0DHJ5.2), OsMTP7 (Q9LDU0.1) and OsMTP8 (Q8H329.2) were retrieved from UniProtKB database (uniprot.org/). These reference sequences were queried against the T. aestivum (TGACv1) genome in Ensembl Plants portal (plants.ensembl.org/Triticum_aestivum/) with a near match search sensitivity using BLASTP and highest hit entries (>e−90 value) for each genome component (A, B and D) was retrieved for use in further analyses. Subsequently, a Hidden Markov Model (HMM) search was applied to retrieved MTP sequences in Pfam (pfam.xfam.org/) to verify the protein domains.
Sequence analysis of MTP genes/proteins
Physico-chemical features of MTP proteins were calculated using ProtParam tool (web.expasy.org/protparam/). Sub-cellular localizations were predicted in Plant-mPLoc server (csbio.sjtu.edu.cn/bioinf/plant-multi/). Membrane protein topologies of MTPs were predicted using TOPCONS (topcons.net/). MTP proteins were aligned by using Bioedit (Hall 2011). Conserved motifs in MTP sequences were predicted by MEME tool with such parameters; max number of motifs to find, 5; and min/max motif width, 6–50 (inclusive) (meme-suite.org/tools/meme; Bailey et al. 2009). Gene duplication events in TaMTPs were investigated using such criteria; (a) length of alignable sequence covers >75% of longer gene, and (b) similarity of aligned regions >75% (Gu et al. 2002; Ozyigit et al. 2016). Phylogenetic tree of MTP proteins was constructed by MEGA 6 using the Maximum likelihood (ML) method with 1000 bootstraps (Tamura et al. 2013). 1000 bp upstream flanks were retrieved using genomic coordinates of TaMTP genes via BioMart-EnsemblPlants (http://www.plants.ensembl.org/biomart/martview/; Kinsella et al. 2011). Cis-regulatory elements in upstream flanks were analyzed in PlantCARE database (http://www.bioinformatics.psb.ugent.be/webtools/plantcare/html/; Lescot et al. 2002). TaMTP coding sequences were scanned for TaMTP-targeted miRNAs in psRNATarget database (plantgrn.noble.org/psRNATarget/; Dai and Zhao 2011) with parameters: max expectation, 3 and target accessibility (UPE), 25. TaMTP proteins were modelled by using Phyre2 server at intensive mode (sbg.bio.ic.ac.uk/phyre2/; Kelley et al. 2015) and structure validation was done by using Ramachandran plot (http://www.mordred.bioc.cam.ac.uk/~rapper/rampage.php/; Lovell et al. 2003). Predicted models were superposed using CLICK server, which calculates the root mean square deviation (RMSD) values based on α-carbon superposition (mspc.bii.a-star.edu.sg/minhn/; Nguyen et al. 2011).
Expression profiles of TaMTP genes
Expression profiles of TaMTP genes were retrieved from a specialized wheat database, WheatExp (http://www.wheat.pw.usda.gov/wheatexp/; Pearce et al. 2015). Coding sequences of TaMTPs were searched using nBLAST algorithm in this database, and highest hit entries (e-value 0.0) complying with genome component (A, B or D genome) of search sequences were downloaded for expression analysis. Expression values were calculated as FPKM (fragments per kilobase of exon per million fragments mapped) from RNA-seq datasets including a developmental time-course in five tissues (spike, root, leaf, grain and stem; Choulet et al. 2014), grain layer developmental time-course (10, 20 and 30 days after anthesis; Pfeifer et al. 2014), and senescing leaf time-course (heading date, 12 and 22 days after anthesis; Pearce et al. 2014).
Results and discussions
Identification of wheat MTP homologs
Wheat is a substantial global cereal grain essential to the human diet that is hexaploid with three genome components A, B and D. The ancestral genomes are thought to be derived from T. urartu (A-genome) and an unknown grass species associated with Aegilops speltoides (B-genome). Reportedly, the first hybridization resulted in tetraploid (AABB) emmer wheat (T. dicoccoides) whose hybridization again with A. tauschii (D-genome) raised the modern wheat T. aestivum (AABBDD) (International Wheat Genome Sequencing Consortium 2014). Currently, availability of an ordered draft genome of wheat allowed the metal tolerance protein (MTP) family genes to be mined by employing various bioinformatics tools and approaches (International Wheat Genome Sequencing Consortium 2014). Herein, using previously identified rice MTP (OsMTP1-8) sequences as a reference, MTP orthologs in the wheat genome were identified using blastp search with near match sensitivity and highest hit entries for each genome component A, B and D were retrieved for further analysis. The homology search and subsequent HMM verification resulted in a total of 20 MTP sequences, that are hereafter annotated as TaMTP1-8A, B or D based on the phylogenetic distribution with known rice MTPs (refer to phylogenetic analysis section; Table 1).
Protein family searches revealed that identified wheat MTPs belong to the cation efflux (cation diffusion facilitator, CDF) family (Table 2) whose members have been reported to transport Co2+, Cd2+, Ni2+ in addition to Fe2+, Mn2+ and Zn2+ (Ricachenevsky et al. 2013). In all wheat MTPs except TaMTP1 and TaMTP8, a zinc transporter dimerization domain, ZT_dimer (PF16916) was identified. This domain may indicate a dimerization region in complete zinc transporters since full-length members have been reported to form a homodimer during transport activity (Lu and Fu 2007; Lu et al. 2009; Kolaj-Robin et al. 2015). Identified MTP genes were distributed on A, B and D genomes of five wheat chromosomes (chr1-4 and 6) and they encoded a protein of 333–698 amino acid residues with 37.0–76.0 kDa molecular weight and 4.94–8.88 pI value, excluding TaMTP8B since it contains some undefined residues. Although the variation of the size of MTP proteins regarding to the number of amino acid residues was found to be quite large in wheat, this is not unusual for MTPs. For example, in Arabidopsis, AtMTP12 is approximately two times the size of AtMTP1; they have 798 and 398 amino acid residues, respectively. In accordance with the literature (Lu and Fu 2007), these proteins were mainly predicted to localize to the tonoplast having six putative TMDs with cytosolic N- and C-terminus. Metal specificity of TaMTP genes was assessed by using ortholog rice genes as a reference for different CDF groups according to Montanini et al. (2007). TaMTP1s and TaMTP8s belonged to Zn-CDFs; TaMTP2s to Fe/Zn-CDFs; and TaMTP3–7s to Mn-CDFs. Since most of the wheat MTPs belong to the group which shows specificity to Mn and Zn, MTPs may play major roles in the Mn and Zn homeostasis of wheat. In summary, putting aside some species-specific variations, primary sequence features of herein identified wheat MTPs mainly seemed to comply with previous reports.
Conserved motifs/residues in TaMTPs
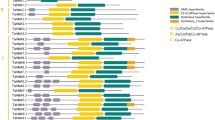
The protein motifs are highly conserved amino acid residues which are considered to possibly have functional and/or structural roles in active proteins (Yamasaki et al. 2013). In this sense, the five most conserved motif sequences in identified TaMTP proteins were searched using MEME tool (Fig. 1). The set of 20 TaMTP sequences analyzed varied between 324 and 851 residues with an average length of 458 residues. Predicted motifs 1–4 were 50 amino acids in length while motif 5 was 41 residues. Motif 1 comprised with residues “VKLALWFYCRTFGNNIVRAYAQDHYF DVITNVVGLVAAVLGDYFYWWIDP”, motif 2 with “LDLMSGFILWFTHFSMKKPNKYK YPIGKKRMQPVGIIVFASVMACLGFQV”, motif 3 with “DTVRAYTFGTHYFVEVDIVLP EDMPLKEAHDIGESLQEKIEQLPEVERAF”, motif 4 with “GAIILAVYTITNWSMTVWE NVVSLVGRSAPPEYLQKLTYLCWNHDKQIRH”, and motif 5 with “KQSEFAMKISN YANMVLFAGKVYATYRSGSMAIIASTLDSL”. From these, motif 2 was related to the cation efflux family (Cation_efflux; PF01545) and motif 3 was associated with the zinc transporter dimerization domain (ZT_dimer; PF16916), while motif 1, 4 and 5 did not relate to any motifs. All TaMTP proteins, except TaMTP1, 2 and 8 members, harbored all five of these conserved motif sequences, implying that MTP variants having the same motifs may possess more similarities in their functional roles. This claim was also corroborated by analysis of the metal specificity of TaMTP genes, according to which TaMTP3-7 s were classified as the Mn-CDF type (also refer to Table 2). In addition, the presence of consecutive preserved motif residues in most TaMTPs, related to either the cation efflux family or zinc transporter dimerization domain, indicate that MTP structures in wheat are well conserved.
The block diagram representation of five most conserved motif sequences in A, B and D genomes of 20 TaMTP proteins. Motif 2 was related with cation efflux family (Cation_efflux; PF01545), motif 3 was associated with zinc transporter dimerization domain (ZT_dimer; PF16916), and motif 1, 4 and 5 did not relate to any motifs
Moreover, to have further insights about this conservation, identified MTP sequences were multiple-aligned by ClustalW, and identical and similar residues respectively were shaded as black and grey (Fig. 2). The approximate location of the zinc transporter dimerization domain (ZT_dimer; PF16916) is specified with a red rectangle on the alignment in which TaMTP3-7 sequences were found to be significantly conserved. On the other hand, it was less conserved in TaMTP2 members. Sites corresponding to these residues were diverged in TaMTP1 and 8 sequences, since they do not contain zinc transporter dimerization domain. In addition, approximate locations of five identified motifs were also specified with different colored rectangles (motif 1 with orange, motif 2 with blue, motif 3 with purple, motif 4 with green and motif 5 with brown). Among these, motives 3 (completely) and 4 (partially) showed higher significance, since they are localized within the dimerization domain. The alignment also revealed that A, B and D genomes of TaMTP3-7 sequences are significantly conserved between themselves, this could be associated with their functional similarities in metal homeostasis as mentioned earlier (refer to Table 2). In contrast, TaMTP1 and 8 members demonstrated a significant divergence from others, inferring their involvement in different processes. Interestingly, Gly (G) in motif 2, and Asp (D) and His (H) in motif 1 were also found to be strictly preserved in all wheat MTP sequences, which may indicate specific functions for these amino acids.
Multiple sequence alignment of identified 20 wheat TaMTP (A, B and D genomes) protein sequences. The relevant identical and similar residues are shaded as black and grey respectively. Red rectangle approximately covers the site of zinc transporter dimerization domain (ZT_dimer; PF16916). The approximate localization of identified motifs is specified with different color rectangles such as motif 1 is with orange, motif 2 with blue, motif 3 with purple, motif 4 with green and motif 5 with brown
Chromosomal locations of TaMTPs
It has been reported that the large size and polyploid nature of the wheat genome have been major constraints in genome analyses. In addition, the hexaploid wheat genome has been highly dynamic with significant reductions in the size of gene families upon domestication and polyploidization, and with the abundance of gene fragments (Brenchley et al. 2012). To reveal further insights into functional diversifications, possible gene duplication events in wheat TaMTP members were analyzed. 20 TaMTP genes were mapped on five wheat chromosomes (chr1–4 and 6; refer to Table 2). The chromosomes 1A and 1D included the TaMTP1A and TaMTP1D genes respectively; chromosomes 2A, 2B and 2D respectively had TaMTP8A, TaMTP8B and TaMTP8D genes; chromosomes 3A, 3B and 3D contained TaMTP5–7A, TaMTP5–7B and TaMTP5–7D genes respectively; chromosomes 4A, 4B and 4D respectively included TaMTP2A and 4A, TaMTP2B and 4B, and TaMTP2D and 4D genes; and chromosome 6 had TaMTP3A, TaMTP3B and TaMTP3D genes respectively. Accordingly, A, B and D genomes of chromosomes 3 and 4 possessed a maximum of 2 TaMTP genes for each, while each genome component of other chromosomes contained only a single TaMTP gene. Genomic duplications are considered to be an essential driving force in evolution of plants. Segmental duplications refer to DNA sequences with an identity rate usually more than >90%, ranging from 1 to 400 kb in length and occur on multiple sites within the genome (Ramsey and Schemske 1998). Segmental duplications occur on different chromosomes, contrasting to tandem duplications which occur on the same chromosomes (Cannon et al. 2004). Herein, the segmental-like duplications are found within each MTP group in A, B and D genomes. This implies that MTPs from each genome donor, T. urartu (A-genome), A. speltoides (B-genome) and A. tauschii (D-genome) either may have been ancestrally similar to each other or originally divergent MTPs could have been stabilized during long domestication process.
Phylogenetic distribution of wheat and rice MTPs
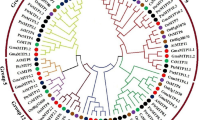
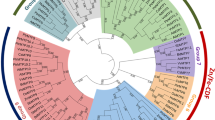
Another main objective of this work was to comparatively investigate the phylogenetic distribution of identified wheat MTP sequences along with known rice MTPs and to infer functional relationships at the cross-species level. In this concept, phylogeny was constructed with the ML method using a total of 28 sequences from eight rice MTPs and 20 wheat homologs (Fig. 3). Based on the distribution of sequences and clustering topology, the tree was divided into seven major groups; group MTP1–4, 5/6, 7 and 8. Each rice MTP had three corresponding homologs in the wheat genome since wheat has three genome components; A, B and D. However, rice MTP1 was only present in A and D genomes but could not be identified in the B genome for the adopted search parameters. In phylogeny, group MTP1 included two TaMTPs and OsMTP1. OsMTP1 has been previously thoroughly characterized. OsMTP1 was recovered from a cDNA library screening in which Cd responsive genes were sought in rice roots (Yuan et al. 2012). Based on yeast complementation assays and elemental analysis of OsMTP1 overexpressor and knock-out mutants, OsMTP1 was proposed to transport a wide range of metals including Zn, Fe, Co, Ni, Arsenic (As)(Yuan et al. 2012; Menguer et al. 2013, Das et al. 2016). In group MTP4, three TaMTPs clustered together with OsMTP4. OsMTP4, also called OsMTP8.1, has been characterized to transport Mn rather specifically to confer Mn tolerance to rice plants by sequestrating excess Mn in the aerial parts (Chen et al. 2013). In group MTP7, three TaMTPs and an OsMTP7 were together. Contrasting to other characterized rice MTPs which localize to the tonoplast, OsMTP7 localizes to the plasma membrane and is involved in the radial transport of Mn in the roots (Ueno et al. 2015). OsMTPs in other groups have not yet been characterized. According to the phylogenetic analysis (Fig. 3), group MTP1 and 8 members were clearly diverged from other MTPs, which is in agreement with alignment and protein domain analyses (refer to Figs. 1, 2). Moreover, a further phylogeny was also constructed using wheat, rice and Arabidopsis MTPs altogether to figure out homologous sequences between monocots and dicots (Suppl. File). Arabidopsis MTPs and their putative transport element specificities were obtained from Ricachenevsky et al. (2013). In phylogeny, Mn-CDFs, Fe/Zn-CDFs and Zn-CDFs showed a clear separation where monocot OsMTP3–7 and dicot AtMTP8–11 were identified as functional Mn-CDF orthologs, OsMTP2 and AtMTP6–7 as functional Fe/Zn-CDF orthologs, and OsMTP1, 8 and AtMTP1–5, 12 as functional Zn-CDF orthologs. Phylogenetic distributions can be employed as a benchmark to infer structure and functional roles across species (Sze et al. 2014; Vatansever et al. 2016). Thus, wheat MTP sequences can be functionally inferred in relation to their respective homologs. Nevertheless, to infer more precise roles to the identified TaMTP variants, further molecular and physiological experimental characterization is required.
Phylogenetic distribution of identified wheat and known rice MTP sequences. Phylogeny was constructed with ML method using a total of 28 sequences from eight rice MTPs (OsMTP1-8) and 20 wheat homologs (marked with red diamonds). Phylogeny was divided into seven major groups based on distribution of sequences such as group MTP1-4, 5/6, 7 and 8. Each rice MTP has three corresponding homologs in wheat genome since wheat has three genome components as A, B and D. However, rice MTP1 was only present in A and D genomes but could not be identified in B genome
Promoter site and miRNA-target analysis of TaMTPs
This study also attempted to understand the regulation of the identified TaMTP genes at the transcriptional level. It was thus imperative to have insights about the upstream regions of genes from the transcription start-site (TSS; Ravel et al. 2015). 1000 bp upstream flanks of TaMTP genes were retrieved using genomic coordinates of genes via EnsemblPlants-BioMart and supplied to the PlantCARE database for cis-regulatory element analysis. Excluding unknown motifs, a total of 64 different cis-elements have been identified in upstream regions of 20 TaMTP genes and to better characterize these large number of motifs a heatmap was constructed based on the availability of elements in each corresponding gene (Fig. 4). Cis-regulatory elements CAAT-box, TATA-box (except for MTP5/6A) and G-box were commonly shared by all TaMTP genes. Notably, CAAT- and TATA boxes are two common cis-regulatory elements in upstream regions of eukaryotic genes. Particularly, CAAT-box, located ~ 80 bp upstream site, forms a binding site for RNA transcription factors and also is a key regulatory motif in modulation of the expression frequency of genes (Laloum et al. 2013). TATA-box is another core element in upstream regions of eukaryotic genes. It forms the binding site for general transcription factors or histone proteins and its binding factor may also involve in the transcription process (Bae et al. 2015). Another cis-element, G-box, was found in the promoter regions of all TaMTPs. G-box, mainly present in promoters of light-responsive genes, is reported to be involved in the light-responsive processes and its binding factors are usually demonstrated to be members of bHLH, bZIP and NAC families (Kircher et al. 1998; Toledo-Ortiz et al. 2003; Liu et al. 2016). Additionally, the promoter regions of most TaMTP genes also harbored cis-regulatory elements such as Sp1 (light responsiveness), CGTCA- and TGACG-motifs (methyl jasmonate responsiveness), ABRE (abscisic acid responsiveness), GCN4- and Skn-1 motifs (endosperm expression) and MBS (drought inducibility). Moreover, many other cis-regulatory elements present in promoter regions of TaMTPs genes were broadly categorized as common cis-acting elements, light responsive elements, hormone responsive elements, tissue-specific elements, and stress responsive and other elements based on their putative functions. Particularly, most cis-regulatory motifs in TaMTP genes were mainly associated with light responsive elements such as ACE, Box I, box II, GATA-motif, GT1-motif, I-box, LS7, TCT-motif, Box 4, CATT-motif, GA-motif, GAG-motif, L-box, MNF1, TCCC-motif, 4 cl-CMA2b, AAAC-motif, ATCT-motif, LAMP-element, rbcS-CMA7a, MRE, chs-CMA1a, 3-AF1 binding site and AE-box. Significantly, many other phytohormone-related cis-elements were also identified in TaMTP promoters including SARE and TCA-element (salicylic acid responsiveness), ERE (ethylene responsiveness), TGA-element and AuxRR (auxin responsiveness), ABRE and motif IIb (abscisic acid responsiveness), and P-box, TATC-box and GARE-motif (gibberellin responsive element). Additionally, tissue-specific cis-regulatory elements such as the CCGTCC-box, dOCT, NON-box and CAT-box (meristem), motif I (root), RY-element (seed), as-2-box (shoot), and HD-Zip 1 and 2 (leaf) were also found in the promotors of TaMTP genes. Furthermore, upstream regions of TaMTP genes also harbored the TC-rich repeats (defense/stress responsiveness), ARE and GC-motif (anaerobic induction), EIRE and Box-W1 (elicitor responsiveness), LTR and HSE (temperature responsiveness), C-repeat/DRE (cold and dehydration responsiveness), circadian (circadian control), and O2-site (zein metabolism regulation) elements. Taken together, the presence of very diverse cis-regulatory elements associated with various metabolic processes indicates the intricate but very dynamic regulation of TaMTP genes.
Distributions of putative cis-regulatory elements in 1000 bp upstream regions of 20 TaMTP genes. Available motifs in corresponding genes are specified with green boxes otherwise with reds (right side). Motifs are also demonstrated with different colors based on their putative functions (left side) such as blue (cis-acting elements), orange (light responsive elements), green (hormone responsive elements), yellow (tissue-specific elements) and grey (stress responsive and other elements). Unknown function motifs are not shown herein
Furthermore, potential miRNAs targeting TaMTP transcripts were also investigated to give insights into the post-transcriptional regulation of wheat genes. miRNAs are short (~21–25 nucleotides) but highly conserved non-coding RNA sequences and they perform their functions either by degrading the target mRNAs or by repressing their translations (Kumar 2014). miRNAs are accounted to regulate about 10–30% of genes in higher eukaryotes (Cui et al. 2006). They are reported to regulate various metabolic processes including development and differentiation, various biotic/abiotic stresses, signal transduction and morphogenesis (Lv et al. 2012). Herein, a scan of the 20 TaMTP coding sequences against all published miRNAs from different species resulted in a total of 14 potential miRNAs (Table 3). TaMTP1A and D were targeted by ath-miR837-3p with translation inhibition; TaMTP2B and D were targeted by mtr-miR319b-5p with translation inhibition; TaMTP7B was targeted by stu-miR8041a/b-5p with cleavage inhibition; TaMTP8A was targeted by gma-miR5772 and mtr-miR5253 with cleavage, and by osa-miR1858a/b with translation inhibition; TaMTP8B was targeted by mtr-miR5253 with cleavage and by osa-miR1858a/b with translation inhibition; and TaMTP8D was targeted by gma-miR5772 with cleavage inhibition. Notably, inferring from alignment and phylogenetic analyses (refer to Figs. 2, 3) the herein predicted miRNAs mainly targeted the members of most diverged groups, implying that their susceptibility for degradation may be related to their functions. In previous works, miR837-3p was reported to be involved in phosphate signal transduction in tomato leaves (Gu et al. 2010). A miR837-3p target was possibly involved in the oxidative pentose phosphate pathway (Meng et al. 2012). miR837-3p was increased in response to nitrogen and sulfur deficiencies but decreased under carbon deficiency (Liang et al. 2015). miR837-3p was significantly downregulated under Cu stress in Paeonia ostii seedlings (Jin et al. 2015). miR319a-5p with most other miRNAs were highly expressed under shading conditions in maize (Yuan et al. 2016). Expression of miR319a-5p was implicated in rhizomes of Oryza longistaminata (Zong et al. 2014) and in the flowering of Oryza rufipogon (Chen et al. 2013a, b). gma-miR319a-5p was reported as one of the most important restorer gene families in plants (Chen and Liu 2014). miR5772 and miR5253 were differentially expressed in tolerant and sensitive cultivars of pepper leaves under high temperatures and high air humidity (Xu et al. 2015). Osa-miR1858a/b was reported to be regulated by southern rice black-streaked dwarf virus (SRBSDV) infection in rice (Xu et al. 2014).
Temporal and spatial expression of TaMTPs
In order to build a basis to predict physiological functions of MTPs in wheat, organ level expression of MTPs in a time-course was analyzed by using publicly available transcriptome data. During a developmental time course, the expression of MTPs showed organ specificity (Fig. 5; Suppl. File). Interestingly, MTP5/6s were expressed mostly in leaves and not in roots, whereas MTP7s expression showed the opposite pattern. MTP4s were expressed in the root as well as in the shoot, unlike its closest ortholog in rice, which was solely expressed in the shoot (Chen et al. 2013). In grains, all analyzed MTPs were first sharply downregulated at 14 DAA (grain_Z71 to grain_Z75) and then upregulated at 30 DAA (grain_Z75 to grain_Z85). The only exception to that was MTP7s, which showed a gradual decrease in their transcript levels. Notably n leaves, all three MTP5/6 showed a clear upregulation as the leaf aged. In root, MTP1s and MTP7s were expressed highly. In spikes the MTP3s, MTP4s and MTP7s were upregulated developmentally. In stems, MTP1s were downregulated, while MTP3s were upregulated gradually during development. As the second node of wheat formed (z32), expression of MTP7s peaked. Overall, the temporal and spatial regulation of MTPs may indicate that MTPs are actively involved in maintaining wheat nutrient homeostasis throughout its life cycle.
Expression profiles of TaMTP genes during developmental timecourse of grain, leaf, root, spike and stem. Expression values are specified as FPKM (Fragments Per Kilobase Of Exon Per Million Fragments Mapped). Developmental timecourses are based on Zadoks cereal developmental scale (refer to Suppl. File)
Senescence is associated with an extensive micronutrient remobilization from leaves to seeds. In accordance with that, in wheat, the disruption of Gpc-B1, a gene encoding a NAC transcription factor involved in the regulation of leaf senescence, resulted in seeds with low concentrations of Fe and Zn (Uauy et al. 2006). This was accompanied by higher concentrations of these elements in the flag leaf, underlining the source-sink relationship between the flag leaf and seed during senescence. In order to investigate MTPs involvement in metal homeostasis in senescencing wheat, MTP activity in both flag leaf and seed was analyzed. Expression analysis of MTPs in the flag leaf revealed that, with an exception of a slight downregulation of TaMTP5/6B, none of the MTPs transcriptionally responded to senescence (Fig. 6; Suppl. File). This data indicated that MTPs are not primarily involved in the remobilization of micronutrients from senescencing leaves.
In seeds of wheat, metal micronutrients are not homogeniously distributed, but instead show organ and tissue level specificity. Most of the Fe, Mn and Zn are found in the aleurone layer of seeds and the embryo; while the endosperm,the largest part of the seed, is poor of these micronutrients (Mazzolini et al. 1985; Borg et al. 2009; Regvar et al. 2011). In order to investigate the possible involvement of MTPs in storage mineral accumulation, activity of MTPs in wheat seeds was analyzed (Fig. 7; Suppl. File). Surprisingly, in general MTP transcript levels were found to be high and regulated throughout development. TaMTPs that were expressed highly in the endosperm included TaMTP1s, TaMTP2s, and TaMTP7A. Expression of TaMTP5/6s in the seeds was high but confined to the aleurone layer. Interestingly, all MTPs were preferentially expressed in the aleurone during grain filling (20DPA and 30DPA). The aleurone layer of the grain stores minerals, subcellularly in the globoids of the vacuoles (Regvar et al. 2011). Sequestration of storage minerals into vacuoles requires tonoplast localized transporter proteins as shown in Arabidopsis for Fe (Kim et al. 2006) and for Mn (Eroglu 2015). Thus, considering that they are usually localized to tonoplasts (Fig. 2), TaMTPs are likely to be involved in making up mineral reserves in the vacuoles of cells in the aleurone layer of wheat grains. In particular, since the closest orthologs of TaMTP1s were characterized as a tonoplast-bound Zn transporter in both Arabidopsis and rice, TaMTP1s may be responsible for higher Zn contents in the vacuoles of the aleurone tissue. In summary, temporal and spatial expression of TaMTPs indicated that they are not likely to play major roles in metal remobilization in senescencing leaves, but instead are likely to be involved in reserve metal accumulation by providing an intercellular sink by effluxing metals out of the cytosol.
Homology modelling of TaMTP proteins
Finally, all 20 wheat TaMTP members, except for TaMTP8B which contained several consecutive undefined residues in its protein sequence, were modelled using Phyre2 server at intensive mode (Fig. 8). Predicted models were based on following templates to heuristically maximize the alignment coverage, percentage identity and confidence score for the tested sequences. Two templates, 2QFI and 3J1Z from structures of zinc transporter YiiPs were used in the modelling of TaMTP1A/D, TaMTP3A/B/D, TaMTP4A, TaMTP5/6A/B/D and TaMTP8A; templates 5ENS (from bacterial efflux pump), 2QFI and 3J1Z in TaMTP2A/D models; templates 2QFI, 3J1Z, 1GHH (from E. coli DinI protein) and 1IWG (from bacterial multidrug efflux transporter AcrB) in TaMTP2B model; templates 2ENK (from human solute carrier family 30/Zn transporter), 2QFI and 3J1Z in TaMTP4B/D and TaMTP7A models; templates 2ENK, 5A39 (from Rad14), 1D4U (from human nucleotide excision repair protein XPA), 2QFI and 3J1Z in TaMTP7B model; templates 2QFI, 3J1Z and 4L6R (from human glucagon G protein coupled receptor) in TaMTP7D model; and templates 2QFI, 3J1Z, and 5GAS (from Thermus thermophilus V/A-ATPase) employed in TaMTP8D model. The quality of models was validated by Ramachandran plot analysis in which >80% of residues were in allowed region indicating the fairly good structures of models. However, it was apparent that to construct more reliable, native-like models the more experimentally solved structures are required from CDF family proteins in particular from plant MTPs. The α-helices primarily constituted the secondary structures of modelled wheat proteins with 48–67% whereas β-strands distributed with a 1−10%.Ttransmembrane (TM) helices possessed a percentage of 19-38 in secondary structure of TaMTPs with mainly six putative TMDs. Moreover, to figure out the similarity or divergence of generated models, structures were superimposed to calculate the percentages of structure overlap. The structure overlap is regarded as e.g, given X and Y proteins the percentage of representative atoms in X protein that is within 3.5 Å of the respective atoms in the superimposed Y protein. The calculations were emphasized on the A, B and D genomes of each TaMTP group within itself (refer to phylogenetic section) to estimate how preserved each genome component during long hybridization and polyploidization processes were structurally, and thereby also functionally. The superimposed TaMTP1A-D models were indicated by 76.88% structure overlap, TaMTP2A-B by 69.12%, TaMTP2A-D by 69.12%, TaMTP2B-D by 44.62%, TaMTP3A-B by 69.51%, TaMTP3A-D by 73.46%, TaMTP3B-D by 80.56%, TaMTP4A-B by 73.75%, TaMTP4A-D by 66.25%, TaMTP4B-D by 68.25%, TaMTP5/6A-B by 73.51%, TaMTP5/6A-D by 72.03%, TaMTP5/6B-D by 77.23%, TaMTP7A-B by 46.58%, TaMTP7A-D by 48.68%, TaMTP7B-D by 60.10%, and TaMTP8A-D by 33.04%. A, B and D genomes of each TaMTP group mainly demonstrated a 66-76% structural overlap. However, some models such as TaMTP2B-D (44.62%), TaMTP7A-B (46.58%), TaMTP7A-D (48.68%) and TaMTP8A-D (33.04%) showed low structure similarity but above the twilight zone (<30%). Taken together, it has been implicated that MTPs from each genome donor, T. urartu (A-genome), A. speltoides (B-genome) and A. tauschii (D-genome) either may have been ancestrally similar to each other or originally divergent MTPs could have been stabilized during long domestication process resulting in changes on protein structures thereby on protein functions.
Predicted 3D models of wheat TaMTP proteins. Models were generated by using Phyre2 server at intensive mode. TaMTP8B structure could not be satisfactorily predicted since it contained some consecutive undefined residues. Models were visualized by rainbow color from N to C terminus and organized in order as TaMTP1-8A, B and D
Conclusion
Wheat is a globally produced cereal grain and plays a crucial role in global food security. However, potential genetic biofortification tools for wheat including the knowledge of MTPs are largely missing due to its large hexaploid genome size. This study identified 20 MTPs in wheat and annotated as TaMTP1-8A, B or D based on their phylogeny. Their preferential expression in the aleurone of the grains, indicated that TaMTPs contribute to build up the rich metal reserves found in this tissue. This study showed potential importance of MTPs in the biofortification of grains with essential micronutrients and will serve as a basis for the physiological characterization of wheat MTPs.
References
Arrivault S, Senger T, Krämer U (2006) The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. Plant J 46(5):861–879
Bae SH, Han HW, Moon J (2015) Functional analysis of the molecular interactions of TATA box-containing genes and essential genes. PLoS ONE 10(3):e0120848
Borg S, Brinch-Pedersen H, Tauris B, Holm PB (2009) Iron transport, deposition and bioavailability in the wheat and barley grain. Plant Soil 325(1–2):15–24
Brenchley R, Spannagl M, Pfeifer M, Barker GL, D’Amore R, Allen AM, Kay S (2012) Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491(7426):705–710
Bailey TL, Boden M, Buske, FA, Frith M, Grant, CE, Clementi L & Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res, gkp335
Cannon SB, Mitra A, Baumgarten A, Young ND, May G (2004) The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4(1):1
Chen L, Liu YG (2014) Male sterility and fertility restoration in crops. Annu Rev Plant Biol 65:579–606
Chen Z, Li F, Yang S, Dong Y, Yuan Q, Wang F, Pei X (2013a) Identification and functional analysis of flowering related microRNAs in common wild rice (Oryza rufipogon Griff.). PLoS ONE 8(12):e82844
Chen Z et al (2013b) Mn tolerance in rice is mediated by MTP8. 1, a member of the cation diffusion facilitator family. J Exp Bot 64(14):4375–4387
Choulet F, Alberti A, Theil S, Glover N, Barbe V, Daron J, Leroy P (2014) Structural and functional partitioning of bread wheat chromosome 3B. Science 345(6194):1249721
Clemens S, Palmgren MG, Krämer U (2002) A long way ahead: understanding and engineering plant metal accumulation. Trends Plant Sci 7(7):309–315
Cubillas C, Vinuesa P, Tabche ML, García-de los Santos A (2013) Phylogenomic analysis of cation diffusion facilitator proteins uncovers Ni2+/Co2+ transporters. Metallomics 5(12):1634–1643
Cui Q, Yu Z, Purisima EO, Wang E (2006) Principles of microRNA regulation of a human cellular signaling network. Mol Syst Biol 2(1):46
Dai X, Zhao PX (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 39(suppl 2):W155–W159
Das N et al (2016) Enhanced cadmium accumulation and tolerance in transgenic tobacco overexpressing rice metal tolerance protein gene OsMTP1 is promising for phytoremediation. Plant Physiol Biochem 105:297–309
Delhaize E, Gruber BD, Pittman JK, White RG, Leung H, Miao Y, Richardson AE (2007) A role for the AtMTP11 gene of Arabidopsis in manganese transport and tolerance. Plant J 51(2):198–210
Eroglu S (2015) Characterization of MTP8 as a tonoplast Fe/Mn transporter essential for Fe efficiency and for Fe and Mn localization in the subepidermis of Arabidopsis embryos (Doctoral dissertation, Halle (Saale), Universitäts-und Landesbibliothek Sachsen-Anhalt, Diss., 2015)
Eroglu S, Meier B, von Wirén N, Peiter E (2016) The vacuolar manganese transporter MTP8 determines tolerance to iron deficiency-induced chlorosis in Arabidopsis. Plant Physiol 170(2):1030–1045
Fujiwara T, Kawachi M, Sato Y, Mori H, Kutsuna N, Hasezawa S, Maeshima M (2015) A high molecular mass zinc transporter MTP12 forms a functional heteromeric complex with MTP5 in the Golgi in Arabidopsis thaliana. FEBS J 282(10):1965–1979
Gu Z, Cavalcanti A, Chen FC, Bouman P, Li WH (2002) Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol Biol Evol 19(3):256–262
Gu M, Xu K, Chen A, Zhu Y, Tang G, Xu G (2010) Expression analysis suggests potential roles of microRNAs for phosphate and arbuscular mycorrhizal signaling in Solanum lycopersicum. Physiol Plant 138(2):226–237
Gustin JL, Zanis MJ, Salt DE (2011) Structure and evolution of the plant cation diffusion facilitator family of ion transporters. BMC Evol Biol 11(1):1
Hall T (2011) BioEdit: an important software for molecular biology. GERF Bull Biosci 2(1):6
International Wheat Genome Sequencing Consortium (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345(6194):1251788
Jin Q, Xue Z, Dong C, Wang Y, Chu L, Xu Y (2015) Identification and characterization of microRNAs from tree peony (Paeonia ostii) and their response to copper stress. PLoS ONE 10(2):e0117584
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10(6):845–858
Kim SA, Punshon T, Lanzirotti A, Li L, Alonso JM, Ecker JR, Kaplan J, Guerinot ML (2006) Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science 314(5803):1295–1298
Kinsella RJ, Kähäri A, Haider S, Zamora J, Proctor G, Spudich G & Kersey P (2011) Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database, bar030
Kircher S, Ledger S, Hayashi H, Weisshaar B, Schäfer E, Frohnmeyer H (1998) CPRF4a, a novel plant bZIP protein of the CPRF family: comparative analyses of light- dependent expression, post-transcriptional regulation, nuclear import and heterodimerisation. Mol Gen Genet MGG 257(6):595–605
Kobae Y, Uemura T, Sato MH, Ohnishi M, Mimura T, Nakagawa T, Maeshima M (2004) Zinc transporter of Arabidopsis thaliana AtMTP1 is localized to vacuolar membranes and implicated in zinc homeostasis. Plant Cell Physiol 45(12):1749–1758
Kolaj-Robin O, Russell D, Hayes KA, Pembroke JT, Soulimane T (2015) Cation diffusion facilitator family: structure and function. FEBS Lett 589(12):1283–1295
Kumar R (2014) Role of microRNAs in biotic and abiotic stress responses in crop plants. Appl Biochem Biotechnol 174(1):93–115
Laloum T, De Mita S, Gamas P, Baudin M, Niebel A (2013) CCAAT-box binding transcription factors in plants: y so many? Trends Plant Sci 18(3):157–166
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30(1):325–327
Liang G Ai Q, Yu D (2015) Uncovering miRNAs involved in crosstalk between nutrient deficiencies in Arabidopsis. Sci Rep, 5
Liu L, Xu W, Hu X, Liu H, Lin, Y (2016) W-box and G-box elements play important roles in early senescence of rice flag leaf. Sci Rep, 6
Lovell SC, Davis IW, Arendall WB, de Bakker PI, Word JM, Prisant MG, Richardson DC (2003) Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins Struct Funct Bioinformat 50(3):437–450
Lu M, Fu D (2007) Structure of the zinc transporter YiiP. Science 317(5845):1746–1748
Lu M, Chai J, Fu D (2009) Structural basis for autoregulation of the zinc transporter YiiP. Nat Struct Mol Biol 16(10):1063–1067
Lv S, Nie X, Wang L, Du X, Biradar SS, Jia X, Weining S (2012) Identification and characterization of microRNAs from barley (Hordeum vulgare L.) by high-throughput sequencing. Int J Mol Sci 13(3):2973–2984
Marcelis LFM (1996) Sink strength as a determinant of dry matter partitioning in the whole plant. J Exp Bot 47:1281–1291
Marschner H (2012) Marschner’s mineral nutrition of higher plants, vol 89. Academic press, London
Mazzolini AP, Pallaghy CK, Legge GJF (1985) Quantitative microanalysis of Mn, Zn and other elements in mature wheat seed. New Phytol 100(4):483–509
Meng Y, Shao C, Ma X, Wang H, Chen M (2012) Expression-based functional investigation of the organ-specific microRNAs in Arabidopsis. PLoS ONE 7(11):e50870
Menguer PK, Farthing E, Peaston KA, Ricachenevsky FK, Fett JP, Williams LE (2013). Functional analysis of the rice vacuolar zinc transporter OsMTP1. J Exp Bot 64(10):2871–2883
Montanini B, Blaudez D, Jeandroz S, Sanders D, Chalot M (2007) Phylogenetic and functional analysis of the Cation Diffusion Facilitator (CDF) family: improved signature and prediction of substrate specificity. BMC Genom 8(1):1
Nguyen MN, Tan KP, Madhusudhan MS (2011) CLICK—topology-independent comparison of biomolecular 3D structures. Nucleic Acids Res 39(suppl 2):W24–W28
Ozyigit II, Filiz E, Vatansever R, Kurtoglu KY, Koc I, Öztürk MX, Anjum NA (2016) Identification and comparative analysis of H2O2-scavenging enzymes (ascorbate peroxidase and glutathione peroxidase) in selected plants employing bioinformatics approaches. Front Plant Sci 7:301
Pearce S, Tabbita F, Cantu D, Buffalo V, Avni R, Vazquez-Gross H, Dubcovksy J (2014) Regulation of Zn and Fe transporters by the GPC1 gene during early wheat monocarpic senescence. BMC Plant Biol 14(1):1
Pearce S et al (2015) WheatExp: an RNA-seq expression database for polyploid wheat. BMC Plant Biol 15(1):1
Peiter E, Montanini B, Gobert A, Pedas P, Husted S, Maathuis FJ, Sanders D (2007) A secretory pathway-localized cation diffusion facilitator confers plant manganese tolerance. Proc Natl Acad Sci USA 104(20):8532–8537
Pfeifer M, Kugler KG, Sandve SR, Zhan B, Rudi H, Hvidsten TR, International Wheat Genome Sequencing Consortium (2014) Genome interplay in the grain transcriptome of hexaploid bread wheat. Science 345(6194):1250091
Qu LQ, Yoshihara T, Ooyama A, Goto F, Takaiwa F (2005) Iron accumulation does not parallel the high expression level of ferritin in transgenic rice seeds. Planta 222(2):225–233
Ramsey J, Schemske DW (1998) Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu Rev Ecol Syst 29:467–501
Ravel C, Fiquet S, Boudet J, Dardevet M, Vincent J, Merlino M, Martre P (2015) Conserved cis-regulatory modules in promoters of genes encoding wheat high-molecular- weight glutenin subunits. Adv Seed Biol, 150
Regvar M, Eichert D, Kaulich B, Gianoncelli A, Pongrac P, Vogel-Mikuš K, Kreft I (2011) New insights into globoids of protein storage vacuoles in wheat aleurone using synchrotron soft X-ray microscopy. J Exp Bot 62:3929–3939
Ricachenevsky FK, Menguer PK, Sperotto RA, Williams LE, Fett JP (2013) Roles of plant metal tolerance proteins (MTP) in metal storage and potential use in biofortification strategies. Front Plant Sci 4:144
Sze H, Chen LQ, Jez JM (2014) Evolutionary relationships and functional diversity of plant sulfate transporters. Evol Transp Plants, 63
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729
Thomine S, Vert G (2013) Iron transport in plants: better be safe than sorry. Curr Opin Plant Biol 16(3):322–327
Toledo-Ortiz G, Huq E, Quail PH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15(8):1749–1770
Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J (2006) A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314(5803):1298–1301
Ueno D, Sasaki A, Yamaji N, Miyaji T, Fujii Y, Takemoto Y, Ma JF (2015) A polarly localized transporter for efficient manganese uptake in rice. Nature plants 1:15170
Vatansever R, Koc I, Ozyigit II, Sen U, Uras ME, Anjum NA, Filiz E (2016) Genome-wide identification and expression analysis of sulfate transporter (SULTR) genes in potato (Solanum tuberosum L.). Planta 244(6):1167–1183
White PJ, Broadley MR (2009) Biofortification of crops with seven mineral elements often lacking in human diets–iron, zinc, copper, calcium, magnesium, selenium and iodine. New Phytol 182(1):49–84
Xu D, Mou G, Wang K, Zhou G (2014) MicroRNAs responding to southern rice black- streaked dwarf virus infection and their target genes associated with symptom development in rice. Virus Res 190:60–68
Xu XW, Li T, Li Y, Li ZX (2015) Identification and analysis of C. annuum microRNAs by high-throughput sequencing and their association with high temperature and high air humidity stress. Int J Bioautom 19:459–472
Yamasaki K, Kigawa T, Seki M, Shinozaki K, Yokoyama S (2013) DNA-binding domains of plant-specific transcription factors: structure, function, and evolution. Trends Plant Sci 18(5):267–276
Yuan L, Yang S, Liu B, Zhang M, Wu K (2012) Molecular characterization of a rice metal tolerance protein, OsMTP1. Plant Cell Rep 31(1):67–79
Yuan L, Tang J, Liu J, Song H, Zhang M, Li H, Li C (2016) Differential miRNA expression in maize ear subjected to shading tolerance. Acta Physiol Plant 38(3):1–12
Zhang Y, Xu YH, Yi HY, Gong JM (2012) Vacuolar membrane transporters OsVIT1 and OsVIT2 modulate iron translocation between flag leaves and seeds in rice. Plant J 72(3):400–410
Zong Y, Huang L, Zhang T, Qin Q, Wang W, Zhao X, Li Z (2014) Differential microRNA expression between shoots and rhizomes in Oryza longistaminata using high- throughput RNA sequencing. The Crop Journal 2(2):102–109
Acknowledgements
We thank Benjamin Gruber for the fruitful discussion and proofreading of the article. Seckin Eroglu thanks Scientific and Technological Council of Turkey (Ankara, Turkey) for the fellowship through BIDEB-2232 program.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Vatansever, R., Filiz, E. & Eroglu, S. Genome-wide exploration of metal tolerance protein (MTP) genes in common wheat (Triticum aestivum): insights into metal homeostasis and biofortification. Biometals 30, 217–235 (2017). https://doi.org/10.1007/s10534-017-9997-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10534-017-9997-x