Abstract
In the present study three species of mackerel, Rastrelliger present in Indian waters were taken for genetic identification using cytochrome c oxidase subunit I and 16s rRNA sequences. Accurate identification of these species is important for fishery management as its morphological characters are very similar. In this study, the sequences of COI and 16S rRNA were determined from 19 individuals of three Rastrelliger species, Rastrelliger kanagurta, Rastrelliger brachysoma and Rastrelliger faughni from Andamans and Indian mainland to study the phylogenetic relationship. The intraspecies and interspecies genetic distance ranged from 0.000 to 0.002 and 0.007 to 0.015 respectively based on 16S rRNA sequences. Using COI data analysis, the intraspecies genetic distance ranged from 0.000 to 0.012, while it varied from 0.039 to 0.086 for interspecies. The present study clearly demarcates three species of mackerel based on the mitochondrial genetic sequences and also showed a non-descriptive genetic distance of R. kanagurta from mainland and Andaman Islands.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The scombrid fish genus Rastrelliger comprises three species: short mackerel (Rastrelliger brachysoma Bleeker, 1851), Indian mackerel (Rastrelliger kanagurta Cuvier, 1817) and Island mackerel (Rastrelliger faughni Matsui, 1967) which inhabits the tropical waters of Indo Pacific regions. These three closely related species are found in Indian waters. The chub mackerel, Rastrelliger is considered to be highly variable with a reported number of ten species but which have now been reduced to synonyms of three species: Rastrelliger kanagurta, Rastrelliger brachysoma and Rastrelliger faughni [3]. They are important food items for the Southeast Asian region. Indian mackerel is the chief target species for both industrial and small-scale fisheries along the Andaman Sea coast [9]. Conventionally, scombrid fishes are identified using morphomeristic characters. Still, significant doubts exist due to morphological resemblance. Correct identification of morphologically similar species is important for fisheries management and population studies.
The application of molecular genetics contributed significantly in species and strain identification. Mitochondrial DNA (mtDNA) is widely used for phylogenetic studies because its evolution is faster than nuclear DNA, and it can be used for differentiating closely related species [16]. Partial sequences of mitochondrial genes especially 16S rRNA and cytochrome-c oxidase subunit I are suitable than other gene sequences to resolve species ambiguity and are used as a “molecular clock” to estimate the chronology of different taxa [7]. Molecular identification tools provide a useful alternative for species identification, especially in cases where conventional methods may result in ambiguities. Hebert et al. [7] demonstrated the utility of mitochondrial COI gene in species delineation. CO1 gene sequence is appropriate for this role because its mutation rate is often fast enough to differentiate closely related species and also because its sequence is conserved among congenerers. The mitochondrial 16S rRNA gene can also be used to explore the phylogenetic relationships of fishes at the generic level [12]; and the species level [2].
In this study, the sequences of COI and 16S were determined from 19 individuals of three Rastrelliger species, R. kanagurta, R. brachysoma and R. faughni from Indian waters to study the phylogenetic relationship.
Materials and methods
Sample collection
Samples of R. kanagurta, R. brachysoma and R. faughni fishes were collected from local fish landings at Andamans (five numbers of each species). R. kanagurta samples were also collected from Indian mainland (four numbers). The fishes were identified up to species level following FAO [1]. A piece of muscle tissue was taken from all the specimens and stored in 95 % ethyl alcohol and stored at 4 °C in a refrigerator.
DNA isolation
DNA extraction was done from tissue samples (muscle) preserved in 95 % alcohol. DNA extraction for all samples followed the protocol of Miller et al. [11]. The extracted DNA was checked through 0.8 % agarose gel electrophoresis with ethidium bromide (5 μg/ml) incorporated in 1× TBE buffer. Final DNA concentration was detected by optical density (OD) reading using a spectrophotometer (SPECORD 205) set at 260 nm.
Amplification and sequencing
Amplification of 16S rRNA region
The mitochondrial 16S rRNA gene was amplified in a 25 µl reactions volume containing 1× assay buffer (100 mM Tris, 500 mM KCl, 0.1 % gelatin, pH 9.0) with 1.5 mM MgCl2 (Genei, Bangalore, India), 5 pmol of each primer, 200 µM of each dNTP (Genei, Bangalore, India), 1.5 U Taq DNA polymerase and 50 ng of template DNA. The primer used for the amplification of the partial 16S rRNA gene were 16SAR (5′-CGCCTGTTTATCAAAAACAT-3′) and 16SBR (5′-CCGGTCTGAACTCAGATCACGT-3′) [13]. The amplification conditions were 95 °C for 7 min followed by 30 cycles at 94 °C for 1 min, annealing 53 °C for 1 min and 72 °C for 1 min, with a final extension of 72 °C for 10 min.
Amplification of COI region
The partial sequence of COI gene was amplified using primers Fish F1 (5′-TCA ACC AAC CAC AAA GAC ATT GGC AC-3′) and Fish R1 (5′-TAG ACT TCT GGG TGG CCA AAG AAT CA-3′) [17] in 25 µl reactions volume containing 1× assay buffer (100 mM Tris, 500 mM KCl, 0.1 % gelatin, pH 9.0) with 1.5 mM MgCl2 (Genei, Bangalore, India), 5 pmol of each primer, 200 µM of each dNTP (Genei, Bangalore, India), 1.5 U Taq DNA polymerase and 20 ng of template DNA. The thermal condition consisted of initial preheat at 95 °C for 3 min, followed by denaturation at 94 °C for 30 s, annealing at 50 °C for 30 s, extension at 72 °C for 35 s, repeated for 29 cycles, followed by a final extension for 3 min at 72 °C.
For each sample, 3 µl of PCR product were electrophoresed through 1.5 % agarose gels following ethidium bromide staining, and visualized under UV illumination in the Gel-Doc system (BIO-RAD, Molecular Imager, Gel DocTM XR). Products were labelled using the Big DyeTerminator V.3.1 Cycle sequencing kit (Applied Biosystems Inc) and sequenced bidirectionally using ABI 3730 capillary sequencer following the manufacturer’s instructions.
Sequence analysis
The raw DNA sequences were edited and aligned using BioEdit sequence alignment editor version 7.0.5.2 [6]. The 16S rRNA sequences of the individuals of each species were aligned to yield a final alignment of 590 bp. The COI sequences of the individuals of each species were aligned to yield a final alignment of 655 bp. The edited sequences were submitted to GenBank (KJ590045–KJ590062, KJ590064–KJ590082). The sequence divergence values within and between species were calculated using Kimura 2 Parameter (K2P) distance model implemented in MEGA V.5.0, molecular evolutionary genetic analysis [15] software. The number of polymorphic sites and nucleotide diversity (Pi), nucleotide composition and number of transition and transversion between species were determined by DnaSpver 3 [14]. UPGMA trees of K2P distance were created to provide graphic representation of divergence with 1,000 replications.
Results
A total of 19 individuals from three species of mackerel (R. kanagurta, R. brachysoma and R. faughni) were used for sequence analysis of partial 16S rRNA and COI genes, which yielded 38 sequences.
Genetic analysis based on 16S rRNA gene
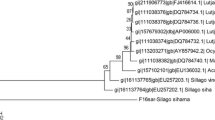
A total of 590 base pairs of aligned sequences of 16S rRNA gene were studied in all the three Rastrelliger species. In this, 575 sites were constant and 15 sites were variable or polymorphic sites (two singleton variable sites and 13 parsimony informative sites). The nucleotide frequencies were found to be A = 30.0 %, T = 22.4 %, G = 23.1 % and C = 24.4 %. The transition/transversion rate ratios are k1 = 6.197 (purines) and k2 = 12.072 (pyrimidines). The overall transition/transversion bias is R = 4.384. The mean genetic divergence value based on 16S rRNA sequences between all these three species was 0.82 %. The mean genetic distance between R. kanagurta (from Andamans, Indian mainland), R. brachysoma and R. faughni was calculated (Table 1). The highest interspecies distance was found between R. kanagurta (Indian mainland) and R. faughni (1.8 %) and the lowest was found between R. kanagurta (Andamans) and R. brachysoma (0.5 %). Pair-wise genetic distance values (K2P) based on 16S rRNA using MEGA V 5.0 are given in Table 2. All the species under Rastrelliger genera were clearly separated by different clusters in the UPGMA tree with a bootstrap value ranging from 96 to 100 % (Fig. 1). All five sequences for each species were incorporated in the phylogenetic analysis. The UPGMA and MP trees provide similar phylogenetic relationship among the species. The haplotype diversity and nucleotide diversity was found to be 0.9123 and 0.00815 respectively.
Genetic analysis based on COI gene
Sequencing of the COI gene created 655 nucleotide base pairs. Insertions, deletions or stop codons were absent in all sequences. Of the 655 sites, 582, 73, 67 and 6 were monomorphic, polymorphic, parsimony informative and singleton respectively. All variable changes within species were third codon position transitional substitutions. The nucleotide frequencies are 22.51 % (A), 27.82 % (T), 30.26 % (C), and 19.41 % (G). The transition/transversion rate ratios are k1 = 18.499 (purines) and k2 = 18.01 (pyrimidines). The overall transition/transversion bias is R = 9.547. The mean genetic divergence value calculated on the basis of COI sequences between all these three species was 4.65 %. The mean genetic distance between R. kanagurta (from Andamans, Indian mainland), R. brachysoma and R. faughniwas calculated (Table 3). Pair-wise genetic distance values (K2P) based on COI using MEGA V 5.0 are given in Table 4. The genetic distance of R. kanagurta from Indian mainland and Andamans was found to be 1.24 %. The UPGMA tree based on K2P genetic distances were created to provide a graphic representation of the patterns of divergences (Fig. 2). The haplotype diversity and nucleotide diversity was found to be 0.9590 and 0.04360 respectively.
Discussion
In traditional taxonomy, it is sometimes complicated to identify species due to lack of criteria for character selection or coding, and sometimes morphomeristic data sets may be quite arbitrary. In these cases, genetic analysis can be used as a complementary tool to establish taxonomic identity. The COI gene has been shown to be useful in species identification and resolve taxonomic ambiguity [7]. Species level identification will be useful in creating precise fishery data and can be used for developing stock structure; this will be helpful in stock specific management measures for conservation and sustainable exploitation of these species.
In the current study, three species of Rastrelliger characterized for generation of COI and 16s rRNA for delineating the taxonomic status of the species. The three species of Rastrelliger from the Indian waters were found genetically discrete from each other and clustered into three groups without any haplotypes sharing. The universal primers amplified the target region in all three species (n = 19), generated 19 sequence of COI gene (655 bp) and 19 sequences of 16s rRNA (590 bp). The primer pairs for amplifying partial sequences of COI used in the present study was developed mainly for invertebrates [17], which could be successfully amplifying an approximately 655 base pair segment in all Rastrelliger species. There was no insertion, deletion or stop codons were observed in the sequences, so it is assumed that all the amplified sequences are derived from a functional mitochondrial COI sequences only. The average length of COI gene (655 bp) and lack of stop codons in amplified sequences clearly indicates that NUMTs (nuclear mitochondrial DNA) were not sequenced. The present result is in accordance with previous studies on COI sequencing of marine fishes [10, 17].
The high degree of K2P nucleotide divergence was found with the analysis of 16S rRNA gene, indicating its ability to adequately describe interrelationship of Rastrelliger species [12] and high genetic divergence among the sciaenid species in the Indian waters was reported [10] using 16S rRNA gene sequences. Because of its slowest mutation and lower substitution rates compared with other mtDNA genes, it has been reported to be useful for analysing families, species and population [5]. The mean genetic divergence value based on 16S rRNA sequences between the Rastrelliger individuals was 0.8 % which can be compared with many other Perciformes [4]. Transitions were more than transversions in the present study in accordance with the preceding studies of mtDNA in fishes [4] which indicates the lower level of divergence. This transition transversion ratio will improve estimates of divergence and phylogenetic relationships and also work as an indirect measure of saturation (loss of phylogenetic information). In COI sequences, the Ti/Tv ratio is nearly 1:1, so that in the present study the Ti/Tv ratio is enough for the phylogenetic study of the species.
The mitochondrial Cyt b gene was used to distinguish the three species of Rastrelliger from Malaysian waters [8]. We examined the utility of the mitochondrial 16s rRNA and COI genes in resolving specific identities in the genus. Estimation of genetic distances based on the 16s rRNA gene placed the population of R. kanagurta from the Andamans closer to R. brachysoma than to R. kanagurta from the mainland. Genetic distances based on the COI gene are closer to what would be expected based on species identity. The results of our study also indicate that R. kanagurta in the Andaman sea possibly represents a subspecies, needing further study. This will be having implication in fishery management and conservation of this species. The genetic information generated from this study can be used to identify larval stages which are difficult to identify morphologically and this will help to reveal the spawning habitat of the candidate species. DNA barcodes developed in the present study can also be utilized for the identification of samples when whole specimens are unavailable.
References
Allen GR (1985) FAO species identification sheets for fishery purposes. Eastern Indian ocean and Western Central Pacific, vol 4. FAO, Rome
Chakraborty A, Iwatsuki Y (2006) Genetic variation at the mitochondrial 16S rRNA gene among Trichiurus lepturus (Teleostei: Trichiuridae) from various localities: preliminary evidence of a new species from west coast of Africa. Hydrobiologia 563:501–513
Chee PE (2000) The status of the Rastrelliger (kembung) fishery on the west coast of Peninsular Malaysia. Supplement to the report of a workshop on the fishery and management of short mackerel (Rastrelliger sp.) on the west coast of Peninsular Malaysia. Food and Agriculture Organization of the United Nations, Rome, pp 6–19
Garcia G, Wlasiuk G, Lessa EP (2000) High levels of mitochondrial cyt b divergence in annual killifishes of the genus Cynolebias (Cyprinodontiformes, Rivulidae). Zool J Linn Soc 129:93–110
Garland ED, Zimmer C (2002) Techniques for the identification of bivalve larvae. Mar Ecol Prog Ser 225:299–310
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hebert PDN, Cywinska A, Ball SL, Ward JR (2003) Biological identifications through DNA barcodes. Proc R Soc Lond B 270:313–322
Jamaluddin JAF, Ahmed AT, Basir S, Masazurah AR, Nor SAM (2010) Rastrelliger systematics inferred from mitochondrial cytochrome b sequences. Afr J Biotechnol 9(21):3063–3067
Krajangdara T, Puntuleng P, Chaleeand P, Hussadee P (2007) Reproductive biology of short mackerel Rastrelliger brachysoma (Bleeker, 1851) and Indian mackerel R. kanagurta (Cuvier, 1816) in Thai water. Technical paper no. 19/2007. Marine Fisheries Research and Development Bureau, Department of Fisheries, Ministry of Agriculture and Cooperatives, Bangkok, Thailand
Lakra WS, Goswami M, Gopalakrishnan A (2009) Molecular identification and phylogenetic relationships of seven Indian sciaenids (Pisces: Perciformes, Sciaenidae) based on 16S rRNA and cytochrome oxidase subunit I mitochondrial genes. Mol Biol Rep 36:831–839
Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cell. Nucleic Acids Res 16(3):1215
Moyer GR, Burr BM, Krajewski C (2004) Phylogenetic relationships of thorny catfishes (Siluriformes: Doradidae) inferred from molecular and morphological data. Zool J Linn Soc (Lond) 140:551–575
Palumbi S, Martin A, Romano S, McMillan WO, Stice L, Grabowski G (1991) The simple fool’s guide to PCR, version 2.0. Department of Zoology and Kewalo Marine Laboratory, University of Hawaii, Honolulu
Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R (2006) DNA sequence polymorphism, version 4.10.9
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Timm J, Figiel M, Kochzius M (2008) Contrasting patterns in species boundaries and evolution of anemone fishes (Amphiprioninae, Pomacentridae) in the centre of marine biodiversity. Mol Phylogenet Evol 49(1):268–276
Ward RD, Zemlac TC, Innes BH, Last PR, Hebert PDN (2005) DNA barcoding Australia’s fish species. Philos Trans R Soc Lond Biol 360:1847–1857
Acknowledgments
The funding support for this study from Central Marine Living Resources and Ecology (CMLRE) and The Ministry of Earth Sciences (MoES), Government of India, and BOBLME/FAO is greatly acknowledged. We express our gratitude to Dr. S Venu, Associate Proffessor Mr. Bitopan, Mr. Ravi Ranjan Kumar and Mr. Santhosh PhD scholars, Department of Ocean Studies & Marine Biology Brookshabad Campus, Pondicherry University for their help in collection of specimens. We also express our thanks to Rahul G. Kumar (NBFGR Cochin Unit) for his support in correction of the manuscript. Thanks also to anonymous reviewers for their helpful comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Basheer, V.S., Mohitha, C., Vineesh, N. et al. Molecular phylogenetics of three species of the genus Rastrelliger using mitochondrial DNA markers. Mol Biol Rep 42, 873–879 (2015). https://doi.org/10.1007/s11033-014-3710-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3710-8