Abstract
Background
Molecular phylogenetic studies of the Asian pit viper genus Gloydius have been widely published in Asia, but Korea population have not been conducted till date.
Objective
This study aimed to analyze the phylogenetic relationships of three Gloydius species (G. saxatilis, G. brevicaudus, and G. ussuriensis) from Korea with other Gloydius species, based on Cytochrome b and ND4.
Methods
We compared 160 samples representing the three species with those of 17 reference species and their phylogenetic status and genetic diversity were analyzed with concatenated sequences of two mitochondrial DNA.
Results
Korean G. brevicaudus and G. saxatilis showed high haplotype diversity and relatively low and moderate nucleotide diversity, respectively. Although G. ussuriensis showed high genetic diversity, it was low in the Baengnyeong Island population. The phylogenetic tree represented two major lineages. One major lineage comprised G. ussuriensis, G. tsushimaensis, G. blomhoffii, and G. brevicaudus. The Chinese G. ussuriensis belonged to the same clade as the Korean G. ussuriensis and was closely related to the Baengnyeong Island population. Moreover, G. tsushimaensis was closely related to G. ussuriensis from southwestern Korean and Jeju Island populations. The other major lineage comprised the remaining 12 species and G. saxatilis. Korean G. saxatilis was closely related to G. saxatilis, G. shedanoensis, and G. intermedius from China.
Conclusion
The phylogenetic status of the Korean Gloydius species in comparison with the other Gloydius species was identified. We suggesting the conservation management unit for the Baengnyeong Island population, while the current conservation status of Korean G. saxatilis is suggested to be revised to a higher level.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Gloydius is a venomous pit viper group of snakes, endemic to Asia (Russia, China, Nepal, and the Korean Peninsula) and the three species, Gloydius saxatilis, Gloydius brevicaudus, and Gloydius ussuriensis are widely distributed in Korea. These three species are divided into two groups based on their morphological characteristics (Guo and Zhang 2002): the brevicaudus group, comprising G. brevicaudus and G. ussuriensis generally having 21 rows of dorsal scales and four palatine teeth; and the intermedius group with G. saxatilis having 23 rows of dorsal scales and three palatine teeth (Gloyd and Conant 1982). These differences in morphological characteristics are also related to the ecological characteristics of the species. G. brevicaudus and G. ussuriensis prefer humid spaces such as valleys, rivers, and wetlands located at warm and low altitudes. In contrast, G. saxatilis prefers dry spaces, such as ridges located in high mountainous areas with relatively low temperatures (Do and Nam 2020; Do 2021).
Phylogenetic studies of the three species have been widely published in Asia and have raised new taxonomic concerns. A phylogenetic relationship study of six Gloydius species in China using mitochondrial DNA suggested that G. shedanoensis, a native species of Shedao Island, is closely related to G. saxatilis as a subspecies (Zhou et al. 2000). In addition, molecular phylogenetic studies of the genus Gloydius in China using mitochondrial DNA (mtDNA) (ND4 and Cytb) and nuclear DNA (nDNA) (c-mos) indicated that G. saxatilis is closely related to G. shedanoensis (Yan et al. 2012). Furthermore, a phylogeographic study revealed that G. brevicaudus, which inhabits China, was divided into three lineages, and G. brevicaudus (one individual) from Korea fell in the same lineage as that in northeastern China (Ding et al. 2011). A phylogenetic study of Gloydius using mtDNA (ND4 and Cytb) and nDNA (c-mos) revealed that the Chinese G. ussuriensis was closely related to G. blomhoffii, which branched into G. blomhoffii and G. ussuriensis from a common ancestor with G. brevicaudus (Yan et al. 2012). A study on the genus Gloydius conducted in China using mitochondrial DNA (12SR, 16SR, Cytb, and ND4), showed that Chinese G. ussuriensis was most closely related to G. tsushimaensis, a Japanese endemic species inhabiting Tsushima Island, followed by G. blomhoffii. It was also found that G. tsushimaensis and G. ussuriensis originated from a common ancestor with G. blomhoffii (Shi et al. 2018; Wang et al. 2019).
In Korea, G. ussuriensis from Gangwon-do Province and Chungcheongnam-do Province was confirmed to be morphologically similar to G. tsushimaensis (Emelianov 1929; Isogawa et al. 1994). However, molecular genetic studies on three Gloydius species (Gloydius saxatilis, Gloydius brevicaudus, and Gloydius ussuriensis) inhabiting Korea have never been published, except for one study on the complete mitochondrial genome of G. saxatilis (Lee et al. 2021).
Therefore, this study was conducted to: (1) confirm the molecular phylogenetic status of three Gloydius species inhabiting South Korea by closely examining their phylogenetic relationship with Gloydius species inhabiting Northeast Asia based on the results of previous studies; (2) compare and analyze the genetic diversity of three Gloydius species from Korea, and (3) propose a conservation unit to establish conservation strategies for Gloydius after confirming the phylogenetically isolated populations.
Materials and methods
Sample collection and DNA analysis
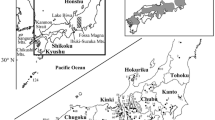
A total of 160 snakes belonging to three species G. saxatilis, G. brevicaudus, and G. ussuriensis were obtained from South Korea in 2020 (Fig. 1, Table 1). All samples were collected from licensed regions (Gangwon-do Province, Gyeonggi-do Province, Incheon Metropolitan city, Chungcheongnam-do Province, Gyeongsangbuk-do Provinces, Gyeongsangnam-do Provinces, jeollabuk-do Provinces, jeollanam-do Provinces Baengnyeong Island, and Jeju Island). Samples were obtained by collecting tail tissue after direct capture or from snakes killed on road. All samples were frozen at − 70 °C in a deep freezer at the National Institute of Biological Resources (NIBR), Incheon, South Korea, until DNA extraction.
Total genomic DNA was extracted from the tissues using QIAamp® DNA Micro Kit (Qiagen, Valencia, CA, USA) following the manufacturer’s protocol and quantified using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). The cytochrome b gene (Cytb, 936 bp) was amplified by polymerase chain reaction (PCR) using primers L14910 (5′–GAC CTG TGA TMT GAA AAC CAY CGT TGT–3′) and H16064 (5′–CTT TGG TTT ACA AGA ACA ATG CTT TA–3′) (Burbrink et al. 2000). The PCR reaction conditions were as follows: 95 °C for 5 min; 35 cycles of 95 °C for 1 min, 60 °C for 1 min (Cytb)/58 °C for 1 min (ND4), and 72 °C for 1 min; and finally, 72 °C for 5 min. The 673 bp fragment of the mtDNA NADH dehydrogenase subunit 4 gene (ND4) was amplified using primers ND4 (5′–CAC CTA TGA CTA CCA AAA GCT CAT GTA GAA GC–3′) and Leu (5′–CAT TAC TTT TAC TTG GAT TTG CAC CA–3′) (Arévalo et al. 1994). The PCR reaction conditions were as follows: 95 °C for 5 min; 35 cycles of 95 °C for 1 min, 58 °C for 1 min, and 72 °C for 1 min; and finally 72 °C for 5 min. Amplification was carried out in 20 µl reaction volumes containing 20–50 ng/template DNA, 2X Bioneer PreMix (100 µM each dNTPs, 1.5 mM MgCl2, 1 unit Taq polymerase), and 10 pmol of each primer. The PCR products were purified by the Ethanol purification method (Genotech Corp, Korea). The purified PCR products were sequenced using an ABI Prism 3730XL Analyzer (Applied Biosystems, Foster City, CA, USA). The sequencing primers for both mtDNA regions were the same as those used for amplification.
Data analysis
All analyses were conducted with concatenated sequences of two mitochondrial DNA region combined sequences. This study used 1609 bp of combined sequences to analyze the diversity of the three Gloydius species. Analysis of genetic distance and phylogenetic tree utilized 1172 bp of combined sequences according to the length of the reference sequence. The Cytb and ND4 sequences obtained from 160 individuals in this study were registered in GenBank (Table 1).
The species of each sequence obtained in this study were identified using BLAST searches (Altschul et al. 1997). Sequences were aligned using Geneious prime v11.0.4 (Kearse et al. 2012). Haplotype diversity (h), nucleotide diversity (π), and polymorphic sites (P) for each species were estimated using DNASP version 6 (Rozas et al. 2017). Pairwise genetic distances among species were calculated using MEGA X v10.1.8 (Kumar et al. 2018). To investigate the evolutionary relationships, phylogenetic trees were constructed using three methods: neighbor-joining (NJ) (Saitou and Nei 1987) using Kimura’s two-parameter distances (Kimura 1980), maximum parsimony (MP), and maximum-likelihood (ML). The reference sequence data corresponded to 46 individuals of the Gloydius species obtained from GenBank (Table 2). Deinagkistrodon acutus and Protobothrops mangshanensis were used as out-groups for phylogenetic tree construction. NJ, MP, and ML trees were constructed using MEGA X v10.1.8 (Kumar et al. 2018). The MP tree was obtained using tree bisection–reconnection (TBR) branch swapping with 10,000 bootstrap replicates. The most appropriate models of sequence evolution for ML trees were selected using MEGA X v10.1.8 (Kumar et al. 2018). The best-fit model for the ML tree was the Tamura-Nei model (TN93) with gamma distribution (+ G) and proportion of invariant sites (+ I). The consensus ML trees were found using Nearest-Neighbor-Interchange (NNI) heuristic searches of 1000 bootstrap replicates.
Results
Mitochondrial DNA diversity and genetic distance
The diversity analysis results showed that G. saxatilis (19 individuals) and G. brevicaudus (22 individuals) had 12 haplotypes, while G. ussuriensis (119 individuals) had 66 haplotypes (Table 1). Unique regional haplotypes were observed among 160 individuals of the three Gloydius species, with the exception of five haplotypes which shared geographical locations: Hap7 (Gangwon and Gyeongnam), Hap23 (Gyeonggi and Chungnam), Hap38 (Gangwon and Gyeongbuk), Hap70 (Jeonnam and Jeonbuk), and Hap79 (Gyeonggi, Gangwon, Jeonnam, Gyeongnam).
In each species, G. saxatilis and G. brevicaudus showed high haplotype diversity (h = 0.936, 0.900) and relatively low (π = 0.164), and moderate nucleotide diversity (π = 0.309), respectively (Table 3). G. ussuriensis showed an overall high haplotype and nucleotide diversity. The regional analysis results of G. ussuriensis revealed that the genetic diversity of four localities with more than 14 individuals showed high haplotype diversity (h = 0.981–0.961) and moderate nucleotide diversity (π = 0.589–0.298). However, only the Baengnyeong Island population showed low genetic diversity (h = 0.545, π = 0.069). There were some other locations with low genetic diversity (i.e., Jeju, Jeonbuk, and Daejeon) but those could not be considered to be representative due to insufficient sample size.
The pairwise genetic distance analysis of species closely related with three Gloydius species showed that G. saxatilis in South Korea was genetically similar to G. saxatilis (0.013), G. intermedius (0.012), and G. shedaoensis (0.018) in China (Table 4). However, it was confirmed that the genetic distances between G. saxatilis and G. intermedius (0.000) and G. shedaoensis (0.010) in China were slightly lower than those in South Korea. The Korean G. brevicaudus was genetically closer to the Chinese G. brevicaudus (0.006), while the Korean G. ussuriensis was genetically similar to the Chinese G. ussuriensis (0.007). In addition, G. tsushimaensis from Tsushima Island, Japan, was slightly closer to G. ussuriensis from Korea (0.020) than that from China (0.033).
Phylogenetic analysis of mitochondrial haplotype
To examine the phylogenetic status of the three Gloydius species in Korea, phylogenetic trees were constructed, including closely related species. Phylogenetic trees using NJ, MP and ML generated similar patterns of the major branches, and therefore ML tree with three bootstrap values was representatively presented in this study. The phylogenetic trees (NJ, MP, and ML) represent a monophyletic Gloydius with two major lineages (Fig. 2). One major lineage (lineage A) consisted of G. ussuriensis, G. tsushimaensis, G. blomhoffii, and G. brevicaudus. The other major lineage (lineage B) comprised the remaining 12 species and G. saxatilis. These two lineages indicated a genetic distance of approximately 0.02.
Phylogenetic haplotype trees [neighbor-joining (NJ), maximum parsimony (MP), and maximum-likelihood (ML)] of Gloydius based on mitochondrial DNA (mtDNA) ND4 and cytochrome b combined sequences (1172 bp). See Table 1 for composition of haplotype. See Table 2 for the abbreviations of scientific names (code)
Lineage A was largely divided into three clades: G. ussuriensis from Korea and China and G. tsushimaensis (clade 1), G. blomhoffii from Japan (clade 2), and G. brevicaudus from Korea and China (clade 3). In clade 1, the Korean G. usuriensis was separated into two groups, Korea main group and southwestern (Jeonnam, Jeonbuk, Jeju) Korea group. The Chinese G. ussuriensis belonged to the Korean G. ussuriensis main group and was most closely related to the Baengnyeong Island population in particular. Moreover, G. tsushimaensis from Tsushima Island was closely related to G. ussuriensis from the southwestern Korea group, especially the Jeju Island population. G. bloomhoffii (clade 2) diverged from a common ancestor of G. usuriensis. The Chinese G. brevicaudus (clade 3) was divided into three groups according to geographical location, and the Korean G. brevicaudus included the Northeastern China group (G.bre5-7). The phylogenetic tree also showed that G. usuriensis has a common ancestor with G. brevicaudus, and gradually branches into G. bloomhoffii, G. tsushimaensis, southwestern Korea G. usuriensis, and Korean and Chinese G. usuriensis.
In lineage B, most species formed species-specific clades, except G. saxatilis, G. shedanoensis, and G. intermedius from China. Despite being different species, these three represented a close genetic relationship. In addition, although Korean G. saxatilis was closely related to these three Chinese species, it was clearly divided into other clades.
Discussion
Phylogenetic status of the three Gloydius species
Molecular genetic studies on Gloydius species in Korea have not been published yet. Until recently, the phylogenetic study of Gloydius published in China was the only reference. Therefore, we established the molecular phylogenetic status of three Gloydius species inhabiting Korea by closely examining the phylogenetic relationship based on the results of previous studies.
Our results were in agreement with the results of previous studies by Zhou et al. (2000) and Yan et al. (2012), which suggested that G. saxatilis was closely related to G. shedaoensis from Shedao Island as a subspecies. However, the Korean G. saxatilis was differentiated from G. shedaoensis and G. saxatilis in China. In addition, the Korean G. saxatilis originated from a common ancestor with the Chinese G. saxatilis, but has the potential to diverge into native species due to its geographical and genetic status.
In a previous study by Ding et al. (2011), G. brevicaudus inhabiting China was divided into three lineages, and G. brevicaudus from Korea was included in the northeastern China lineage. Our study presented the same results as Ding et al. (2011) and supported previous results on G. brevicaudus from Korea. The Chinese G. brevicaudus was divided into three groups according to geographical location, while the Korean G. brevicaudus included the northeastern China group.
According to previous reports from China, G. usuriensis is genetically closely related to G. bloomhoffii and G. tsushimaensis from Japan (Yan et al. 2012; Shi et al. 2018; Wang et al. 2019). These studies also revealed that the species differentiated gradually into G. blomhoffii, G. tsushimaensis, and G. usuriensis from a common ancestor with G. brevicaudus. However, our study showed slightly different results from previous studies, wherein the Chinese G. ussuriensis was closely related to G. tsushimaensis and G. blomhoffii. The Chinese G. ussuriensis belongs to the Korean G. ussuriensis and is particularly most closely related to the Baengnyeong Island population. In addition, G. tsushimaensis was closely related to G. usuriensis from southwestern Korea and Jeju Island. The Jeju Island population has a distinct phylogenetic group and is closely related to G. tsushimaensis from Tsushima Island, Japan. It is predicted that the current phylogenetic status can be attributed to the common ancestor of G. usuriensis, which existed in Jeju and Tsushima Island in the past, due to geographical isolation and low gene flow. Unusual genetic differentiation on the island has been reported not only in Gloydius but also in many other animals (Aquadro and Kilpatrick 1981; Barton 1996; Berry 1996; Lee et al. 2015; Funk et al. 2016) for further research, phylogeographic analysis of Jeju Island and Tsushima Island needs to be conducted with sufficient populations.
Conservation unit and strategy for the Gloydius species
Gloydius saxatilis is distributed throughout the inland and several islands of South Korea, except Jeju Island, and is concentrated around the Taebaek Mountains. (Do et al. 2016). It has been reported that the main habitat of this species is forested areas at high elevations (above 400 m) (Do et al. 2017). G. saxatilis is distributed widely, but is adversely affected by poaching mainly in Korea and has been registered as an endangered species in the past (Bae 2019). Our genetic research showed that the Korean G. saxatilis had low nucleotide diversity and high haplotype diversity. This indicates that most haplotypes have one or two different nucleotides, and also suggests that the Korean G. saxatilis population expanded from a small effective population size (Grant and Bowen 1998). The number of G. saxatilis is smaller than that of other viper species in Korea, and it is highly likely that the population will decrease sharply due to habitat loss (Bae 2019). Moreover, G. saxatilis is a serious ecological threat because it is the most vulnerable to climate change among the three Gloydius species and has the highest possibility of extinction (Do et al. 2021). Our results indicate that, although G. saxatilis in Korea and China were derived from the same ancestor, the Korean G. saxatilis has the potential to diverge into a native species. Therefore, it is necessary to redesignate G. saxatilis from the current least concern (LC) category to a higher level due to its low genetic diversity and ecological vulnerability.
Gloydius brevicaudus is distributed throughout the inland regions of Korea, except for Jeju Island, and it also heavily inhabits the Taebaek Mountains (Do et al. 2016). It is known to inhabit forests, rivers, and paddy wetlands at a lower elevation (≤ 500 m) than the highlands. In addition, G. brevicaudus is exposed to the risk of population decline due to poaching, similar to other viper species in Korea (Bae 2019). The results of this study revealed that the genetic diversity of G. brevicaudus was moderate; therefore, we propose periodic monitoring of poaching, habitat loss, and genetic status.
Gloydius ussuriensis is distributed throughoutthe islands and mainland in Korea, including Jeju Island, and is concentrated around the Taebaek Mountains (Do et al. 2016). Unlike other vipers inhabiting Korea, they live in various environments, such as forests, paddy wetlands, and rivers at altitudes between 0 and 1300 m, preferring valleys in forests (Do and Yoo 2014). G. ussuriensis is designated as a prohibited species rather than an endangered species because it has the largest population among the three Gloydius species in Korea. Nevertheless, G. ussuriensis is also exposed to the risk of population decline due to poaching, similar to other viper species (Bae 2019). The results of this study showed that G. ussuriensis from the inland, which had a large population size, had moderate genetic diversity. Therefore, it appears that the northern part (lineage of the Chinese G. ussuriensis) and the southwestern part (lineage closely related to G. tsushimaensis) of the Korean Peninsula require continuous monitoring. However, G. ussuriensis from the Baengnyeong Island currently has low genetic diversity and is very vulnerable to population decline due to the characteristics of the island. In addition, G. usuriensis of the Baengnyeong Island has unique morphological characteristics (longer tail and more abdominal scales) unlike other localities (An 2020). Consequently, this study proposes the designation of the Baengnyeong Island population as a conservation management unit. Similar to the Baengnyeong Island population, G. usuriensis from Jeju Island has a distinct phylogenetic group and is closely related to G. tsushimaensis from Tsushima Island, Japan. Therefore, it is critical to analyze genetic diversity using sufficient population and establish a conservation strategies for the Jeju Island population based on these results.
In conclusion, this study accurately identified the phylogenetic status of the Korean Gloydius species in comparison with the closely related Gloydius species. Also this study suggested conservation strategies and further management strategies for genetically vulnerable Gloydius species and its isolated population.
References
Altschul SF, Madden TL, Schäffer AA et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. https://doi.org/10.1093/nar/25.17.3389
An J (2020) Genetic diversity of animal resources, vol 4_3. National Institute of Biological Resources, Incheon (In Korean)
Aquadro CF, Kilpatrick CW (1981) Morphological and biochemical variation and differentiation in insular and mainland deer mice (Peromyscus maniculatus). In: Smith MH, Joule J (eds) Mammalian population genetics. University of Georgia Press, Athens, pp 214–230
Arévalo E, Davis SK, Sites JW (1994) Mitochondrial DNA sequence divergence and phylogenetic relationships among eight chromosome races of the Sceloporus grammicus complex (Phrynosomatidae) in central Mexico. Syst Biol 43:387–418. https://doi.org/10.2307/2413675
Bae YJ (2019) Red data book of Republic of Korea. 2. Amphibians and reptiles, 2nd edn. National Institute of Biological Resources, Incheon (In Korean)
Barton NH (1996) Natural selection and random genetic drift as causes of evolution on islands. Philos Trans R Soc Lond B Biol Sci 351:785–794. https://doi.org/10.1098/rstb.1996.0073
Berry RJ (1996) Small mammal differentiation on islands. Philos Trans R Soc Lond B Biol Sci 351:753–764. https://doi.org/10.1098/rstb.1996.0070
Burbrink FT, Lawson R, Slowinski JB (2000) Mitochondrial DNA phylogeography of the polytypic North American rat snake (Elaphe obsoleta): a critique of the subspecies concept. Evolution 54:2107–2118. https://doi.org/10.1554/0014-3820(2000)054[2107:MDPOTP]2.0.CO;2
Ding L, Gan XN, He SP, Zhao EM (2011) A phylogeographic, demographic and historical analysis of the short-tailed pit viper (Gloydius brevicaudus): evidence for early divergence and late expansion during the Pleistocene. Mol Ecol 20:1905–1922. https://doi.org/10.1111/j.1365-294X.2011.05060.x
Do MS (2021) Habitat use and hiding behavior of Central Asian pit viper (Gloydius intermedius), Korean. J Herpetol 12:1–8 (In Korean)
Do MS, Nam KB (2020) Distribution patterns and ecological niches of the red-tongued pit viper (Gloydius ussuriensis) and the Central Asian pit viper (Gloydius intermedius) in Cheonmasan Mountain, South Korea. Russ J Herpetol 28:348–354. https://doi.org/10.13047/kjee.2014.28.6.657
Do MS, Yoo JC (2014) Distribution pattern according to altitude and habitat type of Red-tongue viper snake (Gloydius ussuriensis) in Cheon-ma mountain. J Wetland Res 16:193–204. https://doi.org/10.17663/JWR.2014.16.2.193
Do MS, Lee JW, Jang HJ et al (2016) Interspecific competition and spatial ecology of three species of vipers in Korea: an application of ecological niche-based models and GIS1a. Korean J Environ Ecol 30:173–184. https://doi.org/10.13047/KJEE.2016.30.2.173
Do MS, Nam KB, Yoo JC (2017) Distribution and movement tendencies of short-tailed viper snakes (Gloydius saxatilis) by altitude. Asian Herpetol Res 8:39–47. https://doi.org/10.16373/j.cnki.ahr.160126
Do MS, Choi S, Jang HJ, Suh JH (2021) Predicting the distribution of three Korean pit viper Species (Gloydius brevicaudus, G. ussuriensis and G. intermedius) under climate change. Russ J Herpetol (In press)
Emelianov AA (1929) Snakes of the far eastern district. Men Vladivostok Sec Russ State Geogr Soc 3:1–208 (In Russian)
Fenwick AM, Greene HW, Parkinson CL (2012) The serpent and the egg: unidirectional evolution of reproductive mode in vipers? J Zool Syst Evol Res 50:59–66. https://doi.org/10.1111/j.1439-0469.2011.00646.x
Funk WC, Lovich RE, Hohenlohe PA et al (2016) Adaptive divergence despite strong genetic drift: genomic analysis of the evolutionary mechanisms causing genetic differentiation in the island fox (Urocyon littoralis). Mol Ecol 25:2176–2194. https://doi.org/10.1111/mec.13605
Gloyd HK, Conant R (1982) The classification of the Agkistrodon halys complex. Jpn J Herpetol 9:75–78. https://doi.org/10.5358/hsj1972.9.3_75
Grant WS, Bowen BW (1998) Shallow population histories in deep evolutionary lineages of marine fishes: insights from sardines and anchovies and lessons for conservation. J Hered 89:415–426. https://doi.org/10.1093/jhered/89.5.415
Guo P, Zhang FJ (2002) Phylogenetic and biogeographic studies on Deinagkistrodon and Gloydius in China. J Sichuan Univ Nat Sci 39:378–381 (In Chinese)
Guo P, Liu Q, Li C et al (2011) Molecular phylogeography of Jerdon’s pitviper (Protobothrops jerdonii): importance of the uplift of the Tibetan plateau. J Biogeogr 38:2326-2336. https://doi.org/10.1111/j.1365-2699.2011.02566.x
Isogawa K, Moriya A, Mitsui S (1994) A new snake of the genus Agkistrodon (Serpentes: Viperidae) from Tsushima Island, Nagasaki Prefecture, Japan. Jpn J Herpetol 15:101–111. https://doi.org/10.5358/hsj1972.15.3_101
Kearse M, Moir R, Wilson A et al (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. https://doi.org/10.1007/BF01731581
Kumar S, Stecher G, Li M et al (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Lee YS, Markov N, Voloshina I et al (2015) Genetic diversity and genetic structure of the Siberian roe deer (Capreolus pygargus) populations from Asia. BMC Genet 16:100. https://doi.org/10.1186/s12863-015-0244-6
Lee YS, Do MS, Jeon HS et al (2021) Complete mitochondrial genome of Gloydius saxatilis (Viperidae: Crotalinae) from Korea. Mitochondrial DNA B Resour 6:645–647. https://doi.org/10.1080/23802359.2021.1878957
Malhotra A, Thorpe RS (2004) A phylogeny of four mitochondrial gene regions suggests a revised taxonomy for Asian pitvipers (Trimeresurus and Ovophis). Mol Phylogenet Evol 32:83–100. https://doi.org/10.1016/j.ympev.2004.02.008
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC et al (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol 34:3299–3302. https://doi.org/10.1093/molbev/msx248
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Shi J, Wang G, Chen X et al (2017) A new moth-preying alpine pit viper species from Qinghai-Tibetan Plateau (Viperidae, Crotalinae). Amphib Reptilia 38:517–532. https://doi.org/10.1163/15685381-00003134
Shi J, Yang D, Zhang W et al (2018) New Species of the Gloydius strauchi Complex (Crotalinae: Viperidae: Serpentes) from Qinghai, Sichuan, and Gansu, China. Russ J Herpetol 25:126–138. https://doi.org/10.30906/1026-2296-2018-25-2-126-138
Wang K, Ren J, Dong W et al (2019) A new species of plateau pit viper (Reptilia: Serpentes: Gloydius) from the upper Lancang (=Mekong) valley in the Hengduan Mountain region, Tibet, China. J Herpetol 53:224–236. https://doi.org/10.1670/18-126
Yan J, Li H, Zhou K (2008) Evolution of the mitochondrial genome in snakes: gene rearrangements and phylogenetic relationships. BMC Genom 9:569. https://doi.org/10.1186/1471-2164-9-569
Yan X, Qin L, Edward AM et al (2012) Molecular phylogeny of the genus Gloydius (Serpentes: Crotalinae). Asian Herpetol Res 3:127–132. https://doi.org/10.3724/SP.J.1245.2012.00127
Zhou JL, Yao YG, Huang MH et al (2000) Phylogenetic relationships among Viperidae, Crotalinae based on mitochondrial 12S rRNA sequence variations. Yi Chuan Xue Bao 27:283–289 (In Chinese)
Acknowledgements
This work was supported by a grant [NIBR202005102] from the National Institute of Biological Resources (NIBR), funded by the Ministry of Environment (MOE), Republic of Korea. We would like to thank Editage (www.editage.co.kr) for English language editing.
Author information
Authors and Affiliations
Contributions
All authors contributed to the concept and design of the study. Sample collection andpreparation of materials were done by MSD, WK, HSJ, S-CL and J-HJ. YSL performed the data analysis and also wrote the initial draft of the manuscript; and all authors commented and added on it. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors report that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lee, Y.S., Do, M.S., Kim, W. et al. Phylogenetic relationships between three Korean pit viper Gloydius (Serpentes: Crotalinae) species using mitochondrial DNA genes. Genes Genom 44, 517–526 (2022). https://doi.org/10.1007/s13258-022-01222-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-022-01222-3