Abstract
In order to extend the knowledge of rabies pathogenesis, a two-dimensional electrophoresis/mass spectrometry based postmortem comparative proteomics analysis was carried out on human brain samples. Alteration in expression profile of several proteins was detected. Proteins related to cytoskeleton, metabolism, proteasome and immune regulatory systems showed the most changes in expression levels. Among these groups, the cytoskeleton related proteins (dynein light chain, β-centractin, tubulin alpha-1C chain and destrin) and metabolism associated proteins (fatty acid-binding protein, macrophage migration inhibitory factor, glutamine synthetase and alpha enolase) were the main altered proteins. These alterations may be considered as an evidence of disturbances in neuronal key processes including axonal transport, synaptic activity, signaling and metabolic pathways in rabies virus infected human brain.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rabies is an acute and still untreatable viral encephalitis [1]. Globally, it kills over 55,000 people each year and more than 3 billion people are at risk of the infection in over 100 countries. Most human deaths occur in developing countries [2, 3]. In Iran, rabies is an endemic and also the most important zoonotic disease with very high economic burden. Post exposure prophylaxis containing prompt and proper wound cleansing, vaccination and administration of rabies immune globulin could be lifesaving [3]. However, after the invasion of virus to the central nervous system and the development of clinical symptoms, the disease will be untreatable [4]. Hence, the elucidation of underlying mechanisms in rabies pathogenicity may facilitate the development of new approaches in treatment of this fatal infection. In this context, several lines of research have been considered. For instance, the proteomics analysis on different models of rabies virus (RV) infection has been carried out to find the molecular events lead to rabies-induced neuronal dysfunction [5–9]. The results have demonstrated the expression modulation of some host proteins with crucial roles in various cellular functions. Since the RV-infected human brain may be considered as the most informative model for investigating the neuronal pathways affected by RV, we have used a two dimensional electrophoresis/mass spectrometry (2-DE/MS) based comparative proteomics approach to examine the protein expression pattern of brain tissues from those individuals who were accidentally infected with street rabies virus.
Materials and methods
Human brain tissue samples
Human brain samples were provided by WHO-Collaborating Center for Reference & Research on Rabies, Pasteur Institute of Iran. These samples were referred to the center by Forensic Medicine organization of Islamic Republic of Iran to confirm rabies infection. The positive samples had been taken from the occipital lobe of seven individuals who had died of rabies (postmortem interval: 48 h). Seven negative samples were also obtained from the same organization but from individuals who died of cardiac complications and had no clinical history of neurological disorders, head trauma or alcoholism. All Sample transfer agreements and handling procedures were approved by Ethical committee of Pasteur institute of Iran.
Protein extraction
Protein extraction was performed as described previously [10]. Briefly, aliquots of 150 mg of each individual brain sample was homogenized separately in 720 μl of ice-cold lysis buffer consisting of 7 M urea (Bio-Rad, CA, USA), 2 M thiourea (Merck, Germany), 100 mM dithiotreitol (Bio-Rad), 1 % Sigma anti-protease cocktail (USA), 4 % 3-[(3-cholamidopropyl)-dimethyl ammonio]-1-propane sulfonate (MP Biomedicals, CA, USA), and 2 % ASB14 (CalBiochem, UK) using a HeidolphSilentcrusher S device (Germany). The lysates were separated by centrifugation at 14,000×g for 15 min at 4 °C. Protein concentration in the resulting supernatants was determined using the Bradford method [11]. Aliquots (1,600 μg of protein) were stored at −80 °C until use.
Two-dimensional gel electrophoresis
1,600 micrograms of the protein extract from each brain sample was applied separately to immobilized pH gradient (IPG) gel strips (pH 3–10 NL, 17 cm; BioRad, USA) in a total volume of 350 μl of rehydration solution containing 7 M urea, 2 M thiourea, 2 % 3-[(3-cholamidopropyl)-dimethyl ammonio]-1-propane sulfonate, 2 % carrier ampholytes pH 3-10 (Bio-Rad, USA), 70 mM dithiotreitol and 0.001 % bromophenol blue. After rehydration for 12 h, protein separation by isoelectrofocusing (IEF) was carried out at 20 °C using 50 μA/strip for 50,000 Vh at the maximum of 10,000 V in the PROTEAN IEF cell (BioRad, USA). Following the IEF, IPG strips were equilibrated for 15 min in the equilibration solution (50 mM Tris–HCl pH 8.8, 6 M urea, 20 % glycerol, 2 % sodium dodecyl sulfate, 0.01 % bromophenol blue) containing 2 % dithiotreitol and then alkylated for a further 15 min in the equilibration solution containing 2.5 % iodoacetamide. The second dimensional separation was performed on the 12 % sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gels and electrophoresis was performed at 16 mA/gel for 30 min and 24 mA/gel for 5 h at 20 °C using a Protean II xi electrophoresis cell (Bio-Rad Laboratories, Hercules, CA, USA). Separated protein spots were stained with modified colloidal coomassie blue [12].
Image analysis
The images of stained gels were scanned at a resolution of 300 dpi with the densitometer GS-800 (BioRad, USA) and imported into the image analysis software program (PG200, Nonlinear Dynamics, UK) for quantitative analysis of differentially expressed proteins. The amount of each protein spot was expressed in terms of its volume. To reflect the quantitative variations in the protein spot volumes, the spot volumes were normalized as a percentage of the total volume of all the spots present in a gel. Statistical analysis of protein variations was carried out using the student t test on vol% of matched spots (p < 0.05).
Protein identification by mass spectrometry
Gel pieces were washed two times with 50 % (v:v) aqueous acetonitrile containing 25 mM ammonium bicarbonate, then once with acetonitrile and dried in a vacuum concentrator for 20 min. Gel pieces were rehydrated by adding 10 μl of trypsin solution and incubation for 30 min. Subsequently, an appropriate amount of 25 mM ammonium bicarbonate solution was added to cover the gel pieces and digests were incubated overnight at 37 °C. A 1 μl aliquot of each peptide mixture was applied directly to the ground steel matrix-assisted laser desorption/ionization (MALDI) target plate, followed immediately by an equal volume of a freshly prepared 5 mg/ml solution of 4-hydroxy-α-cyano-cinnamic acid (Sigma) in 50 % aqueous (v:v) acetonitrile containing 0.1 %, trifluoroacetic acid (v:v). Positive-ion MALDI mass spectra were obtained using a Bruker ultraflex III in reflectron mode, equipped with a Nd:YAG smart beam laser. MS spectra were acquired over a mass range of m/z 800–4,000. Tandem mass spectral data were submitted to database searching using a locally running copy of the Mascot program (Matrix Science Ltd., version 2.1), through the Bruker ProteinScape interface (version 2.1). Search criteria included: Enzyme, Trypsin; Fixed modifications, Carbamidomethyl (C); Variable modifications, Oxidation (M); Peptide tolerance, 250 ppm; MS/MS tolerance, 0.5 Da; Instrument, Matrix-Assisted Laser Desorption/Ionization-Time of Flight-Time of Flight (MALDI-TOF-TOF). Statistical confidence limits of 95 % were applied for protein identification.
Western blot analysis
Western blot analysis was performed as described in details [6]. Aliquots of 35 μg protein extracts from seven control and seven rabid brain tissues were loaded and separated by 12 % SDS-PAGE at 100 V. Separated proteins were then electrotransferred to nitrocellulose membrane blots (Hybind ECL, GE healthcare, UK), using semidry trans-blot cell (Bio-Rad). After blocking of non-specific sites in tris-buffered saline containing 5 % skim milk, blots were incubated with monoclonal antibodies to Alpha enolase (1 μg/ml or 1:500, Abcam, UK), TUB A6 (1 μg/ml, Abcam, UK), Clathrin light chain (1:250, Abcam, UK), β-actin as a control protein (1:1,000, Sigma-Aldrich, UK) and polyclonal antibodies to ACTR1B (1:500, Abcam, UK) and NDUFS8 (1:200, Abcam, UK). The incubation period was 2 h at room temperature for all primary antibodies except for NDUFS8 in which the proteins were incubated at 4 °C overnight. The membranes were then incubated with peroxidase-conjugated anti-mouse IgG (1:500 and 1:1,000) and peroxidase-conjugated anti-rabbit IgG (1:1,000) secondary antibodies (Razi Biotech, Iran). The immunoreactive bands were then detected by ECL plus kit (GE healthcare, UK) using Kodak Image Station 4000MM Pro. Densitometric analysis was performed using the Quantity One software version 4.6.3 (Bio-Rad). The statistical analysis was performed by the use of F test two sample for variances, t test: two-sample assuming equal variances and t test: two-sample assuming unequal variances data analysis tools using Microsoft Excel 2007 software and p value <0.05 was considered statistically significant.
Results
2-DE was used to highlight any possible alteration in the protein expression profile of rabies virus-infected human brain tissue (Fig. 1). As protein extraction and solubilization are critical steps for successful gel-based proteomics analysis, ASB-14 in combination with 3-[(3-cholamidopropyl)-dimethyl ammonio]-1-propane sulfonate was used to solubilise human brain proteins [10]. Totally, twenty-seven protein spots showed significant differences in expression level (more than 1.3-fold) between rabid and control groups (Fig. 2). Fourteen proteins were successfully identified through MALDI-TOF-MS analysis and MASCOT searching against the IPI human database. Among these identified proteins, thirteen proteins showed decreased level and only one protein showed increase in abundance in rabid group (Table 1).
Representative 2D gels show protein maps from the brains of a non-rabid (left) and rabid (right) human. 14 identified protein spots indicated by arrows, are numbered according to Table 1
a Comparison of 2D profiles between rabid and non-rabid human brain tissues. Five representative differentially expressed protein spots in rabid human brain tissues are shown: TUBA6 (1), ENO1 (3), ACTR1B (5), NDUFS8 (9), DYNLRB1 (14). Spot numbers are the same as those in Fig. 1 and Table 1. b Comparison of selected five protein levels (normalized spot volume) in brains of rabid and non-rabid human determined by PG 200 software. Bars represent mean ± SD, p < 0.05, n = 7 samples from rabid and seven samples from non-rabid groups
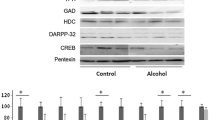
The identified proteins were classified into seven categories including metabolic enzymes, cytoskeleton and cytoskeleton associated proteins, proteasome related proteins, redox related proteins, immune regulatory proteins, apoptosis related proteins and signalling proteins. Metabolic enzymes (included 5 identified proteins) and cytoskeletal related proteins (included 4 identified proteins) were the two major categories with most changes in expression level. Western blot analysis of some of the differentially expressed proteins confirmed all the alterations identified by 2-DE/MS except for mitochondrial NADH dehydrogenase [ubiquinone] iron-sulfur protein 8 (Fig. 3). In our previous report, we detected a marked under expression of clathrin light chain in neuroblastoma cell line infected by the Challenge Virus Standard [9]. Here we examined the expression pattern of this protein by western blotting and a significant decrease was observed.
a Western blot images of TUBA1C, ENO1, ACTR1B, NDUFS8 and Clathrin of conrol and rabid human brains. β-actin was used as a control protein. Densities of immunoreactive bands were measured by using the Quantity One software version 4.6.3 (Bio-Rad). Statistical analysis using t test is shown in (b), in which bars represent mean ± SD, n = 7 samples from rabid and seven samples from non-rabid groups. Asterisk represents the statistical difference between rabid and control brains (p < 0.05)
Discussion
Comparative proteomics analysis is a useful strategy to elucidate viral pathogenesis mechanisms [13]. In order to understand the molecular mechanisms involved in rabies pathogenesis, different proteomics approaches have been used including proteome analysis of several in vitro [6, 7, 9] and in vivo [5, 8] host models infected with different virulent rabies strains. This study was designed to analyze the effects of rabies virus infection on the human brain proteome profile. 2-DE technique was used to compare seven infected and seven non-infected brain samples. Among twenty-seven differentially expressed proteins, fourteen were successfully identified by mass spectrometry.
Dynein light chain, centractin (β isoform) and tubulin alpha-1C chain (TUBA1C) were the three under expressed microtubule associated proteins. The dynein/dynactin complex has critical role in retrograde axonal transport of binding cargos and signaling pathways in neurons [14, 15]. In this protein complex, direct cargo binding is attributed to light intermediate and light chains of dynein [16]. Dynactin, the dynein activator complex, mediates cytoplasmic dynein binding to selected cargos [15]. Defects of dynein/dynactin function due to mutations or expression alterations of its components have been shown to affect many key processes such as retrograde axonal transport in neurons. In the present study, the decreased expression of dynein/dynactin subunits including dynein light chain and centractin β isoform (a major component of dynactin) together with one alpha tubulin isoform, TUBA1C in rabid group, may be considered as a sign for axonal trafficking damage and microtubular instability of neurons [14, 15, 17]. Raux et al. [18] suggested a dynein/dynactin-mediated mechanism for axonal transport of rabies virus along microtubules in neurons which could support a role for affected dynein/dynactin complex in rabies pathogenesis. Furthermore, the proteome profiling of RV-infected mouse neuro N2a cells and the brains of naturally RV-infected dogs, have shown some alterations in tubulin pattern [7, 8].
Actin filament dynamics play an important role in all aspects of neurons life including synaptogenesis and synaptic activity [19]. In confocal microscopic examination of RV-infected neuroblastoma cells, alteration of the actin-based cytoskeleton with decrease in filamentous actin (F-actin) fluorescent staining has been observed [20]. Under expression of destrin as a major actin dynamics regulatory protein [21–23] in rabid human brains may indicate the disturbance of normal balance between F-actin and G-actin pools of cytoplasmic actin and consequent defects or abnormalities in neuronal actin dynamics and related biological processes. Application of fluorescent-based approaches [24, 25] in order to study the G and F-actin contents of RV-infected human brain tissue could be helpful.
In the present study, under expression of FABP3 (a fatty-acid binding protein) [26] and Macrophage migration inhibitory factor (MIF) as a pro-inflammatory cytokine [27] was observed. FABP3 has vital role in arachidonic acid uptake, metabolism, energy production, long-chain polyunsaturated fatty-acid-related signaling and dopamine D2 receptor (D2R) regulation in brain [28, 29]. MIF involves in cellular immune responses and inflammation development in the CNS [27], induction of arachidonic acid metabolism [30] and modulating the release of signaling molecules in brain physiology [31, 32]. Therefore, decreased amount of these two proteins could be considered as an evidence for disturbances in metabolism and signaling of rabid human brains. Moreover, under expression of MIF in human brain infected with rabies virus would be probably interpreted as one of the modulatory mechanisms or immune invasion responses by which the virus could be able to escape from immune system recognition in the brain.
Glutamine synthetase (GS), another decreased metabolic enzyme in rabid human brain group involves in modulation of glutamate excitotoxicity through catalyzing the rapid amidation of glutamate to form non-toxic glutamine [33]. Decreased expression of GS and subsequent glutamate excitotoxicity injury of neurons have been reported in some neurodegenerative diseases and viral infections [34, 35]. Study of cultured neuronal cells infected with the CVS strain of rabies virus showed no evidence of excitotoxicity induction. [36]. Regarding the observed under expression of GS in our experiment, further studies are needed to investigate excitotoxicity induction by street rabies virus variants which can affect neuronal function.
Under expression of proteasome subunit alpha type-1(PSMA1) is another significant finding in the present study. Proteasome, as the major protein quality control of eukaryotic cells, controls the assembly, connectivity, function and signaling of the synapse in neurons [37, 38]. Mitochondrial dysfunction, oxidative modification of proteasome subunits and the alteration of proteasome subunit levels can affect proteasome activity [39–41] which results in neuronal dysfunction [42]. Down regulation of the proteasome subunit alpha type-3 has been reported previously [7]. Proteasome dysfunction has also been implicated in pathogenesis of some neurodegenerative diseases [43–45]. Interestingly, a mitochondrial and proteasome hypofunctioning of hipocampal neurons due to decreased expression of mitochondrial and proteasome genes including mitochondrial NADH dehydrogenase [ubiquinone] iron-sulfur protein 4 (NDUFS4) and proteasome subunit alpha type-1(PSMA1), has been suggested to contribute to Schizophrenia pathogenesis [45]. According to our data, decreased levels of PSMA1 in rabid group may be an evidence of proteasomal dysfunction in RV-infected human brains.
Alpha enolase (ENO1) was the only over expressed metabolic protein identified in rabid group. The over expression of alpha enolase in proteomics analysis of mouse neuro N2a cells infected with variant virulence of RVs has also been noticed before [7]. Over expression of this important glycolytic enzyme in brain metabolism [46] may indicate a compensatory response to decreased activity in other metabolic pathways of neurons during human brain rabies infection.
Interestingly, we investigated the expression alteration of the clathrin light chain protein in rabid human brains. Although the under expression of this protein was not detected by 2-DE analysis, western blotting confirmed this modification. This result is consistent with other study carried out on neuroblastoma cell line [9] which could be considered as a sign of affected synaptic vesicle recycling process in synaptic ends.
In summary, the comparative proteomics analysis of accidentally infected human brains with rabies virus, has provided interesting results indicating significant expression alteration of some host proteins involved in cytoskeletal stability, microtubule associated axonal transport, signaling and metabolic pathways which may affect respective neuronal key processes. Detailed analysis of interactions between virus and brain cytoskeleton as well as targeted metabolic pathways may help in elucidation of molecular pathogenesis of rabies virus.
References
Jackson AC (1991) Biological basis of rabies virus neurovirulence in mice: comparative pathogenesis study using the immunoperoxidase technique. J Virol 65(1):537–540
Wunner WH, Briggs DJ (2010) Rabies in the 21 century. PLoS Negl Trop Dis 4(3):e591
Leung AK, Davies HD, Hon KL (2007) Rabies: epidemiology, pathogenesis, and prophylaxis. Adv Ther 24(6):1340–1347
Durai R, Venkatraman R (2006) Human rabies and its prevention. Br J Hosp Med (Lond) 67(11):588–593
Dhingra V, Li X, Liu Y, Fu ZF (2007) Proteomic profiling reveals that rabies virus infection results in differential expression of host proteins involved in ion homeostasis and synaptic physiology in the central nervous system. J Neurovirol 13(2):107–117
Zandi F, Eslami N, Soheili M, Fayaz A, Gholami A, Vaziri B (2009) Proteomics analysis of BHK-21 cells infected with a fixed strain of rabies virus. Proteomics 9(9):2399–2407
Wang X, Zhang S, Sun C, Yuan ZG, Wu X, Wang D, Ding Z, Hu R (2011) Proteomic profiles of mouse neuro N2a cells infected with variant virulence of rabies viruses. J Microbiol Biotechnol 21(4):366–373
Thanomsridetchai N, Singhto N, Tepsumethanon V, Shuangshoti S, Wacharapluesadee S, Sinchaikul S, Chen ST, Hemachudha T, Thongboonkerd V (2011) Comprehensive proteome analysis of hippocampus, brainstem, and spinal cord from paralytic and furious dogs naturally infected with rabies. J Proteome Res 10(11):4911–4924
Zandi F, Eslami N, Torkashvand F, Fayaz A, Khalaj V, Vaziri B (2013) Expression changes of cytoskeletal associated proteins in proteomic profiling of neuroblastoma cells infected with different strains of rabies virus. J Med Virol 85(2):336–347
Martins-de-Souza D, Menezes de Oliveira B, dos Santos Farias A, Horiuchi RS, Crepaldi Domingues C, de Paula E, Marangoni S, Gattaz WF, Dias-Neto E, Camillo Novello J (2007) The use of ASB-14 in combination with CHAPS is the best for solubilization of human brain proteins for two-dimensional gel electrophoresis. Brief Funct Genomics Proteomics 6(1):70–75
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Candiano G, Bruschi M, Musante L, Santucci L, Ghiggeri GM, Carnemolla B, Orecchia P, Zardi L, Righetti PG (2004) Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 25(9):1327–1333
Maxwell KL, Frappier L (2007) Viral proteomics. Microbiol Mol Biol Rev 71(2):398–411
Levy JR, Holzbaur EL (2006) Cytoplasmic dynein/dynactin function and dysfunction in motor neurons. Int J Dev Neurosci 24(2–3):103–111
Gulesserian T, Kim SH, Fountoulakis M, Lubec G (2002) Aberrant expression of centractin and capping proteins, integral constituents of the dynactin complex, in fetal down syndrome brain. Biochem Biophys Res Commun 291(1):62–67
Vallee RB, Williams JC, Varma D, Barnhart LE (2004) Dynein: an ancient motor protein involved in multiple modes of transport. J Neurobiol 58(2):189–200
Weng YQ, Qiu SJ, Liu YK, Fan J, Gao Q, Tang ZY (2008) Down-regulation of beta-centractin might be involved in dendritic cells dysfunction and subsequent hepatocellular carcinoma immune escape: a proteomic study. J Cancer Res Clin Oncol 134(2):179–186
Raux H, Flamand A, Blondel D (2000) Interaction of the rabies virus P protein with the LC8 dynein light chain. J Virol 74(21):10212–10216
Sarmiere PD, Bamburg JR (2004) Regulation of the neuronal actin cytoskeleton by ADF/cofilin. J Neurobiol 58(1):103–117
Ceccaldi PE, Valtorta F, Braud S, Hellio R, Tsiang H (1997) Alteration of the actin-based cytoskeleton by rabies virus. J General Virol 78(Pt 11):2831–2835
Vartiainen MK, Mustonen T, Mattila PK, Ojala PJ, Thesleff I, Partanen J, Lappalainen P (2002) The three mouse actin-depolymerizing factor/cofilins evolved to fulfill cell-type-specific requirements for actin dynamics. Mol Biol Cell 13(1):183–194
Yeoh S, Pope B, Mannherz HG, Weeds A (2002) Determining the differences in actin binding by human ADF and cofilin. J Mol Biol 315(4):911–925
Verdoni AM, Aoyama N, Ikeda A, Ikeda S (2008) Effect of destrin mutations on the gene expression profile in vivo. Physiol Genomics 34(1):9–21
Kiuchi T, Nagai T, Ohashi K, Mizuno K (2011) Measurements of spatiotemporal changes in G-actin concentration reveal its effect on stimulus-induced actin assembly and lamellipodium extension. J Cell Biol 193(2):365–380
Wang WH, Abeydeera LR, Prather RS, Day BN (2000) Polymerization of nonfilamentous actin into microfilaments is an important process for porcine oocyte maturation and early embryo development. Biol Reprod 62(5):1177–1183
Owada Y, Yoshimoto T, Kondo H (1996) Spatio-temporally differential expression of genes for three members of fatty acid binding proteins in developing and mature rat brains. J Chem Neuroanat 12(2):113–122
Bacher M, Meinhardt A, Lan HY, Dhabhar FS, Mu W, Metz CN, Chesney JA, Gemsa D, Donnelly T, Atkins RC, Bucala R (1998) MIF expression in the rat brain: implications for neuronal function. Mol Med 4(4):217–230
Murphy EJ, Owada Y, Kitanaka N, Kondo H, Glatz JF (2005) Brain arachidonic acid incorporation is decreased in heart fatty acid binding protein gene-ablated mice. Biochemistry 44(16):6350–6360
Shioda N, Yamamoto Y, Watanabe M, Binas B, Owada Y, Fukunaga K (2010) Heart-type fatty acid binding protein regulates dopamine D2 receptor function in mouse brain. J Neurosci 30(8):3146–3155
Mitchell RA, Liao H, Chesney J, Fingerle-Rowson G, Baugh J, David J, Bucala R (2002) Macrophage migration inhibitory factor (MIF) sustains macrophage proinflammatory function by inhibiting p53: regulatory role in the innate immune response. Proc Natl Acad Sci USA 99(1):345–350
Bozza M, Satoskar AR, Lin G, Lu B, Humbles AA, Gerard C, David JR (1999) Targeted disruption of migration inhibitory factor gene reveals its critical role in sepsis. J Exp Med 189(2):341–346
Bernhagen J, Mitchell RA, Calandra T, Voelter W, Cerami A, Bucala R (1994) Purification, bioactivity, and secondary structure analysis of mouse and human macrophage migration inhibitory factor (MIF). Biochemistry 33(47):14144–14155
Poon HF, Vaishnav RA, Getchell TV, Getchell ML, Butterfield DA (2006) Quantitative proteomics analysis of differential protein expression and oxidative modification of specific proteins in the brains of old mice. Neurobiol Aging 27(7):1010–1019
Robinson SR (2001) Changes in the cellular distribution of glutamine synthetase in Alzheimer’s disease. J Neurosci Res 66(5):972–980
Darman J, Backovic S, Dike S, Maragakis NJ, Krishnan C, Rothstein JD, Irani DN, Kerr DA (2004) Viral-induced spinal motor neuron death is non-cell-autonomous and involves glutamate excitotoxicity. J Neurosci 24(34):7566–7575
Weli SC, Scott CA, Ward CA, Jackson AC (2006) Rabies virus infection of primary neuronal cultures and adult mice: failure to demonstrate evidence of excitotoxicity. J Virol 80(20):10270–10273
de Vrij FM, Fischer DF, van Leeuwen FW, Hol EM (2004) Protein quality control in Alzheimer’s disease by the ubiquitin proteasome system. Prog Neurobiol 74(5):249–270
Pak DT, Sheng M (2003) Targeted protein degradation and synapse remodeling by an inducible protein kinase. Science 302(5649):1368–1373
McNaught KS, Belizaire R, Isacson O, Jenner P, Olanow CW (2003) Altered proteasomal function in sporadic Parkinson’s disease. Exp Neurol 179(1):38–46
McNaught KS, Jackson T, JnoBaptiste R, Kapustin A, Olanow CW (2006) Proteasomal dysfunction in sporadic Parkinson’s disease. Neurology 66(10 Suppl 4):S37–S49
McNaught KS, Olanow CW (2006) Protein aggregation in the pathogenesis of familial and sporadic Parkinson’s disease. Neurobiol Aging 27(4):530–545
Sherman MY, Goldberg AL (2001) Cellular defenses against unfolded proteins: a cell biologist thinks about neurodegenerative diseases. Neuron 29(1):15–32
Cassarino DS, Bennett JP Jr (1999) An evaluation of the role of mitochondria in neurodegenerative diseases: mitochondrial mutations and oxidative pathology, protective nuclear responses, and cell death in neurodegeneration. Brain Res 29(1):1–25
Shamoto-Nagai M, Maruyama W, Kato Y, Isobe K, Tanaka M, Naoi M, Osawa T (2003) An inhibitor of mitochondrial complex I, rotenone, inactivates proteasome by oxidative modification and induces aggregation of oxidized proteins in SH-SY5Y cells. J Neurosci Res 74(4):589–597
Altar CA, Jurata LW, Charles V, Lemire A, Liu P, Bukhman Y, Young TA, Bullard J, Yokoe H, Webster MJ, Knable MB, Brockman JA (2005) Deficient hippocampal neuron expression of proteasome, ubiquitin, and mitochondrial genes in multiple schizophrenia cohorts. Biol Psychiatry 58(2):85–96
Tholey G, Ledig M, Mandel P (1982) Modifications in energy metabolism during the development of chick glial cells and neurons in culture. Neurochem Res 7(1):27–36
Acknowledgments
This study was supported by grant No. 438 from Pasteur Institute of Iran. Authors acknowledge gratefully Dr. Adam Dowle (Proteomics Technology Facility, Department of Biology, University of York, England) for his critical review on matrix-assisted laser desorption/ionization-time of flight/time of flight mass spectrometry analysis. We also thank, Fatemeh Torkashvand, Elmira Haghighatjoo, Atefeh Mirzakhani and Ahmad Adeli for technical and supportive help.
Author information
Authors and Affiliations
Corresponding author
Additional information
Firouzeh Farahtaj and Fatemeh Zandi have equally contributed to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Farahtaj, F., Zandi, F., Khalaj, V. et al. Proteomics analysis of human brain tissue infected by street rabies virus. Mol Biol Rep 40, 6443–6450 (2013). https://doi.org/10.1007/s11033-013-2759-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-013-2759-0