Abstract
A gene encoding the carboxymethylcellulase (CMCase) of a marine bacterium, Bacillus subtilis subsp. subtilis A-53, was cloned in Escherichia coli JMB109 and the recombinant strain was named as E. coli JMB109/A-53. The optimal conditions of rice bran, ammonium chloride, and initial pH of the medium for cell growth, extracted by Design Expert Software based on response surface methodology, were 100.0 g/l, 7.5 g/l, and 7.0, respectively, whereas those for production of CMCase were 100.0 g/l, 7.5 g/l, and 8.0. The optimal temperatures for cell growth and the production of CMCase by E. coli JM109/A-53 were found to be and 40 and 35 °C, respectively. The optimal agitation speed and aeration rate of a 7 l bioreactor for cell growth were 400 rpm and 1.5 vvm, whereas those for production of CMCase were 400 rpm and 0.5 vvm. The optimal inner pressure for cell growth was 0.06 MPa, which was the same as that for production of CMCase. The production of CMCase by E. coli JM109/A-53 under optimized conditions was 880.2 U/ml, which was 2.9 times higher than that before optimization. In this study, rice bran and ammonium chloride were developed as carbon and nitrogen source for production of CMCase by a recombinant E. coli JM109/A-53 and the productivity of E. coli JM109/A-53 was 5.9 times higher than that of B. subtilis subp. subtilis A-53.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the last 50 years much effort has gone into the study on enzymes with cellulolytic activity as a potential means to obtain energy, chemicals, and single-cell proteins from an abundant and renewable resource, i.e., cellulosic biomass [1]. Highly efficient cellulolytic bacteria and fungi generally produce one or more types of enzymes; endoglucanase (1,4-β-d-glucan-4-glucanohydrolase; carboxymethylcellulase (CMCase), EC 3.2.1.4), exocellobiohydrolase (1,4-β-d-glucan glucohydrolase; avicelase, EC 3.2.1.91), and β-glucosidase (β-d-glucoside glucohydrolase, EC 3.2.1.21) to grade microcrystalline cellulose to glucose [2]. The enzymatic saccharification of cellulosic materials such as rice hulls was performed by commercial cellulases, in which the major one was CMCase [3]. However, a major constrain in enzymatic saccharification of cellulosic materials is the cost of cellulases and low productivity [4].
One approach to increasing productivity of cellulases by cellulolytic organisms would be to isolate hyper-producers or constitutive mutants for cellulases. Another approach would be to clone genes coding for cellulases and then enhance expression of these genes by current molecular genetic techniques [5]. Several endo-1,4-glucanase from Bacillus species have been isolated and their genes were sequenced [6, 7]. These could include coupling genes to strong promoters, eliminating operators sensitive to repression, and increasing the efficiency of translation. Genes coding for CMCase have been also cloned from Apriona sp., Bacillus subtilis, Cellulomonas fimi, Clostridium thermocellum, and Streptomyces sp. [7–10].
We had reported production and characterization of the CMCase by a newly isolated marine bacterium, B. subtilis subsp. subtilis A-53 [11, 12]. In this study, we described cloning of the CMCase gene of B. subtilis subsp. subtilis A-53 and its expression in Escherichia coli JM109. The optimal conditions for the production of CMCase of B. subtilis subsp. subtilis A-53 by a recombinant E. coli JM109/A-53 were established using response surface methodology (RSM). The optimization of culture medium by the traditional ‘one-factor-at-a-time’ technique requires a considerable amount of work and time [13]. An alternate strategy is a statistical approach, for example, orthogonal array method (OAM) based on factorial experimental design and response surface method (RSM) involving a minimum number of experiments for a large number of factors [14–16].
Materials and methods
Bacterial strain and medium
B. subtilis subsp. subtilis A-53 was isolated from seashore of the Kyungsang Province in Korea and identified in the previous study [11]. It utilized cellulosic materials such as carboxymethylcellulose (CMC), rice bran, and rice hulls and produced CMCase. The strain was maintained on agar medium containing 20.0 g/l glucose, 2.5 g/l yeast extract, 5.0 g/l K2HPO4, 1.0 g/l NaCl, 0.2 g/l MgSO4·7H2O, 0.6 g/l (NH4)2SO4 and 15.0 g/l agar [12].
Cloning and sequence analysis of CMCase
The full-length CMCase gene of B. subtilis subsp. subtilis A-53 was amplified by polymerase chain reaction (PCR) using two specific primers, 5′-AGGAGGAAAAGATCAGATATGAAACGGTCAATC-3′ (forward, B. sub cmc P1) and 5′-TCCAGTATTTCATCCACAACGCAAACCTCC-3′ (reverse, B. sub cmc P2). These primers were designed on the basis of DNA sequences of Bacillus CMCase genes cloned previously: CMCase genes of B. subtilis AY044252.1 (NCBI-gi: 15375077), B. subtilis Z29076.1 (gi: 509266), and B. subtilis X67044.1 (gi: 39776). The chromosomal DNA of B. subtilis subsp. subtilis A-53 extracted using a Wizard Genomic DNA Prep. Kit (Promega Co., Madison, USA) was used as a template. PCR amplification was performed as described previously [17]. PCR for amplification was run for 35 cycles in a DNA thermal cycler (Model No. 9700, Perkin-Elmer Co. Wellesley, USA). The following thermal profile was used for the PCR: denaturation at 94 °C for 1 min, primer annealing at 60 °C for 1 min, and extension at 72 °C for 2 min. The final cycle included extension for 10 min at 72 °C to ensure full extension of the products. The amplified PCR products were ligated with the T-tail site of pGEM-T Easy Vector System (Promega Co., Madison, USA) and constructed plasmids were transformed into E. coli JM109. The plasmid contained PCR product was isolated from transformed E. coli JM109 and was sequenced in both directions using ALF Red automated DNA sequencer (Pharmacia, Sweden). Analysis of sequence data and sequence similarity searches were performed using the BLAST(N) program of the National Center for Biotechnology Information (NCBI) [18]. The transformed E. coli JM109 harboring the CMCase gene of B. subtilis subsp. subtilis A-53 was named as E. coli JM109/A-53.
Production of CMCase by E. coli JM109/A-53
E. coli JM109/A-53 was grown at 37 °C in LB medium containing 100 μg/ml ampicillin. Starter cultures were prepared by transferring cells from agar slants to 50 ml of LB medium in 250 ml Erlenmeyer flasks. The resulting cultures were incubated at 37 °C for 2 days under aerobic conditions. Each starter culture was used as an inoculum for 150 ml of medium in 500 ml Erlenmeyer flasks. The main culture for production of CMCase by E. coli JM109/A-53 was carried out in the medium containing 20.0 g/l CMC, 2.5 g/l yeast extract, 5.0 g/l K2HPO4, 1.0 g/l NaCl, 0.2 g/l MgSO4·7H2O, and 0.6 g/l (NH4)2SO4 for 72 h under aerobic conditions [11]. Batch fermentations for the production of CMCase by E. coli JM109/A-53 were performed in 7 and 100 l bioreactors (Ko-Biotech Co., Korea). Working volumes of the 7 and 100 l bioreactors were 5 and 70 l, respectively, and inoculum size of batch fermentations for the production of CMCase was 5 % (v/v). Agitation was provided by three six-flat-blade impellers in 7 and 100 l bioreactors.
Experimental design and optimization for production of CMCase
The rice bran (X 1), ammonium chloride (X 2), and initial pH of the medium (X 3) were chosen as the independent variables and cell growth (Y 1) and CMCase (Y 2) were used as a dependent output variable. The model constructed as a response function of the variables on cell growth and production of CMCase was a second-order polynomial as follows (Eq. 1):
where y is the measured response (cell growth as measured dry cells weight or production of CMCase), β 0, β i , and β ij are the regression coefficients, and X i and X j are the factors under study. For three variable systems, the model equation is given below (Eq. 2)
Regression analysis and estimation of the coefficient were performed using the statistical software, Design-Expert (Version 7.1.6, Stat-Ease Inc., Minneapolis, USA). The contribution of individual parameters and their quadratic and interactive effects on cell growth and production of CMCase were determined.
The agitation speed (X 1) and aeration rate (X 2) were also chosen as the independent variables and cell growth \( (Y_{1}^{'} ) \) and CMCase \( (Y_{2}^{'} ) \) were used as a dependent output variable. The interrelationships of the variables were also determined by fitting the second-order polynomial equation. Statistical analysis of the model was performed to evaluate the analysis of variance (ANOVA).
Analytical methods
CMCase activity was measured by the DNS (3,5-dinitrosalicylic acid) method, through the determination of the amount of reducing sugars liberated from CMC solubilized in 50 mM Tris–HCl buffer, pH 8.0 [12]. This mixture was incubated for 20 min at 50 °C and the reaction was stopped by the addition of DNS solution. The treated samples were boiled for 10 min, cooled in water for color stabilization, and the optical density was measured at 550 nm. CMCase activity was determined by using a calibration curve for glucose (Sigma-Aldrich Co., UK). One unit of enzyme activity was defined as the amount of enzyme that released 1 μmol of glucose per min. Cell growth was measured as an absorbance at optical density of 600 nm, and the dry cells weight was measured by directly weighing the biomass after drying to constant weight at 100–105 °C after collection of cells by centrifugation at 12,000×g for 10 min. Reducing sugars were determined by using DNS method [11].
Results and discussion
Cloning and sequence analysis of the gene encoding CMCase
The full-length CMCase gene of B. subtilis subsp. subtilis A-53 was cloned by PCR with primers based on DNA sequences of the CMCase gene of Bacillus species cloned previously [17]. As shown in Fig. 1, the open reading frame (ORF) of the cloned gene consists of 1,498 nucleotides encoding a protein of 499 amino acids with a predicted molecular weight of 55,110 Da. Analysis of the putative cleavage site of signal peptide by using the SignalP program was located between Ala-29 and Ala-30 [19]. Comparison of the deduced amino acid sequence of the cloned gene with those of proteins registered in the BLAST database showed the highest identity to the CMCase of Bacillus sp. strain AAX54913.1 (NCBI-gi:61742183), B. subtilis AAA22307.1 (gi:142671), and B. subtilis AAZ22322.1 (gi:71063308) with 99 % identity as shown in Fig. 2.
The amino acid sequence alignment of the CMCase from B. subtilis subsp. subtilis A-53 was compared with those of other CMCase using Clustal W, a multiple alignment program. Amino acid sequence alignments of CMCase from B. subtilis subsp. subtilis A-53 (A-53), Bacillus sp. (AAA22307), Bacillus sp. (AAZ22322), Bacillus sp. (ABV45393), Bacillus sp. (AAC43478), and Bacillus lichenformis (AAU23613)
Most CMCases like xylanase and β-1,3-1,4-glucanases are modular enzymes, which consist of two or more functional modules, such as catalytic and carbohydrate-binding modules (CBMs), connected to each other via a linker sequence [18, 20]. On the basis of amino acid sequence similarities, catalytic modules, which are engaged in the hydrolysis of cellulose, are classified into 106 groups in glycoside hydrolase families and CBMs, which bind preferentially to cellulose, are also classified into 45 groups (http://afmb.cnrs-mrs.fr/CAZY/index.html). Computer analysis with the deduced mature amino acid sequence of the CMCase of B. subtilis subsp. subtilis A-53 using CDART program of NCBI revealed a modular enzyme composed of two discrete domains in the following order: a catalytic domain (Gly-31 through Asn-332) of glycoside hydrolase family 5 (GH5), and a family 3 CBM (Glu-350 through His-499). Like the modular organization (GH5/CBM3) in many Bacillus endoglucanases (http://afmb.cnrs-mrs.fr/CAZY/index.html), this enzyme has its catalytic domain in the N-terminal portion and CBM in the C-terminal portion [17].
Effect of carbon and nitrogen sources on production of CMCase
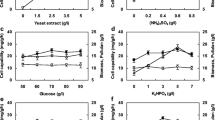
Effect of carbon and nitrogen sources on cell growth and production of the CMCase by E. coli JM109/A-53 was investigated at flask scales. Carbon sources tested for production of CMCase were 20.0 g/l glucose, fructose, maltose, sucrose, rice bran, and rice hulls. Nitrogen sources tested were 2.5 g/l malt extract, peptone, tryptone, yeast extract, ammonium chloride, and ammonium nitrate. The initial pH of the medium and cultural temperature were 7.0 and 37 °C. Sucrose and malt extract were found to be the best combination of carbon and nitrogen sources for cell growth of E. coli JM109/A-53, whereas rice bran and tryptone were the best on for production of CMCase, as shown in Fig. 3. However, a combination of rice bran and ammonium chloride was chosen for next examination based on their cost and availability as well as productivity for the industrial scaled production. Cell mass measured as dry cells weight and production of CMCase from 20.0 g/l rice bran and 5.0 g/l tryptone were 2.14 g/l and 305.2 U/ml, whereas those from 20.0 g/l rice bran and 5.0 g/l ammonium chloride were 0.70 g/l and 206.1 U/ml.
The best combinations for cell growth of B. subtilis subsp. subtilis A-53 had been reported to be rice bran and yeast extract, whereas that for production of CMCase was rice bran and yeast extract [12]. Sucrose and malt extract were found to be the best combination of carbon and nitrogen sources for the cell growth of Psychrobacter aquimaris LBH-10 isolated from seawater, whereas rice bran and peptone were the best one for production of CMCase [21]. The best carbon and nitrogen sources for the production by B. amyloliquefaciens DL-3 were rice hulls and peptone [22]. Major carbon sources for the production of cellulases by Aspergillus and Trichoderma species were reported to wheat bran [4]. All strains investigated to date were inducible by cellulose, lactose or sophorose, and all are repressible by glucose, which can explain why most carbon sources for production of cellulases are rice hulls, rice bran or wheat bran [23].
Optimization of rice bran, ammonium nitrate, and initial pH for production of CMCase
The optimal conditions of rice bran, ammonium chloride, and initial pH of the medium for cell growth and the production of CMCase by E. coli JM109/A-53 were investigated using one-factor-at-a time experiments. Composition of basic medium was 50.0 g/l rice bran, 5.0 g/l ammonium chloride, and initial pH of 7.5. Cell mass and production of CMCase ranged from 1.14 to 5.46 g/l and 148.2 to 419.9 U/ml. The optimal conditions of rice bran, ammonium chloride, and initial pH for cell growth of E. coli JM109/A-53 were 125.0 g/l, 10.0 g/l, and 7.5, respectively, whereas those for production of CMCase were 125.0 g/l, 5.0 g/l, and 8.0, as shown in Fig. 4.
Based on results from one-factor-at-a-time experiments, the simultaneous effect of rice bran, ammonium chloride, and initial pH of the medium on cell growth and production of CMCase was also investigated using RSM. The coded values of minimum and maximum ranges of rice bran (X 1), ammonium chloride (X 2), and initial pH of the medium (X 3) were 50.0 and 100.0 g/l, 2.5 and 7.5 g/l, and 7.0 and 8.0, respectively. Cell growth and production of CMCase from 20 different conditions ranged from 2.68 to 4.03 g/l and from 262.5 to 340.8 U/ml, as shown in Table 1. Multiple regression analysis of the experimental data gave the following second-order polynomial equation to represent cell growth of E. coli JM109/A-53 (Eq. 3). The optimal conditions of rice bran, ammonium chloride, and initial pH of the medium for cell growth extracted by Design Expert Software were 100.0 g/l, 7.5 g/l, and 7.0, respectively. The maximum cell mass of 3.94 g/l was predicted by this model
Adequacy of the model was tested by the fisher’s statistical test for the analysis of variance (ANOVA), as shown in Table 2. The model F-value of 28.88 from the analysis of variance (ANOVA) of cell growth implied that this model was significant. There was only a 0.01 % chance that a “Model F-value” could occur to die to noise. The P values were used as a tool to check the significance of each of the coefficients, which, in turn were necessary to understand the pattern of the mutual interactions between the test variables. The smaller the magnitude of the P value, the more significant is the corresponding coefficient. The ANOVA indicated that this model and the model term of X 1 were highly significant (“probe > F” less 0.0001) and the model term of X 2 was significant (“probe > F” less 0.0500) for cell growth of E. coli JM109/A-53. The regression equation obtained from ANOVA indicated that the multiple correlation coefficient of R 2 was 0.9630, which could explain 96.30 % variation in the response. The value of the adjusted determination coefficient (Adj. R 2 = 0.9296) was very high to advocate for a high significance of this model [16]. The predicted determination of coefficient of 0.8841 was in reasonable agreement with the Adj. R 2 of 0.9296. From the statistical results obtained, it was shown that the above models were adequate to predict the cell growth of E. coli JM109/A-53 within the range of variables studied.
Multiple regression analysis of the experimental data also gave the following second-order polynomial equation to represent the production of CMCase by E. coli JM109/A-53 (Eq. 4). The optimal conditions of rice bran, ammonium chloride, and initial pH of the medium for production of CMCase were 100.0 g/l, 7.5 g/l, and 8.0, respectively. The maximum production of CMCase of 333.8 U/ml was predicted by this model
The model F-value of 29.58 from the ANOVA of production of CMCase implied that this model was also significant. The ANOVA indicated that this model and model terms of X 21 were highly significant and those of X 3 and X 22 were significant. The regression equation obtained from ANOVA indicated that the multiple correlation coefficient of R 2 was 0.9638. The value of the adjusted determination coefficient (Adj. R 2 = 0.9312) was high to advocate for a high significance of this model. The predicted determination of coefficient of 0.8723 was also in reasonable agreement with the Adj. R 2 of 0.9312. From the statistical results obtained, it was shown that the above models were adequate to predict the production of CMCase by E. coli JM109/A-53 within the range of variables studied.
The optimal conditions of rice bran, yeast extract, and initial pH of the medium for cell growth of B. subtilis subsp. subtilis A-53 were 100 g/l, 10 g/l, and 7.3, whereas those for production of CMCase were 50 g/l, 1.0 g/l, and 6.8. [12]. The optimal conditions of rice bran, peptone and initial pH for the production of avicelase and CMCase by P. aquimaris LBH-10 were 50.0, 3.0, and 8.0 g/l, respectively, which were also different from those for filter paperase [21]. The optimal conditions for cell growth of E. coli JM109/A-53 as well as B. subtilis subsp. subtilis A-53 were found to be different from those for production of CMCase.
Effect of temperature on production of CMCase
The effect of temperature on cell growth and the production of CMCase by E. coli JM109/A-53 was investigated. The carbon and nitrogen source and initial pH of the medium were 100.0 g/l rice bran, 7.5 g/l ammonium chloride, and 8.0, respectively. The temperature for cell growth and production of CMC ranged from 25 to 45 °C. The optimal temperatures for cell growth and the production of CMCase by E. coli JM109/A-53 were found to be 40 and 35 °C, respectively, as shown in Fig. 5. Cell mass and production of CMCase at 40 °C were 4.05 g/l and 517.7 U/ml, whereas those at 35 °C were 3.59 g/l and 584.0 U/ml. Significance of each value was analyzed by MYSTAT software (Systat Software, Inc., Chicago, USA). The optimal temperatures for cell growth and the production of CMCase by B. subtilis subsp. subtilis A-53 were 35 and 30 °C, respectively. Those for production of CMCases by bacterial and fungal stains ranged from 25 to 37 °C, except for thermophilic microorganisms such as Thermoascus aurantiacu, which optimal temperature for production of CMCase is 50 °C [4]. The optimal temperature for cell growth of B. amyloliquefaciens DL-3 was 32 °C, whereas that for production of CMCase was 37 °C [22]. The optimal temperature for cell growth of E. coli JM109/A-53 was also different from that for production of CMCase as production of other CMCases.
Effects of agitation speed and aeration rate on production of CMCase
The effect of agitation speed on cell growth of CMCase by E. coli JM109/A-53 in a 7 l bioreactor was investigated using one-factor-at-a-time. Carbon and nitrogen sources for production of CMCase were 100.0 g/l rice bran and 7.5 g/l ammonium chloride. The initial pH of the medium and temperature were 8.0 and 35 °C. Agitation speed ranged from 200 to 500 rpm and aeration rate was 1.0 vvm. Higher agitation speeds and aeration rates, which resulted in an increase of dissolved oxygen in the medium, enhanced cell growth as well as production of CMCase, as shown in Fig. 6. The optimal agitation speeds for cell growth and production of CMCase were 400 rpm. The effect of aeration rate on cell growth and production of CMCase also was investigated. Aeration rate ranged from 0.5 to 2.0 vvm and the agitation speed was fixed to 400 rpm. The optimal aeration rate for cell growth was 1.5 vvm, whereas that for production of CMCase was 0.5 vvm.
Based on results from one-factor-at-a-time experiments, the simultaneous effects of agitation speed and aeration rate on cell growth and the production of CMCase by E. coli JM109/A-53 were investigated using RSM. The coded values of minimum and maximum ranges of agitation speed (X 1) and aeration rate (X 2) were 300 and 500 rpm and 0.5 and 1.5 vvm. Cell mass and production of CMCase from 13 different conditions ranged from 2.58 to 3.61 g/l and from 427.1 to 647.0 U/ml, as shown in Table 3. Multiple regression analysis of the experimental data gave the following second-order polynomial equation to represent cell growth of E. coli JM109/A-53 (Eq. 5). The optimal agitation speed and aeration rate for cell growth were 395 rpm and 1.38 vvm. The maximum cell mass of 3.73 g/l was predicted by the model
The model F-value of 74.43 from ANOVA of cell growth implied that this model was significant, as shown in Table 4. The ANOVA indicated that this model and the model terms of X 2 and X 21 were highly significant for cell growth. The regression equation obtained from ANOVA indicated that the multiple correlation coefficient of R 2 is 0.9815. The value of the adjusted determination coefficient (Adj. R 2 = 0.9684) is very high to advocate for a high significance of this model.
Multiple regression analysis of the experimental data also gave the following second-order polynomial equation to represent the production of CMCase by E. coli JM109/A-53 (Eq. 6). The optimal agitation speed and aeration rate for production of CMCase were 396 rpm and 0.55 vvm. The maximum production of CMCase of 650.9 U/ml was predicted by the model.
The model F-value of 53.46 from ANOVA of production of CMCase implied that this model was also significant. The ANOVA indicated that this model and the model terms of X 2 and X 21 was also highly significant model terms. The regression equation obtained from ANOVA indicated that the multiple correlation coefficient of R 2 is 0.9745. The value of the adjusted determination coefficient (Adj. R 2 = 0.9563) is very high to advocate for a high significance of this model. The predicted determination of coefficient of 0.8629 is also in reasonable agreement with the Adj. R 2 of 0.9563.
The three-dimensional response surface plots were generated to investigate the interaction among agitation speed and aeration rate and to visualize the combined effects on the response of cell growth and the production of CMCase by E. coli JM109/A-53, as shown in Fig. 7. The three-dimensional response surfaces and their respective contour plots are the graphical representation of Eqs. 5 and 6. In contrast to the circular shapes, the elliptical nature of curves indicates more significant mutual interactions between variables. The interactive effect of agitation speed and aeration rate on production of CMCase (Probe > F = 0.0100) was found to be higher than those on cell growth (Probe > F = 0.0497).
The concentration of dissolved oxygen in the medium is influenced by agitation speed, aeration rate, and the inner pressure of bioreactors [24]. Variation in agitation speed and aeration rate results in a change in the concentration of dissolved oxygen in the medium, which in turn affects cell growth and the production of microbial metabolites [25]. The optimal agitation speed and aeration rate for cell growth of B. subtilis subsp. subtilis A-53 were 400 rpm and 1.5 vvm, whereas those for the production of CMCase were 300 rpm and 1.0 vvm [12]. The optimal agitation speed for cell growth was the same as that for the production of CMCase by B. subtilis subsp. subtilis A-53, whereas the optimal aeration rate for cell growth was different from that for production of CMCase. It seems that cell wall of E. coli JM109/A-53 was stronger than that of B. subtilis subsp. subtilis A-53 and higher than optimal concentration of dissolved oxygen for production of CMCase leads the biosynthetic pathway to cell growth, but not to production of CMCase.
Effect of inner pressure on production of CMCase
The effect of inner pressure on cell growth and the production of CMCase by E. coli JM109/A-53 was investigated in a 100 l bioreactor. The inner pressure ranged from 0.00 to 0.08 MPa. The agitation speed and aeration rate of a 100 l bioreactor were 250 rpm and 0.5 vvm. The radius of the impeller in a 100 l bioreactor was bigger than that in a 7 l bioreactor. The angular velocity of a 100 l bioreactor at 250 rpm is almost the same as that of a 7 l bioreactor at 400 rpm. The concentration of the dissolved oxygen in the medium with an inner pressure decreased until 36 h after cultivation, whereas that in the medium without an inner pressure decreased until 24 h after cultivation, as shown in Fig. 8a. The production of CMCase by E. coli JM109/A-53 was paralleled with cell growth, as shown in Fig. 8b, c. The optimal inner pressures for cell growth and production of CMCase by E. coli JM109/A-53 were 0.06 MPa. Productions of CMCase by E. coli JM109/DL-3 with inner pressure of 0.00, 0.02, 0.04, 0.06, and 0.08 MPa after 72 h cultivation were 616.3, 697.7, 737.9, 880.2, and 841.3 U/ml, respectively. The production of CMCase with an inner pressure of 0.06 MPa was 1.4 times higher than that without an inner pressure. The production of CMCase by E. coli JM109/A-53 under optimized conditions was 880.2 U/ml, which was 2.9 times higher than that before optimization.
The factors affected by scale-up are the number of generations, the mutation probability, the quality of temperature, and pH controls, agitation, aeration, and inner pressure [26]. Oxygen transfer often can be most important upon scale-up due to its low solubility in medium [27]. Due to higher concentrations of dissolved oxygen, the production of pullulan by A. pullulans with an inner pressure of 0.04 MPa was 1.3 times higher than that without inner pressure [28]. Increasing the driving force for the diffusion from the air bubbles to the medium by elevating the inner pressure of a bioreactor increased the oxygen transfer rate (OUR), which resulted in enhanced production of β-glucosidase by P. pastoris, while cell growth did not increase [29]. The optimal inner pressure for the production of CMCase by E. coli JM109/DL-3 was higher than those by Bacillus species. It seems that cell wall of E. coli is more resistant to inner pressure than other species. Higher agitation speeds and inner pressures, which could afford for higher concentration of dissolved oxygen, did not seem to damage to cells, which resulted in enhanced cell growth and the production of CMCase by E. coli JM109/A-53.
Conclusion
Various carbon sources have been used for the production of cellulases, but their prices are too expensive to apply commercially [30, 31]. The use of cheap resources as substrates can reduce the cost of cellulase production [32]. Rice bran from the rice processing industry is produced in large amounts in Korea, as well as other rice producing countries. Low-cost ammonium chloride as a nitrogen source is also available in large quantities. In this study, rice bran and ammonium chloride were developed as carbon and nitrogen sources and optimal conditions for the production of CMCase by E. coli JM109/A-53 were established using RSM. Maximal production of CMCase by a recombinant E. coli JM109/A-53 was 5.9 times higher than that of B. subtilis subsp. subtilis A-53, as shown in Table 5. Most commercial cellulases are produced by solid state fermentations of fungal species [23, 32]. Time for production of cellulases by fungal species in solid-state fermentation normally takes 7 to 10 days [30, 31]. Reduced time for production of CMCase from 7 to 3 days using a recombinant E. coli JMB109/A-53 with submerged fermentations also resulted in increase of productivity and decrease in their production cost.
References
Sukumaran RK, Singhania RR, Mathew GM et al (2009) Cellulase production using biomass feed stock and its application in lignocellulose saccharification for bio-ethanol production. Renew Energy 34:421–424
Yi JC, Sandra JC, John AB et al (1999) Production and distribution of endoglucanase, cellobiohydrolase, and β-glucosidase components of the cellulolytic system of Volvariella volvacea, the edible straw mushroom. Appl Environ Microbiol 65:553–559
Wei GY, Gao W, Jin IH et al (2009) Pretreatment and saccharification of rice hulls for the production of fermentable sugars. Biotechnol Bioprocess Eng 14:828–834
Kalogeris E, Christakopoulos P, Katapodis P et al (2003) Production and characterization of cellulolytic enzymes from the thermophilic fungus Thermoascus aurantiacus under solid state cultivation of agricultural waste. Process Biochem 38:1099–1104
Chang JC, Park IH, Lee YS et al (2010) Cloning, expression, and characterization of ß-glucosidase from Exiguobacterium sp. DAU5 and transglycosylation activity. Biotechnol Bioprocess Eng 16:97–106
Koide Y, Nakamura A, Uozumi AT et al (1986) Molecular cloning of a cellulase gene from Bacillus subtilis and its expression in Escherichia coli. Agric Biol Chem 50:233–237
Lee YJ, Kim HJ, Gao W et al (2012) Statistical optimization for production of carboxymethylcellulase of Bacillus amyloliquifaciens DL-3 by a recombinant Escherichia coli JM109/DL-3 from rice bran using response surface method. Biotechnol Bioprocess Eng 17:227–235
Lam TL, Wong RSC, Wong WK (1997) Enhancement of extracellular production of a Cellulomonas fimi exoglucanase in Escherichia coli by the reduction of promoter strength. Enzyme Microb Technol 20:482–488
Park CS, Kawaguchi T, Sumitani JL et al (2005) Cloning and sequencing of exoglucanase gene from Strptomyces sp. M23 and its expression in Strptomyces lividans TK-24. J Biosci Bioeng 4:434–436
Wei YD, Lee KS, Gui ZZ et al (2006) Molecular cloning, expression, and enzymatic activity of a novel endogenous cellulase form the mulberry longicorn beetle, Apriona germari. Comp Biochem Physiol 145:220–229
Kim BK, Lee BH, Lee YJ et al (2009) Purification and characterization of carboxymethylcellulase isolated from a marine bacterium, Bacillus subtilis subsp. subtilis A-53. Enzyme Microb Technol 44:411–416
Lee BH, Kim BK, Lee YJ et al (2010) Industrial scale of optimization for the production of carboxymethylcellulase from rice bran by a marine bacterium, Bacillus subtilis subsp. subtilis A-53. Enzyme Microb Technol 46:38–42
Park EY, Ikeda Y, Okuda N (2002) Empirical evaluation of cellulase on enzymatic hydrolysis of waste office paper. Biotechnol Bioprocess Eng 7:268–274
Gao W, Kim YJ, Chung CH et al (2010) Optimization of mineral salts in medium for enhanced production of pullulan by Aureobasidium pullulans HP-2001 using orthogonal array method. Biotechnol Bioprocess Eng 15:837–845
Kim HJ, Gao W, Chung CH et al (2011) Statistical optimization for production of carboxymethylcellulase from rice hulls by a new isolated marine microorganism Bacillus licheniformis LBH-52 using response surface method. J Life Sci 21:1083–1093
Kim HJ, Lee YJ, Gao W et al (2012) Optimization of salts in medium for production of carboxymethylcellulase by a psychrophilic marine bacterium, Psychrobacter aquimaris LBH-10 using two statistical methods. Korean J Chem Eng 29:384–391
Lee YJ, Kim BK, Lee BH et al (2008) Purification and characterization of cellulase produced by Bacillus amyloliquefaciens DL-3 utilizing rice hull. Bioresour Technol 99:378–386
Brich PR, Sims PF, Broda P (1995) Substrate dependent differential splicing of introns in the regions encoding the cellulose binding domains of two exocellobiohydrolase-like genes in Phanerochaete chryscsporium. Appl Environ Microbiol 61:3741–3744
Nielsen H, Engelbrecht J, Brunak S et al (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10:1–6
Ohmiya K, Sakka K, Karita S et al (1997) Structure of cellulases and their applications. Biotechnol Genet Eng Rev 14:365–414
Kim HJ, Lee YJ, Gao W et al (2011) Statistical optimization of fermentation conditions and comparison of their influences on production of cellulases by psychrophilic marine bacterium, Psychrobacter aquimaris LBH-10 using orthogonal array method. Biotechnol Bioprocess Eng 16:542–548
Jo KI, Lee YJ, Kim BK et al (2008) Pilot-scale production of carboxymethylcellulase from rice hull by Bacillus amyloliquefaciens DL-3. Biotechnol Bioprocess Eng 13:182–188
Hanif A, Yasmeen A, Rajoka MI (2004) Induction, production, repression, and de-repression of exoglucanase synthesis in Aspergillus niger. Bioresour Technol 94:311–319
Seo JH, Li H, Kim MJ et al (2007) Determination of agitation and aeration conditions for scale-up of cellulolytic enzymes production by Trichoderma inhamatum KSJ1. Korean J Chem Eng 24:800–805
Zhong JJ (2010) Recent advances in bioreactor engineering. Korean J Chem Eng 27:1035–1041
Song HJ, Li H, Seo JH et al (2009) Pilot-scale production of bacterial cellulose by a spherical type bubble column bioreactor using saccharified food waste. Korean J Chem Eng 26:141–146
Junker BH (2004) Scale-up methodologies for Escherichia coli and yeast fermentation process. J Biosci Bioeng 97:347–364
Seo HP, Chung CH, Kim SK et al (2004) Mass production of pullulan with optimized concentrations of carbon and nitrogen sources by Aureobaisdium pullulans HP-2001 in a 100L bioreactor with the inner pressure. J Microbiol Biotechnol 14:237–242
Charoenrat T, Ketudat-Cairns M, Jahic M et al (2006) Increased total air pressure versus oxygen limitation for enhanced oxygen transfer and product formation in a Pichia pastoris recombinant protein process. Biochem Eng J 30:205–211
Domingues FC, Queiroz JA, Cabral JMS et al (2000) The influence of culture conditions on mycelial structure and cellulase production by Trichoderma reesei Rut C-30. Enzyme Microb Technol 26:394–401
Lee SM, Koo YM (2001) Pilot-scale production of cellulose using Trichoderma reesei Rut C-30 in fed-batch mode. J Microbiol Biotechnol 11:229–233
Mekala NK, Singhania RR, Sukumaran RK et al (2008) Cellulase production under solid-state fermentation by Trichoderma reesei RUT C30: statistical optimization of process parameters. Appl Biochem Biotechnol 151:122–131
Acknowledgments
This paper was financially supported by Dong-A University Research Fund.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lee, EJ., Lee, BH., Kim, BK. et al. Enhanced production of carboxymethylcellulase of a marine microorganism, Bacillus subtilis subsp. subtilis A-53 in a pilot-scaled bioreactor by a recombinant Escherichia coli JM109/A-53 from rice bran. Mol Biol Rep 40, 3609–3621 (2013). https://doi.org/10.1007/s11033-012-2435-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-012-2435-9