Abstract
UDP-Glucose Pyrophosphorylase (EC 2.7.7.9, UGPase) plays an important role in Streptococcus equi subsp. zooepidemicus (S. zooepidemicus) cell envelope Hyaluronic acid (HA) biosynthesis and it is also recognized as a virulence determinant in several bacterial species. HA is valuable biopolymer used in the pharmaceutical and cosmetic industry. In addition, encapsulation by HA is considered an important virulence factor in other streptococci. Research UGPase will contribute to the vaccine development of S. zooepidemicus and the production of HA. In this study, The UGPase gene fragment (789 bp) obtained from previous research was amplified using PCR, and located by Genome walking technology (Genebank No.GQ423507). The UGPase was expressed, purified and identified using UGPase antibody. The enzyme kinetic parameters were determined, the temperature and pH of the highest activity for the cloned UGPase were 37°C, pH 7.5. The K m and K cat value against UTP and G-1-P was 8.5 μM, 69.05 s−1 and 36.41 μM, 48.81 s−1, respectively. The homology-modeling was operated. Overexpression of the UGPase in S. zooepidemicus, its virulence was slightly affected, and HA yield reduced. Real-time PCR was carried out to determine the UGPase expression levels of both SEZp and SEZugp in different grow period, the level is high in logarithmic phase and low in Decline phase.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
UDP-Glucose Pyrophosphorylase is involved in cell envelope biosynthetic pathway. An Escherichia coli mutant with an knock out in the UGPase gene had deformed cell wall and membrane, since the enzyme UGPase could not participate in the Leloir pathway [1]. UDP-Glucose Pyrophosphorylase is involved in carbohydrate metabolism, catalyzing a reversible reaction that produces UDPG and pyrophosphate (PPi) from Glc-1-P and UTP. The UDPG is a high energy substrate for cellulose biosynthesis in both bacteria and plants. The UGPase monomer is the active form of the protein, its activity can be regulated by reversible oligomerization [2, 3] The ability of UGPase to oligomerize in vivo has recently been confirmed [4], It plays an important role in the pathway of HA synthesis [5].
Streptococcus equi subsp. zooepidemicus (S. zooepidemicus) belongs to the Lancefield group C, it has no host preference. It mutates easily from one host to another, therefore the bacterium has diverse antigenicity. The bacteria infects wide range of animal species through the alimentary canal and open wounds [6, 7] causing septicemia, meningitis, endocarditis, arthritis and mammitis. Humans could be infected through consumption of contaminated food or contact wounds from the infected animals [8]. This bacteria also has advantage, it can be used as producer of HA, which is very useful at cosmetics and medicine. Most part of the capsule of S. zooepidemicus is formed by HA, and the capsule plays an important role in S. zooepidemicus infection.
Two-dimensional electrophoresis, combined with Western blot and matrix-assisted laser desorption tandem time-of-flight mass spectrometry, determined that the UDP-Glucose Pyrophosphorylase serves a vital role in the pathogenicity and reproduction of the bacterium. Therefore, has a potential for use in vaccine research [9] and may become an attractive target in combating bacterial diseases [10].
The present study represents the first successful preparation of a recombinant UGPase from S. zooepidemicus CY, and provides biochemical characterization of this enzyme. With the homology-modeling methed [11–13], we identify the active site of this enzyme monomer. Overexpression of UGPase in S. zooepidemicus CY was used to identify the influence of this enzyme. This work can be useful for further studies of the UGPase. It is also helpful in the production of HA and pathogenic mechanism of S. zooepidemicus.
Methods and materials
Analysis and clone of the UGPase gene from S. zooepidemicus CY
Streptococcus equi subsp. zooepidemicus CY was grown in THB media (BactonTM, USA). The bacterial cells were harvested at late-log phase and the genomic DNA was isolated using Genomic DNA mini Kit (Geneaid, Taiwan). The UGPase gene PCR primers were designed by Primer Premier 5.0 software (Premier, Canada). The PCR amplified products were purified from 1% agarose gel using Gel/PCR DNA Fragments Extraction Kit (Geneaid, Taiwan) and ligated to pMD™ 18-T Vector (TAKARA, Japan). The sequencing was performed by Invitrogen (USA). The 3′-UTR and 5′-UTR of the UGPase were identified using the Genome Walking Kit (TAKARA, Japan) following the manufacturer’s instructions. The UGPase gene specific primers are listed in Table 1; Walking-LSP1, 2, 3 were used to get unknown upstream 5′ UTR sequence and Walking-RSP1, 2, 3 were used to get unknown downstream sequence 3′ UTR sequence. All sequencing reactions are operated by Invitrogen. (USA).
Expression, purification and immunoblotting analysis of UGPase expressed in E. coli
The expression of UGPase in E. coli was performed by ligating the gene containing EcoRI and Xhol sites into the pET32a expression vector (Takara, Japan). E. coli BL21 was transformed with the pET32a expression construct containing UGPase and grown at 37°C in LB agar containing 50ug/ml ampicillin for selection. The selected transformants were then cultured in LB broth. Once the OD600 values reached between 0.5 and 0.6, IPTG (Sigma, USA) was added to induce UGPase expression. The cells were cultured for another 14 h at 20°C and 80 rpm. The cells were then lysed and centrifuged to separate soluble and insoluble proteins and stored at 4°C till further use.
This His-tagged UGPase protein was then purified from the soluble fraction under native conditions using Ni-NTA columns according to the manufacturer’s instructions (GE, USA). The purified protein was then filtered in an Amicon® Ultra-15 Centrifugal Filter Device (Millipore, USA) following manufacturer’s instructions. The filtered protein content was determined using a spectrophotometer (Bio-Rad Smartspec3000, USA) at 595 nm using the Bio-Rad Assay Kit (Bio-Rad, USA) based on a Bradford method, with bovine serum albumin as standard.
The purified protein was visualized using SDS-PAGE. Following the SDS-PAGE, the proteins in the gel were transferred to a PVDF membrane for western blot analysis. The blotted membranes were treated with mouse anti-CY IgG and the antigen–antibody complex was visualized using anti-mouse HRP-labeled secondary antibody.
Enzyme kinetic assays
UDP-Glucose Pyrophosphorylase was diluted 200-fold in 100 mM Hepes (pH 7.5) containing 1 mg/ml bovine serum albumin. The pyrophosphorolytic activity of UGPase, was assayed using UDP-glucose and PPi as substrates measured at 340 nm. The reaction was coupled with the production of glucose-1-P and NADPH in the presence of NADP, catalysed by phosphoglucomutase (PGM) (Sigma) and glucose-6-P dehydrogenase (GDH) (Sigma). An 1 ml standard reaction mixture contained, 100 mM Hepes (pH 7.5), 0.85 mM UDP-glucose, 0.5 mM PPi, 5 mM MgCl2, 0.3 mM NADP, and 5 units each of PGM and GDH. The reaction was allowed for 12 min, and then terminated by heating up to 100°C for 5 min, followed by cooling on ice. Approximately 3–7 μg of purified protein was used in the assay. One unit of UGPase was defined as the amount of the enzyme required to reduce 1 μmol of NADP, equivalent to the production of 1 μmol of glucose-1-P, per minute.
Construction of recombinant S. zooepidemicus strain
The gene encoding S. zooepidemicus UGPase was amplified by PCR. The PCR product was blunted with T4 DNA polymerase T4. The fragment was introduced into SfiI-digested pSLCP25 (Derived from pSL5.28 [14] which was given by Vaccine and Infectious Disease Organization, University of Saskatchewan), the primers are listed in Table 1 (O, P). The recombinant plasmid and original pSLCP25 were separately electroporated into S. zooepidemicus CY competent cells [15] with a electroporator (BTX ECM630) at 12.5 kV/cm and 200 Ω. Transformed cells were selected with 200 mg/ml spectinomycin in THB medium. The transformants of S. zooepidemicus containing the recombinant plasmid and original pSLCP25 were designated as the SEZugp and SEZp strains, respectively.
Real-time PCR to detect the expression level of UGPase in SEZugp and SEZp
Total RNA from SEZugp and SEZp growing in THB media at 1, 5, 9, 12, and 16 h were extracted using Trizol reagent (Invitrogen), according to the manufacturer’s protocol. SYBR® Premix Ex Taq™ (TAKARA) was used for real time PCR experiments in an ABI PRISM 7300 Fast Real-time PCR. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was a housekeeping internal control [16]. At the end of each cycle, the fluorescence emitted by SYBR Green was measured. The relative change in gene expression was recorded as comparative CT (2−ΔΔCT) [17].
Analysis HA yield and virulence of SEZugp and SEZp
Bitter–Muir method was taken to detect HA yield of SEZugp and SEZp which were cultured in THB medium for 16 h. Eight-week-old BALB/c mice were assigned to six groups, six mice per group. SEZugp and SEZp were separately inoculated in THB and cultured at 37°C without shaking until log phase. The bacteria were spun and resuspended in sterile 10 mmol PBS (pH 7.2). The concentration of bacteria was then adjusted to 5 × 108 CFU/ml and a 10× dilution serial with the lowest concentration being 5 × 104 CFU/ml was prepared. Mice were injected with 0.2 ml bacteria intramuscularly and death of mice was recorded. LD50 was calculated using Karber method.
Other methods
The homology-modeling was accomplished by Swiss-model website and all data was analysed with MOE software (CCG).
Results
UGPase gene location, homology analysis and homology-modeling
We used Genome walking technology to locate the UGPase gene with its upstream (1,805 bp) and downstream (1,959 bp) regions. The two UTR were combined with the full length gene sequence to form a 4,667 bp length sequence (Fig. 1, Genebank No. GQ423507). The promoter and RBS sites in the 5′ UTR sequence was determined using the software obtained from www.softberry.com (Table 2). Every RBS site was corresponding to at least one ORF except the integral membrane protein gene, in our known sequence, no RBS site of this gene was found. There was a shared promoter in the upstream involved in the regulation of UGPase gene, the Glycerol-3-phosphate dehydrogenase [NAD(P)+] gene and the 5-formyl-THF cyclo-ligase gene [18]. These 3 ORFs belonged to the same operon [19].
ORF found in upstream and downstream sequence of UGPase gene. (DNAStar software used), filled circle means the site of promoter, filled square means the site of RBS, Table 3 shows the exactly location of promoter and RBS (Genebank No. GQ423507)
A phylogenetic tree displaying the comparisons of the deduced amino acid sequence from the S. zooepidemicus CY UGPase and 8 of other known sequences are shown in Fig. 2a. We have recently homology-modeled the primary sequence for S. zooepidemicus UGPase, based on crystal structure of Sphingomonas Elodea ATCC 31461 UGPase (PDB 2ux8 chain G), to produce a predicted 3D structure. The central domain is based on a 9-stranded β-sheet which is located at the core of the domain and surrounded by helices and loops. This domain contains the active site of the enzyme. The active site pocket is made of the β-sheet on one side from which protrudes a loop and a helix, both covering the pocket from the top [20]. In Fig. 3a, the conjectured active site of S. zooepidemicus UGPase is similar to Sphingomonas Elodea. Ramachnadran plot shows that the conformation of this model is stable.
a Phylogenetic tree based on the sequence alignment of the UGPase proteins in NCBI Database by using the CLUSTAL V alignment program MEGA4. b Homologous align the primary sequence of S. zooepidemicus UGPase to 8 other known UGPase. The pane shows conserved amino acid residues which are relative to formation of the enzyme active site
a The left figure is predicted Ribbon 3D structure of S. zooepidemicus UGPase monomer and active site for substrate catalysis, The right figure is Ribbon 3D structure derived from crystal structure of Sphingomonas Elodea ATCC 31461 UGPase monomer and its active site with ligand G-1-P. b Structural alignment of S. zooepidemicus UGPase using MOE, regions of secondary structure (line-helixes, dashed-sheets, dotted-loop). “*” denotes amino acid residues which are relative to formation of the enzyme active site
Protein expression, purification and immunoblotting
In our previous research [9], using matrix assisted laser desorption tandem time-of-flight mass spectrometry (MALDI-TOF/TOF–MS), we obtained the amino acid sequence of UGPase and searched using BLAST in NCBI database to obtain UGPase’s nucleotide sequence of S. zooepidemicus. Sequence specific primers were designed on this template, the genome extracted from S. zooepidemicus CY was used as the PCR template. The nucleotide sequence obtained was then compared to sequences from the NCBI database. The results indicated that the UGPase sequence from S. zooepidemicus CY had high homology to other S. zooepidemicus strains (Table 3).
Primers for expressing the UGPase without signal sequence in E.coli were designed. PCR product codes for a polypeptide comprising 256 amino acids with a calculated molecular mass of 29,044 Da. Protein was expressed using pET32a vector that has 18 kD His-Tag, therefore, the UGPase protein’s molecular weight is about 47 kD. The pET-32a vector was transformed into E. coli BL21 for protein expression (Fig. 4a). Ultrafiltration is an effective way to remove ions in the protein solution and avoid any noises and artifacts in the enzyme activity studies. Meanwhile, it increases the activity and concentration of the enzyme [21]. Western-blot analysis was showed in Fig. 4b.
SDS-PAGE analysis of total cellular proteins and purified fusion proteins from Escherichia coli cells and Immunoblotting analysis result. a Lane M, marker. Lane 1, negative control, Lanes 2–5, the UGPase protein expression change from 2 to 5 h; b Lane M, marker. Lane 1, the purified UGPase protein after ultrafiltration. Lane 2, the purified UGPase protein without ultrafiltration. Lane 3, Western-blot result
Enzyme kinetic analysis of the purified recombinant UGPase
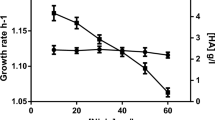
It was observed that recombinant UGPase achieved maximal activity at around pH 7.5, and the protein was not stable at high temperature and lost the most of activity after treated with 60°C for 10 min (Fig. 5). The kinetic analysis of UGPase in the forward reaction showed a typical Michaelis–Menten substrate saturation pattern (Fig. 6). In the presence of 10 mM Mg2+, the K m value against UTP or G-1-P was 8.5 and 36.41 μM, respectively. The K cat value was 69.05 and 48.81 s−1 against UTP or G-1-P, respectively (Table 4). The UGPase of S. equi subsp. zooepidemicus exhibited K m values comparable to previously described for E. coli which are 70 μM for UTP and 10 μM for G-1-P.
Over-expression of UGPase in S. zooepidemicus CY
We constructed a plasmid pSLCP25, this plasmid is derived from pSL5.28, it contains a powerful promoter CP25 [22], UGPase gene was connected behind this promoter. The expression level is displayed in Fig. 7, SEZugp has higher expression level about UGPase than SEZp in all period. In both SEZp and SEZugp, UGPase expression level is higher during logarithmic growth phase (before 9 h) than plateau phase (12 h) and decline phase (16 h) (Fig. 7).
Analysis HA yield and virulence of SEZugp and SEZp
Overexpression of UGPase in SEZugp led to a 26.3% decrease in HA yield, and this phenomenon had been reported by Chen et al. [23]. Death of mice was recorded within 3 days after the challenge. The LD50 was 9.1 × 104 cfu for SEZp. In contrast, the LD50 for SEZugp was 2.2 × 105 cfu, suggesting the virulence of the UGPase overexpression strain was slightly reduced (Table 5).
Discussion
We have characterized S. zooepidemicus UGPase in this research. UGPase gene appears to have been duplicated into the S. zooepidemicus CY has operon from remotely located but near-identical paralogues most likely to improve HA productivity by gene dosage in this Streptococcus [19]. A detailed understanding of the flanking sequence of UGPase gene may assist in studying evolution of streptococcal has operons. The UGPase amino acid sequence in Streptococci is highly conserved, and it shows relatively lower sequence homologies with eukaryotic pyrophosphorylases. But all those amino acid residues, which are relative to formation of the enzyme active site conjectured by the MOE, are strictly conserved between eukaryote and prokaryote (Fig. 2b), this indicate that the function of UGPase in biochemical analysis evolution is highly conserved.
To express the protein in abundance without the accumulation of inclusion bodies, the cultivation conditions were changed to lower temperature and shaking after addition of IPTG. This could enable recombinant E. coli BL21 for processing primary structure peptide chain to complete its secondary structure, so that the enzyme can be obtained and maintained in an active state. Western-blot analysis indicated that recombinant UGPase still maintained its antigenicity, it is a potential target for vaccine development.
Because of the duplication of UGPase gene in the S. zooepidemicus CY genome, it is hard to research the molecular mechanism behind the enzyme’s involvement in the virulence of the bacteria with gene knock-out experiments. Overexpression seems to be the best way in studying the function of UGPase in S. zooepidemicus CY. In most Gram-positive bacteria, UDP-glucose serves as a substrate for glycosylation of cell wall teichoic acids (WTA) and for biosynthesis of the glycolipid diglucosyl-dialcylglycerol (Glc2-DAG), which is the predominant membrane anchor moiety of lipoteichoic acids (LTA) [24]. The expression level indicated that at logarithmic growth phase bacterial mitosis is vigorous, and cell wall needs to be synthesized rapidly, so relatively higher levels of UGPase is expressed to meet the bacteria’s requirements. During the decline phase, the expression of UGPase did not terminate nor reduce drastically, which indicated that UGPase probably has an alternative function besides envelope biosynthesis pathways in S. zooepidemicus, maybe relative to the virulence of the bacteria[10].
This virulence attenuate of SEZugp maybe induced by decrease of HA production. Most part of the capsule of S. zooepidemicus is formed by HA, and the HA capsular material contributes to adherence properties of S. zooepidemicus and might help the bacteria to resist phagocytosis by macrophages [25]. Some reports indicate UGPase plays an important role in virulence factor biosynthetic pathway. In Mesophilic Aeromonas, UGPase was involved in the synthesis of O34-antigen lipopolysaccharide (LPS) which is related to adhere and during infection [26]. In S. zooepidemicus, we believe that the UGPase affects bacterial virulence though participating biosynthetic pathway of HA. It may represent an important target in fighting bacterial infectious diseases.
References
Weissbom AE, Liu Q, Rumley MK (1994) UTP: alpha-d-glucose-l-phosphate uridylytransferase of Escheichia coli: isolation and DNA sequence of the galU gene and purification of the enzyme. J Bacteriol 176:2611–2618
Kleczkowski LA, Martz F, Wilczynska M (2005) Factors affecting oligomerization status of udp-glucose pyrophosphorylase. Phytochemistry 66(24):2815–2821
Martz F, Wilczynska M, Kleczkowski LA (2002) Oligomerization status, with the monomer as active species, defines catalytic efficiency of UDP-glucose pyrophosphorylase. Biochem J 367(Pt 1):295–300
Gandhi TK, Zhong J, Mathivanan S, Karthick L, Chandrika KN, Mohan SS, Sharma S, Pinkert S, Nagaraju S, Periaswamy B, Mishra G, Nandakumar K, Shen B, Deshpande N, Nayak R, Sarker M, Boeke JD, Parmigiani G, Schultz J, Bader JS, Pandey A (2006) Analysis of the human protein interactome and comparison with yeast, worm and fly interaction datasets. Nat Genet 38(3):285–293
Crater DL, Dougherty BA, van de Rijn I (1995) Molecular characterization of hasC from an operon required for hyaluronic acid synthesis in group A streptococci. Demonstration of UDP-glucose pyrophosphorylase activity. J Biol Chem 270(48):28676–28680
Las Heras A, Vela AI, Fernandez E, Legaz E, Dominguez L, Fernandez-Garayzabal JF (2002) Unusual outbreak of clinical mastitis in dairy sheep caused by Streptococcus equi subsp. Zooepidemicus. J Clin Microbiol 40(3):1106–1108
Salasia SI, Wibawan IW, Pasaribu FH, Abdulmawjood A, Lammler C (2004) Persistent occurrence of a single Streptococcus equi subsp. Zooepidemicus clone in the pig and monkey population in indonesia. J Vet Sci 5(3):263–265
Ural O, Tuncer I, Dikici N, Aridogan B (2003) Streptococcus zooepidemicus meningitis and bacteraemia. Scand J Infect Dis 35(3):206–207
Mao Y, Fan H, Lu C (2008) Immunoproteomic assay of extracellular proteins in Streptococcus equi ssp. Zooepidemicus. FEMS Microbiol Lett 286(1):103–109
Lai X, Wu J, Chen S, Zhang X, Wang H (2008) Expression, purification, and characterization of a functionally active Mycobacterium tuberculosis UDP-glucose pyrophosphorylase. Protein Expr Purif 61(1):50–56
Arnold KBL, Kopp J, Schwede T (2006) The swiss-model workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201
Kiefer FAK, Kunzli M, Bordoli L, Schwede T (2009) The swiss-model repository and associated resources. Nucleic Acids Res 37:D387–D392
Peitsch MC (1995) Protein modeling by e-mail. Bio/Technology 13:658–660
Lun S, Willson PJ (2004) Expression of green fluorescent protein and its application in pathogenesis studies of serotype 2 Streptococcus suis. J Microbiol Methods 56(3):401–412
Zhang J, Hao N, Chen GQ (2006) Effect of expressing polyhydroxybutyrate synthesis genes (phbcab) in Streptococcus zooepidemicus on production of lactic acid and hyaluronic acid. Appl Microbiol Biotechnol 71(2):222–227
Silveira EDA-FM, Guimaraes LA, da Silva FR, Carneiro VT (2009) Selection of reference genes for quantitative real-time PCR expression studies in the apomictic and sexual grass Brachiaria brizantha. BMC Plant Biol 9(1):84
Carey CMKM, Thompson S (2009) Escherichia coli o157:H7 stress and virulence gene expression on romaine lettuce using comparative real-time PCR. J Microbiol Methods 77(2):235–242
Gao JGA, Scott JR, Churchward G (2005) Binding of the global response regulator protein covR to the sag promoter of Streptococcus pyogenes reveals a new mode of covR-DNA interaction. J Biol Chem 280(47):38948–38956
Blank LMHP, Nielsen LK (2008) Evolution of the hyaluronic acid synthesis (has) operon in Streptococcus zooepidemicus and other pathogenic streptococci. J Mol Evol 67(1):13–22
Geisler M, Wilczynska M, Karpinski S, Kleczkowski LA (2004) Toward a blueprint for UDP-glucose pyrophosphorylase structure/function properties: Homology-modeling analyses. Plant Mol Biol 56(5):783–794
Gottschalk LM, Bon EP, Nobrega R (2008) Lignin peroxidase from Streptomyces viridosporus T7A: enzyme concentration using ultrafiltration. Appl Biochem Biotechnol 147(1–3):23–32
Jensen PR, Hammer K (1998) The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Appl Environ Microbiol 64(1):82–87
Chen WY, Marcellin E, Hung J, Nielsen LK (2009) Hyaluronan molecular weight is controlled by UDP-N-acetylglucosamine concentration in Streptococcus zooepidemicus. J Biol Chem 284(27):18007–18014
Chassaing DAF (2007) The lmo1078 gene encoding a putative UDP-glucose pyrophosphorylase is involved in growth of Listeria monocytogenes at low temperature. FEMS Microbiol Lett 275(1):31–37
Wibawan IW, Pasaribu FH, Utama IH, Abdulmawjood A, Lammler C (1999) The role of hyaluronic acid capsular material of Streptococcus equi subsp. Zooepidemicus in mediating adherence to Hela cells and in resisting phagocytosis. Res Vet Sci 67(2):131–135
Vilches S, Canals R, Wilhelms M, Salo MT, Knirel YA, Vinogradov E, Merino S, Tomas JM (2007) Mesophilic aeromonas UDP-glucose pyrophosphorylase (galU) mutants show two types of lipopolysaccharide structures and reduced virulence. Microbiology 153(Pt 8):2393–2404
Acknowledgments
This study was supported by grants from the National Basic Research Program of China (2006CB504403), and the Program for New Century Excellent Talents (NCET) in University (No. NCET-08-0794) of China.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ma, Z., Fan, Hj. & Lu, Cp. Molecular cloning and analysis of the UDP-Glucose Pyrophosphorylase in Streptococcus equi subsp. zooepidemicus . Mol Biol Rep 38, 2751–2760 (2011). https://doi.org/10.1007/s11033-010-0420-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-010-0420-8