Abstract
Fruit flesh color in tetraploid sour cherry (Prunus cerasus) is an important market-driven trait in the USA where the fruit from the dominant cultivar has brilliant red skin but clear/yellow flesh. This brilliant red color in the processed products differentiates products from sour cherries grown in the USA compared to those in Europe where the cultivars predominantly have dark purple-red flesh. In sweet cherry (P. avium), red skin and flesh colors were shown to be controlled by a major MYB10-associated locus. Sour cherry, which is derived from sweet cherry and ground cherry (P. fruticosa), also exhibits a range of flesh colors, but the genetic control of flesh color is not known. Our objectives were to test the hypothesis that the MYB10 locus controls flesh color in sour cherry and develop a predictive DNA test for dark purple-red flesh color. Pedigree-linked sour cherry plant materials were phenotyped for flesh color. Thirteen haplotypes for the sour cherry MYB10 region were distinguished based on markers scored from the use of the cherry 6K Infinium® II SNP array. Six haplotypes were significantly associated with variation in flesh color, supporting a role for MYB10 in controlling flesh color variation in sour cherry. A simple sequence repeat primer pair, designed from the peach genome sequence near MYB10, amplified a fragment that uniquely identified the haplotype that was associated with the darkest purple-red flesh color. This marker can be used for marker-assisted breeding to identify individuals that are predicted to have dark purple-red flesh.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The US sour cherry industry is based on the cultivar ‘Montmorency’ that has yellow flesh and a brilliant red skin color. This is in contrast to the sour cherry cultivars grown in other major sour cherry-producing countries that typically have dark red, almost purple, flesh color. The brilliant red color of the processed products from ‘Montmorency’ provides a unique market class for US producers and is the recognizable color of cherry pie. Therefore, one of the major goals of the Michigan State University (MSU) sour cherry breeding program is to develop new improved ‘Montmorency’ types that retain the characteristic ‘Montmorency’ red color. Breeding efficiency would be significantly increased if progeny populations could be enriched for individuals that have the desired flesh color through the use of a DNA test that would allow the elimination prior to field planting of those individuals predicted to have dark purple-red flesh.
The genetic control of the red anthocyanin pigment in sour cherry is not known; however, in apple (Malus spp.) an anthocyanin MYB transcription factor, MdMYB10, was shown to control red fruit flesh and foliage (Espley et al. 2007; Chagné et al. 2007). In sweet cherry, a quantitative trait locus (QTL) associated with the locus that has the highest homology with MdMYB10 (PavMYB10) was identified as the major determinant of red coloration in sweet cherry skin and flesh (Sooriyapathirana et al. 2010). This major locus, located on Prunus linkage group 3, controlled as much as 94 % of the variation for sweet cherry flesh color with red flesh dominant to yellow flesh. Since diploid sweet cherry is one of the two progenitor species of tetraploid sour cherry (2n = 4x = 32), the ground cherry being the other progenitor species (Olden and Nybom 1968), we hypothesized that this gene, MYB10, may also be the major determinant of flesh color in sour cherry.
To determine the genetic control of flesh color in sour cherry, we took advantage of existing sour cherry breeding populations segregating for fruit flesh color and the ability to obtain genetic data for the parents and progeny individuals using the cherry 6K Infinium® II SNP array as part of the RosBREED project (www.rosbreed.org; Peace et al. 2012). As sour cherry is a segmental allotetraploid that exhibits irregular pairing at meiosis, the generation of a linkage map and a subsequent QTL analysis were not possible (Beaver and Iezzoni 1993; Iezzoni et al. 2005). Therefore, a targeted approach based on the hypothesis that MYB10 is the major determinant of flesh color was used. The objective was to test this hypothesis and develop a DNA test that would be predictive of flesh color for use in marker-assisted breeding.
Materials and methods
Plant populations and phenotyping
A total of 273 cultivars and seedlings from five bi-parental populations including parents were used in this study (Fig. 1). Populations were as follows: ‘Újfehértói Fürtös’ (‘UF’) × ‘Surefire’ (n = 69), M172 × 25-02-29 (n = 78), 25-14-20 × 25-02-29 (n = 58), ‘Montmorency’ × 25-02-29 (n = 44), and ‘Rheinische Schattenmorelle’ (‘RS’) × ‘Englaise Timpurii’ (‘ET’) (n = 24). These individuals were planted at the MSU Clarksville Research Station, Clarksville, Michigan.
Pedigrees of plant materials that were used for validating the MYB10-associated flesh color QTL in sour cherry visualized using Pedimap Software (Voorrips et al. 2012). The five full sib families used in this study represent a large portion of the diversity found in the breeding germplasm at Michigan State University. N is the number of progeny individuals in each population
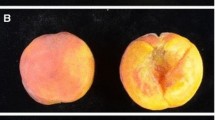
A fruit flesh color rating for each individual was taken in 2011 according to the Sweet Cherry Flesh Color Index from Washington State University (WSU) (Supplementary Fig. S1). A visual rating was given after observing five fruits. The 1–5 score represented clear/yellow flesh color (score of 1), through shades of red (scores 2–4) to dark purple-red color (score of 5). Results from the previous analysis conducted in sweet cherry supported this phenotyping strategy (Sooriyapathirana et al. 2010) as the color index values were highly significantly correlated with quantitative color measurements obtained using a spectrophotometer, and the color index values were highly significantly correlated across 3 years of evaluation.
Genotyping and haplotype construction
Four hundred and two sour cherry individuals, including founders, seedlings, and all 273 individuals in the five bi-parental populations, were genotyped using the 6K Infinium® II SNP array developed as part of the RosBREED project (www.rosbreed.org; Peace et al. 2012). The Illumina® Genome Studio software was used to determine the SNP genotype. Available SNP data from a subset of sweet cherry selections and hybrid seedlings (105 individuals) were included to aid in the determination of dosage by showing the two homozygous (AAAA and BBBB for sour cherry corresponding to AA and BB in sweet cherry) and balanced heterozygous (AABB for sour cherry corresponding to AB in sweet cherry) classes.
A total of 47 SNP markers that spanned the location of the MYB10 were used to build the sour cherry haplotypes (Supplementary Table S1). These SNPs spanned the physical map location of the three MYB10 homologs identified on the Peach v1.0 linkage group 3 assembly (scaffold 3, 12.84–12.91 Mb, Verde et al. 2013). For each parent and progeny individual, four haplotypes (to represent the four chromosomes in a tetraploid) were built for the target regions of the genome in an Excel spreadsheet by hand based on progeny inheritance and segregation in each of the bi-parental populations. The haplotypes were initially arbitrarily named with alphabetical letters; however, they were subsequently renamed based upon the presence or absence of a significant phenotypic effect, whereby ‘D’ and ‘d’ were used to indicate a significant increase or decrease, respectively, in dark red color associated with the presence of the haplotype, and an ‘x’ was used to indicate that no significant effect on color was associated with this haplotype.
Statistical analysis
ANOVA calculations were used to determine whether the MYB10 haplotypes identified in sour cherry were significantly associated with flesh color. A linear additive model test was implemented with a user-defined design matrix to consider the presence or absence of each haplotype as well as the number of times each haplotype was present (to account for dosage), using a modified R-script 2.15.1 (R Development Core Team 2011). The Proc Mixed least squares means statement in SAS 9.2® (SAS Institute Inc., Cary, NC, USA) was used to determine whether the means of the different haplotypes were significantly different. To confirm the ANOVA calculations and determine whether the MYB10 haplotypes had a positive or negative effect on the trait, Student’s t tests were performed comparing progeny individuals within each family with and those without each of the haplotypes that were significant in the linear additive model.
DNA test for flesh color
SSR markers for flesh color that tagged the MYB10 were designed based on the peach genome v1.0 (Verde et al. 2013) available at www.rosaceae.org. Microsatellite repeats were found using Microsatellite Repeats Finder (www.biophp.org/minitools/microsatellite_repeats_finder/demo.php). The criteria for SSR selection were the same as in Stegmeir et al. (2014): 2-bp minimum repeat length and 5-bp maximum repeat length with a minimum number of 10 repeats. If multiple SSRs were found in the region of interest, those with the largest number of tandem repeats were selected. Primer pairs were designed using Primer3web (bionifo.ut.ee/primer3/), with product size limited to 150–250 bp, and primers selected on either side of the repeat region with a CG clamp. Primer sequences were compared to the Peach genome v1.0 scaffolds (Verde et al. 2013) using NCBI’s BLAST. Primers that co-located to multiple scaffolds with at least 75 % sequence similarity were not used.
To test the ability of the SSR to distinguish between the MYB10 haplotypes, DNA was extracted from a subset of the parents and hybrid seedlings known to segregate for the dark red-purple flesh haplotype using the procedure described by Edge-Garza et al. (2014). The PCR mixture was as in Olmstead et al. (2008). A touchdown PCR was used for the flesh color diagnostic SSR primer pair called LG3_13.146, which has a forward primer sequence of 5′ ATG TGG CCA AAG GTC AGC 3′ and reverse primer sequence 5′ TGA TCC CAA TCA CGT TTT CC 3′. The conditions were as follows: 94 °C for 5 min followed by nine cycles of 94 °C for 30 s, 60 °C for 45 s (−1 °C per cycle), 72 °C for 1 min, and then 24 cycles of 94 °C for 30 s, 55 °C for 45 s, 72 °C for 1 min with an elongation step of 72 °C for 5 min. PCR fragments were separated on a 6 % polyacrylamide gel and visualized with silver staining as in Olmstead et al. (2008).
MASS was implemented using this SSR for five seedling populations generated from crosses made in 2013 to identify those individuals predicted to have dark red-purple flesh color, and the numbers and percentages of progeny individuals culled were recorded. All crosses had the dark purple-red-fleshed MSU selection 25-14-20 as the seed parent with five different red- and clear/yellow-fleshed MSU selections as pollen parents (27-03-08, 27e-04-54, 27e-05-33, 27e-15-38, and 27e-16-47 (Supplementary Table S2).
Results
Phenotypic variation
The phenotypic distributions for flesh color in all populations ranged from 1 (no red color in the flesh) to 5 (very dark purple-red flesh), but did not show a normal distribution in any of the populations (Supplementary Fig. S2). The three populations that had a very dark purple-red-fleshed parent (underlined), ‘UF’ × ‘Surefire,’ 25-14-20 × 25-02-29, and ‘RS’ × ‘ET,’ tended to skew toward the darker red flesh colors. In contrast, those populations that did not have a very dark purple-red-fleshed parent, M172 × 25-02-29, and ‘Montmorency’ × 25-02-29, were slightly skewed to lighter flesh color. The use of 25-02-29 as a parent in three of the five populations allowed a comparison of the combining ability of this selection with parents with color index scores of 5 (25-14-20), 4 (M172), and 2 (‘Montmorency’). When 25-02-29 was crossed with 25-14-20, the progeny were skewed toward the dark purple-red color class, possibly due to a dominant allele for red color contributed from 25-14-20. In contrast, when 25-02-29 was crossed with the lighter-colored M172 and then the clear/yellow-fleshed ‘Montmorency,’ there were increased percentages of lighter-fleshed progeny individuals.
Haplotypes for the MYB10 region
A total of 47 SNPs that spanned the MYB10 region (9.73–15. 46 Mb) were used to build 13 haplotypes that represented the alleles identified in the eight sour cherry parents (Supplementary Table S1). These haplotypes could be distinguished using a subset of the constructed SNP region that ranged from 10.57 to 14.02 Mb and represented approximately 10 cM (Table 1). Progeny segregation data for the 13 haplotypes were used to determine the haplotype genotypes of the parents, including whether the haplotype was in single or double dose, such as the presence of two x2 haplotypes in 25-02-29 (Table 2). The genotypes deduced were consistent with the known parent–offspring relationships. ‘UF’ and 25-14-20 share the x1, x2, and D1 haplotypes which is expected as ‘UF’ is the maternal parent of 25-14-20 (Fig. 1). Likewise ‘RS’ and 25-02-29 share the x2 and D2 haplotypes as ‘RS’ is the maternal parent of 25-02-29 (Fig. 1).
Haplotypes associated with fruit color
ANOVA indicated that seven of the thirteen MYB10 haplotypes contribute significantly to flesh color in sour cherry (Supplementary Table S2). Within the five bi-parental populations, t tests confirmed that six of the thirteen MYB10 haplotypes were significantly associated with flesh color (Table 2). Progeny that inherited the D1 haplotype from the dark purple-red-fleshed ‘UF’ or its dark purple-red-fleshed offspring 25-14-20 had significantly darker red flesh color than those progeny that did not inherit the D1 haplotype. Progeny that inherited the D2 haplotype from ‘RS’ or its offspring 25-02-29 also had significantly darker red flesh than those that did not inherit the D2 haplotype. Inheritance of the D3 haplotype from ‘Surefire’ or ‘ET’ also resulted in progeny that had significantly redder flesh than those without the D3 haplotype. Finally, the inheritance of the D4 haplotype from M172 or ‘ET’ also resulted in significantly redder flesh color. Collectively, the effect of these haplotypes on color was greatest for the D1 haplotype followed by the D2, D3, and D4 haplotypes.
Two haplotypes were significantly associated with lighter flesh color in at least one population (Table 2). The presence in the offspring of the d1 haplotype from 25-02-29 and ‘Montmorency’ was significantly associated with lighter flesh color. The presence of the d2 haplotype derived from ‘UF’ was significantly associated with light flesh color, but the effect of d2 derived from ‘M172’ was nonsignificant. The significance of the d2 haplotype from ‘UF’ may be due to the absence of inheriting the D1 haplotype, whereas the lighter red-fleshed ‘M172’ does not have the very purple-red D1 haplotype.
Since no parent had more than one significant dark red flesh haplotype (with the exception of ‘ET’ which had two), the effect of an individual having two dark red-fleshed haplotypes was examined in the three largest progeny populations. For ‘UF’ × ‘Surefire,’ progeny individuals that had neither the D1 nor D3 haplotypes had light flesh (mean = 1.9; Fig. 2a). The presence of a D3 haplotype, in the absence of the D1 haplotype, resulted in a significant increase in flesh color (mean = 3.0). However, any progeny individual that had a D1 haplotype had significantly darker flesh color (means of 4.8 and 4.5) irrespective of the presence of the D3 haplotype. The effect of the D1 haplotype was similar when compared to the D2 haplotype, suggesting that the D1 haplotype by itself results in the maximum purple-red flesh color (Fig. 2b). This is consistent with the ‘UF’ × ‘Surefire’ and 25-02-29 × 25-24-20 progeny phenotypic data that are highly skewed to the dark purple-red color class when one parent contains the D1 haplotype (Fig. 3). The vast majority of those progeny that had a color index score of 5 had the D1 haplotype (Fig. 3). In contrast, the two ‘weaker’ red-fleshed haplotypes (D2 and D4) exhibited an additive effect on flesh color with the presence of both haplotypes having a significantly increased red flesh color compared to the presence of only one haplotype (Fig. 2c). The interaction of these flesh color haplotypes is consistent with the transgressive segregation for flesh color (scores 1–5) seen in the cross M172 × 25-02-29, where both parents have a color index score of 4 (Supplementary Fig. S2).
Within population mean comparisons of dark flesh haplotypes D1, D2, D3, and D4 for populations, a ‘UF’ × ‘Surefire,’ b 25-14-20 × 25-02-29, and c M172 × 25-02-29. Each parent contributes one dark red-fleshed haplotype. Within each population, means that were significantly different (P < 0.05) are identified with different letters
Flesh color ratings for the 69 progeny from the ‘UF’ × ‘Surefire’ population (a) and 58 progeny from the 25-14-20 × 25-02-29 population (b). Those progeny individuals that have the D1 haplotype are identified with white dots on a black background and those that do not have the D1 haplotype are identified with black dots on a white background. Arrows mark the flesh color ratings of the parents
Marker-assisted seedling selection (MASS)
The presence of one D1 haplotype was significantly associated with the darkest flesh color class (purple-red) that is not preferred for the US sour cherry markets. Therefore, the effects of implementing MASS against the D1 haplotype were examined in the ‘UF’ × ‘Surefire’ and the 25-14-20 × 25-02-29 populations (Fig. 3). If MASS had been implemented in these two populations, 41 % (52/127) of the progeny would have been discarded. None of the progeny discarded would have had color index scores of 1 and 2. The progeny that would have been discarded would have overwhelmingly had a color index score of 5 (71 %), followed by a score of 4 (23 %) and a score of 3 (6 %). Of those progeny that did not have the D1 haplotype and would not have been discarded, 11 % (8/75) had the color index score of 5. Therefore, MASS against the D1 haplotype collectively in these two populations would have reduced the frequency of progeny individuals with the dark purple-red flesh color from 35 to 11 %. This enrichment of the remaining seedlings was sufficiently compelling to implement in the sour cherry breeding program.
To enable MASS for the presence or absence of the D1 haplotype in a progeny individual, a DNA marker had to be developed that uniquely identified the D1 haplotype. The D1 haplotype differs from all the other MYB10 haplotypes by the presence of BBBA for four consecutive SNPs (Table 1, see bold). A SNP diagnostic marker, however, could not be developed because none of the SNPs were unique to this haplotype. Instead, it was the phase of the SNP markers (coupling versus repulsion for the A and B alleles) that distinguished this haplotype. Therefore, the strategy was to design an SSR marker that had a repeat length that uniquely distinguished the D1 haplotype.
Taking advantage of the synteny between peach and cherry, forty SSRs were identified within 1 Mb of the MYB10 and tested for their ability to uniquely distinguish the D1 haplotype (Supplementary Fig. S3). Of the forty SSRs, only one that was located ~200,000 kb from MYB10 at position 13.146 Mb was found to uniquely differentiate the D1 haplotype. A 218-bp fragment amplified with this SSR uniquely co-segregated with the presence/absence of the D1 haplotype. This SSR also amplified a 220-bp fragment that uniquely co-segregated with the presence/absence of the D2 haplotype (Supplementary Fig. S4). Therefore, the SSR marker, LG3_13.146, fit the criteria for use for MASS for the D1 haplotype.
MASS against the D1 haplotype was implemented using the LG3_13.146 marker in progeny populations where the maternal parent was the dark purple-red-fleshed selection 25-14-20 that has one copy of the D1 haplotype (Supplementary Table S3). As none of the pollen parents have the D1 haplotype, 50 % of the progeny would be expected to have the D1 haplotype resulting in the potential of discarding a half of the seedlings. Across all populations, 619 progeny were screened and the 358 progeny (58 %) were found to have the D1 haplotype. These plants were subsequently discarded; therefore, the number of plants advanced to the breeding orchard totaled 261 as opposed to 619.
Discussion
Thirteen unique SNP haplotypes were identified spanning the MYB10 locus from analyses of eight sour cherry parents. For seven of these haplotypes, no significant effect on fruit flesh color was identified. However, four and two of the haplotypes were significantly associated with dark purple-red versus light red/yellow flesh colors in sour cherry, respectively, validating the MYB10-associated fruit flesh color QTL previously identified in sweet cherry (Sooriyapathirana et al. 2010). This supports the previous finding that the MYB10 locus on Prunus linkage group 3 is a major factor contributing to sweet cherry red flesh color. However, even the presence of the darkest purple-red-fleshed haplotype D1 for MYB10 was not 100 % predictive of dark red flesh color. For example, in the cross ‘UF’ × ‘Surefire,’ three out of 21 progeny with the darkest color index score of 5 did not have the D1 haplotype, and two of the 13 with the D1 haplotype had the color index value of 3. Although MYB10 is likely the major determinant of flesh color in sour cherry, these results indicate that other loci are contributing to flesh color. This is similar to the findings in sweet cherry and peach. In sweet cherry, alleles at loci on chromosomes 6 and 8 influenced the fruit color predicted based on PavMYB10 alone (Sooriyapathirana et al. 2010). In peach (P. persica), a major QTL for red blush color was significantly associated with the linkage group 3 MYB10 region; however, minor QTLs were identified on linkage groups 4 and 7 (Frett et al. 2014). Identifying these additional loci influencing fruit anthocyanin pigmentation will be critical to increase the predictive ability of marker-assisted breeding for red flesh color in cherry.
This study was unable to determine how multiple copies of the same haplotype would influence flesh color, since no populations studied had more than one copy of any dark red flesh haplotype. For example, there were no individuals that had more than one copy of the D1 haplotype. However, based on the finding that the moderate flesh color haplotypes D3, D4, and D2 tended to have an additive affect when found together in the same individual, it could be inferred that two copies of the same dark flesh haplotype would have a similar result.
The D1 haplotype was determined to be an excellent candidate for marker-assisted breeding in sour cherry as it was very significantly associated with the dark purple-red fruit flesh color that is not preferred in the US markets and it tended to exhibit dominant gene action. Reducing the frequency of the D1 haplotype in the MSU breeding seedling populations would likely result in enrichment of progeny for the desired lighter flesh colors, resulting in significant cost savings. For example, those seedlings that were discarded due to the presence of the D1 haplotype would otherwise have been planted and grown in the breeding orchard for three or four years before the flesh color would have been determined. In Europe, where a very dark purple-red flesh color is preferred for cherry juice and certain other processed products, a DNA test for the D1 haplotype would also potentially be useful. However, in their case, MASS would likely be used to identify those progeny individuals predicted to have very dark red-purple flesh color so that they could be maintained.
Our ability to develop a DNA test that provided sufficient confidence for use in the breeding program was due in a large part to the examination of a wide range of germplasm that represented the diversity in the breeding program. This allowed the identification of multiple haplotypes segregating in the genepool and the characterization of the functional haplotypes in different genetic backgrounds. A DNA test that met the specific breeding goal could then be designed. This approach of QTL validation in pedigree-linked germplasm is a central strategy used in the ‘RosBREED’ project (Peace et al. 2014) to enable marker-assisted breeding in rosaceous crops.
As sweet cherry is one of the two progenitor species of sour cherry, the sour cherry haplotypes were further compared to the sweet cherry haplotypes identified to date in a diverse set of 268 sweet cherry individuals in the ‘RosBREED’ sweet cherry Crop Reference Set (Peace et al. 2012) using three common SNPs that flanked the MYB10 region (ss490551678, ss490551684, and ss490551723) (sweet cherry data provided by P. Sandefur and C. Peace, per. comm.). Based on these three SNPs, the sour cherry haplotypes D1, D2, D3, x1, x2, x3, and x7 were not identified in sweet cherry, suggesting that these haplotypes may be unique to sour cherry, possibly contributed by the ground cherry subgenome. In particular, the dark purple-red flesh color haplotype D1 was not identified in sweet cherry. For the six other sour cherry haplotypes that had SNP markers in common with those in sweet cherry, it was not possible to confirm identity based on the SNPs alone. Additionally, it is possible that the SNP haplotypes overrepresent the number of MYB10 alleles. Further characterization of the MYB10 alleles would require sequence data spanning the MYB10 region. However, the different effects of the four red color haplotypes in sour cherry suggest that there are multiple functional variants of the MYB10 locus that result in differences in the presence and intensity of red flesh color.
References
Beaver JA, Iezzoni AF (1993) Allozyme inheritance in tetraploid sour cherry (Prunus cerasus L.). J Am Soc Hortic Sci 118:873–877
Chagné D, Carlisle CM, Blond C, Volz RK, Whitworth CJ, Oraguzie NC, Crowhurst RN, Allan AC, Espley RV, Hellen RP, Gardiner SE (2007) Mapping a candidate gene (MdMYB10) for red flesh and foliage colour in apple. BMC Genom 8:212
Edge-Garza D, Rowland T, Haendiges S, Peace C (2014) A high-throughput and cost-efficient DNA extraction protocol for the tree fruit crops apple, sweet cherry, and peach relying on silica beads during tissue sampling. Mol Breed 34:2225–2228
Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 49:414–427
Frett TJ, Reighard GL, Okie WR, Gasic K (2014) Mapping quantitate trait loci associated with blush in peach [Prunus persica (L.) Batsh]. Tree Genet Genomes 10:367–381
Iezzoni A, Sebolt AM, Wang D (2005) Sour cherry breeding program at Michigan State University. Acta Hortic 667:131–134
Klagges C, Campoy JA, Quero-Garcia J, Guzman A, Mansur L, Gratacos E, Silva H, Rosyara UR, Iezzoni A, Meisel LA, Dirlewanger E (2013) Construction and comparative analyses of highly dense linkage maps of two sweet cherry intra-specific progenies of commercial cultivars. PLoS ONE 8(1):e54743
Olden EJ, Nybom N (1968) On the origin of Prunus cerasus L. Hereditas 59:327–345
Olmstead JW, Sebolt AM, Cabrera A, Sooriyapathirana SS, Hammar S, Iriarte G, Wang D, Chen CY, van der Knaap E, Iezzoni AF (2008) Construction of an intra-specific sweet cherry (Prunus avium L.) genetic linkage map and synteny analysis with the Prunus reference map. Tree Genet Genomes 4:897–910
Peace C, Bassil N, Main D, Ficklin S, Rosyara UR, Stegmeir T, Sebolt A, Gilmore B, Lawley C, Mockler TC, Bryant DW, Wilhelm L, Iezzoni A (2012) Development and evaluation of a genome-wide 6K SNP array for diploid sweet cherry and tetraploid sour cherry. PLoS ONE 7(12):e48305
Peace CP, Luby JJ, van de Weg WE, Bink MCAM, Iezzoni AF (2014) A strategy for developing representative germplasm sets for systematic QTL validation, demonstrated for apple, peach, and sweet cherry. Tree Genet Genome 10(6):1679–1694
R Development Core Team (2011) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. ISBN 3-900051-07-0, www.R-project.org
Sooriyapathirana SS, Khan A, Sebolt AM, Wang D, Bushakra JM, Lin-Wang K, Allan AC, Gardiner SE, Chagné D, Iezzoni AF (2010) QTL analysis and candidate gene mapping for skin and flesh color in sweet cherry fruit (Prunus avium L.). Tree Genet Genomes 6:821–832
Stegmeir T, Schuster M, Sebolt A, Rosyara U, Sundin G, Iezzoni A (2014) Cherry leaf spot resistance in cherry (Prunus) is associated with a quantitative trait locus on linkage group 4 inherited from P. canescens. Mol Breed 34(3):927–935
Verde I, Abbott AG, Scalabrin S, Jung S, Shu S, Marroni F, Zhebentyayeva T, Dettori MT, Grimwood J, Cattonaro F, Zuccolo A, Rossini L, Jenkins J, Vendramin E, Meisel LA, Decroocq V, Sosinski B, Prochnik S, Mitros T, Policriti A, Cipriani G, Dondini L, Ficklin S, Goodstein DM, Xuan P, Del Fabbro C, Aramini V, Copetti D, Gonzalez S, Horner DS, Ralchi R, Lucas S, Mica E, Maldonado J, Lazzari B, Bielenberg D, Pirona R, Miculan M, Barakat A, Testolin R, Stella A, Tartarini S, Tonutti P, Arús P, Orellana A, Wells C, Main D, Vizzotto G, Silva H, Salamini F, Schmutz J, Morgante M, Rokhsar DS (2013) The high-quality draft of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat Genet 45:487–494
Voorrips RE, Bink MCAM, van de Weg WE (2012) Pedimap: software for the visualization of genetic and phenotypic data in pedigrees. J Hered 103:903–907
Acknowledgments
The authors thank Paul Sandefur and Cameron Peace for their suggestions during manuscript preparation. This work was partially funded by USDA’s National Institute of Food and Agriculture (NIFA) Hatch project number MICL02332 and the USDA-NIFA Specialty Crop Research Initiative Competitive Grant entitle ‘RosBREED: Combining disease resistance and horticultural quality in new rosaceous cultivars’ (Grant No. 2014-51181-22378).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Stegmeir, T., Cai, L., Basundari, F.R.A. et al. A DNA test for fruit flesh color in tetraploid sour cherry (Prunus cerasus L.). Mol Breeding 35, 149 (2015). https://doi.org/10.1007/s11032-015-0337-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-015-0337-y