Abstract
Sharka is a severe apricot viral disease caused by the plum pox virus (PPV) and is responsible for large crop losses in many countries. Among the known PPV strains, both PPV-D (Dideron) and PPV-M (Marcus) are virulent in apricot, the latter being the most threatening. An F1 apricot progeny derived from Lito, described in the literature as resistant, crossed to the susceptible selection BO81604311 (San Castrese × Reale di Imola) was used to study the genetic control of resistance to PPV. A population of 118 individuals was phenotyped by inoculating both PPV-D and PPV-M strains in replicated seedlings and scored for 3 years. An additional set of 231 seedlings from the same cross was also phenotyped for 2 years. SSR-based linkage maps were used for quantitative trait locus (QTL) analysis. A major QTL of resistance to both PPV-M and PPV-D strains was found in the top half of the Lito linkage group 1, where a QTL was previously described in Stark Earli-Orange, the donor of Lito resistance. The LOD score was considerably enhanced when the recovery of plants from infection was taken into account. The results obtained in Lito were compared with those observed in a second apricot cross progeny (Harcot × Reale di Imola) in which QTL of resistance to sharka were also mapped in the same linkage group 1 for both PPV strains. Several models of resistance to sharka disease are discussed considering the segregation frequencies, the QTL alignment in the two maps and the information gathered from the literature.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sharka is currently the most serious disease of apricots and other stone fruit crops. The disease, caused by the plum pox virus (PPV), has spread rapidly throughout the world during the last 20–30 years and thousands of hectares of orchards have been cleared in an attempt at control. The cost worldwide of sharka management during the last 30 years has been estimated to exceed EUR10 billion (Cambra et al. 2006). Several sources of resistance have been identified in apricot germplasm and are currently being used both to understand the genetic control of the disease and to produce resistant selections. In spite of the large body of literature available, the genetic basis of sharka resistance in apricots is still under debate. Individual reports indicate that a single gene (Dicenta et al. 2000), two genes (Moustafa et al. 2001) or three genes (Guillet-Bellanguer and Audergon 2001) are responsible. In addition, polygenic resistance has been described in P. davidiana (Decroocq et al. 2005; Marandel et al. 2009a), a species closely related to peach [Prunus persica (L.) Batsch.]. In a more detailed study Soriano et al. (2008), analysed the segregation of resistance in four progenies (two F1 progenies with Goldrich and two F2 progenies with Lito as donors of resistance). They concluded that all progenies differed significantly from the expected ratio for a single dominant gene (1:1 in the F1 and 3:1 in the F2) and that the data better fitted a digenic model with two dominant independent loci controlling PPV resistance (expected and observed segregations were 1:3 in the F1 and 9:7 in the F2), except for the cross Goldrich × Canino whose progeny better fitted a three gene-based model.
The main problem in studying sharka resistance in apricot is that the phenotypic evaluation of disease symptoms in segregating progenies, beside being expensive, tedious and time-consuming, sometimes does not allow a reliable assignment to discrete classes of resistance/susceptibility. The determination of the number and localisation of the genetic determinants of sharka resistance is therefore a priority in apricot breeding programs, particularly in order to take advantage of marker-assisted selection (MAS).
Mapping in stone fruit species is made easier by the Prunus Reference Map based on the F2 progeny from Texas (almond) × Earlygold (peach) cross T × E (Joobeur et al. 1998; Aranzana et al. 2003; Dirlewanger et al. 2004; Howad et al. 2005). This reference map allows several maps of Prunus species, such as peach, plum, and apricot, to be aligned given the strict colinearity of genomes of those species (Dirlewanger et al. 2004, http://www.bioinfo.wsu.edu/gdr/MapView.php).
Several apricot genetic maps have recently been produced (Hurtado et al. 2002; Vilanova et al. 2003; Lambert et al. 2004, 2007; Dondini et al. 2007; Lalli et al. 2008; Soriano et al. 2008; Marandel et al. 2009a, b) with the aim of mapping the genetic determinants of sharka resistance in this species.
A first determinant was mapped on linkage group 1 (LG1) by using an F1 progeny of Goldrich × Valenciano (Hurtado et al. 2002). Goldrich is known to be tolerant to the pathogen while Valenciano was described as susceptible (Martinez-Gomez et al. 2000; Dicenta et al. 2000). This preliminary result was recently confirmed by a quantitative trait locus (QTL) analysis carried out on another F1 progeny, Goldrich × Currot (Soriano et al. 2008).
A major QTL was also identified in LG1 by the analysis of F1 and F2 progenies of Stark Earli-Orange (SEO; Lambert et al. 2004) and its offspring Lito (Vilanova et al. 2003; Soriano et al. 2008). Minor QTL were identified in the Polonais × SEO progeny in LG3 and LG5 of both SEO and Polonais (Lambert et al. 2007). The main QTL on LG1 was confirmed by Sicard et al. (2008), who developed new microsatellite (SSR) markers flanking the QTL, and by Lalli et al. (2008) in a backcross population of SEO × Vestar, and was again confirmed by Soriano et al. (2008) in the extended F2 Lito selfed progeny. Most of the resistance determinants described above were characterised using the PPV-D strain as a source of inoculum and, at present, the only resistances specific to the PPV-M strain are those reported in the BC1 SEO × Vestar (Lalli et al. 2008) and in P. davidiana (Marandel et al. 2009a).
The presence of QTL for sharka resistance in LG1 and LG3 has been also reported by Marandel et al. (2009b) in the cultivar Harlayne (by analysing an F1 progeny derived from the cross Harlayne × Marlén). In contrast to the above reports, a recombinant strain of PPV (PPV-rec; Glasa et al. 2004) was used.
A recent survey of apricot germplasm of different geographical origins (Zhebentyayeva et al. 2008) suggested an introgression of Chinese germplasm into the North American PPV-resistant gene pool. These authors also suggested a different breeding history of SEO and Goldrich-Harlayne lineages and a possible introgression of genetic material from non-domesticated apricots P. mandshurica, P. sibirica var. davidiana and P. mume.
Several preliminary results have been published concerning the genes involved. A cDNA-AFLP approach pointing to changes in gene expression profiles following PPV-D infection (Schurdi-Levraud Escalettes et al. 2006) revealed a putative class III chitinase being repressed in infected plants of the resistant cultivar Goldrich compared to the susceptible Screara, while a transketolase, a kinesin-like and an ankyrin-like protein were clearly associated with the susceptible interaction. SEO revealed very little differential expression even in the early stages after inoculation (Schurdi-Levraud Escalettes et al. 2006).
Marandel et al. (2009a, b) correlated the presence of mRNA of genes coding for eukaryotic translation initiation factors eIF4E and eIF(iso)4G with the resistance QTL found in P. davidiana P1908. This result seems to confirm previous data reported by Decroocq et al. (2005) who described a co-segregation of a Prunus eIF(iso)4E with a major QTL of resistance to sharka in peach. Similar findings were reported by Sicard et al. (2008) in apricot. Several other reports indicate that eIF4E and eIF(iso)4E genes, which seem critical for plant growth (Duprat et al. 2002), are as well required by potyvirus to carry on the infection (Ruffel et al. 2002; Diaz-Pendon et al. 2004). Such an occurrence has been found, for instance, in Arabidopsis (Duprat et al. 2002; Nicaise et al. 2007), lettuce (Nicaise et al. 2003) and Capsicum (Kang et al. 2005). This resistance model that deals with the inability of the virus to find one or several plant genes required for infection implies a recessive condition. If the evidence reported by Sicard et al. (2008) and Marandel et al. (2009a, b) is confirmed, the recessive condition of several sources of resistance could explain part of this trait, although this does not seem in agreement with the data collected in other segregating progenies which clearly indicate that PPV resistance is controlled by at least one dominant gene.
In the present work, both PPV-M and PPV-D strains were used to phenotype two F1 apricot progenies, namely Lito × BO81604311 (already described in Dondini et al. 2007) and one from Harcot × Reale di Imola. Their genetic maps were used to identify QTL of resistance to both virus strains and to discuss the nature of sharka resistance in apricot.
Materials and methods
Plant material
The apricot cultivars Lito and Harcot were selected because of their different genetic origins and resistance responses to PPV. Lito came from the cross SEO (the donor of sharka resistance) × Early of Tyrinthos. Harcot’s pedigree, as reported by Brooks and Olmo (1997), is [(Geneva × Naramata) × Morden 604 (Scout × McClure)] × NJA1 (Phelps × Perfection) and the resistance to sharka should be inherited from Perfection.
Two segregating F1 progenies were produced:
-
Lito × BO81604311 (hereinafter L × B and L × B extended) with 118 seedlings and 231 seedlings respectively;
-
Harcot × Reale di Imola (H × R) with 98 seedlings.
Inoculation with PPV
A phenotyping protocol developed at the Dipartimento di Protezione delle Piante e Microbiologia Applicata, Università di Bari (DPPMA) was used to evaluate resistance of the apricot populations to PPV (Amenduni et al. 2004). All seedlings were grafted onto Myrobalan 29C or GF305 rootstocks with 14 replicates, seven for the analyses with PPV-M strain and seven for the PPV-D strain. Five of the seven replicates were inoculated on the rootstock by double chip budding, while the other two plants were not inoculated, but retained as healthy controls. The virus strains used as inocula were isolate 0019-GR of PPV-M, found in Greece (Boscia et al. 1997), and isolate 0025-IT of PPV-D, found in Italy (Di Terlizzi et al. 1988). Both isolates are maintained on GF 305 in an in vivo collection of PPV isolates at the DPPMA. The trial took place in an insect-proof screen house at the Centro di Ricerca e Sperimentazione in Agricoltura “Basile Caramia” in Locorotondo (Apulia, Italy) until the end of June, at 368 m above sea level, in a hilly area characterised by a cool climate, suitable for expression of visual viral symptoms and for analysis for detection of PPV infections. Symptoms of the disease were visually scored and ELISA analyses were also performed. The success of the virus inoculations was confirmed by the presence of symptoms and by ELISA tests on the rootstocks. L × B and H × R progeny were phenotyped for 3 years and the L × B extended progeny for 2 years.
Visual inspections were carried out weekly during each growth cycle and leaf symptoms (vein clearing, yellow rings, spots and blotches) were scored as 0 (absence of symptoms), 1 (very mild symptoms, mainly in the basal part of plant), 2 (moderate symptoms on several leaves) and 3 (severe symptoms on whole plant). Based on the scoring system reported in the literature (Kegler et al. 1998; Dicenta et al. 2000), the seedlings were classified as: (a) resistant, that is a plant always negative to ELISA and showing no symptoms; (b) tolerant, that is a plant always positive to ELISA but showing no symptoms; (c) susceptible, that is a plant in which PPV readily multiplies and spreads (both rootstock and apricot are symptomatic).
Resistance/tolerance of the seedlings was also confirmed by top-grafting the apricot scions with GF305 since the second vegetative cycle. In this case the effective presence of the virus in the GF 305 was visually checked and confirmed by both ELISA and PCR.
DNA extraction, marker analysis and map construction
Genomic DNA was extracted from 200 mg of freeze-dried young leaves collected in spring following the protocol by Mercado et al. (1999) as modified by Dondini et al. (2007). The Lito and BO81604311 maps described in Dondini et al. (2007) were upgraded with the analysis of six new SSRs that filled the largest gaps on LG1. The new markers were aprigms 1, 2, 3, 10, and 16 (Vilanova et al. 2006) and MA067 (Yamamoto et al. 2005). A framework map of the LG1 based on 18 well-distributed SSR markers was then transferred to the L × B extended population.
A framework map was constructed for the H × R population by genotyping the 98 individuals with a set of regularly distributed markers from the series UDP (Cipriani et al. 1999; Testolin et al. 2000), UDAp (Messina et al. 2004), BPPCT (Dirlewanger et al. 2002), CPPCT (Aranzana et al. 2003), PaCITA (Lopes et al. 2002), MA (Yamamoto et al. 2005), aprigms (Vilanova et al. 2006; Lalli et al. 2008), UDA (Testolin et al. 2004) and CPDCT (Mnejja et al. 2005).
Segregation analysis of individual loci, identification of linkage groups and the linear position of markers were carried out with JoinMap 3.0 (van Ooijen and Voorrips 2002), which allows analysis of markers showing different segregation patterns in the one mapping population. Markers were first grouped using a minimum log of odds (LOD) score of 5 for the reference maps of Lito and BO81604311 and LOD 3 for the Harcot and Reale di Imola maps; all markers were included even if they showed skewed segregations. Kosambi’s function was used for calculating map distances. Maps were drawn using MapChart 2.0 (Voorrips 2002).
QTL analysis
The Kruskal–Wallis test and the Interval Mapping analysis were performed using MapQTL 4.0 (Van Ooijen et al. 2002) for QTL identification. Two sets of phenotypic data were used for both PPV strains: the first data set recorded the highest level of susceptibility found in each seedling during the 3 years of observation. The second data set was created with the scoring limited to the third season and therefore took into account those seedlings that had recovered. In LG1, only markers with the same statistical bases (no missing data) were used for QTL analysis.
For the L × B extended population, phenotyping for both PPV strains was limited to two seasons and in this case recovery was not taken into account. The “permutation test” (1000 permutations) was used to determine the significance threshold of the LOD score (corresponding to a probability higher than 95%) for a QTL to be considered significant after Interval Mapping analysis.
Results and discussion
Phenotypic analysis of PPV resistance: scoring classes and differences among PPV strains and crosses
During the 3 years of phenotypic observations, seedlings were consistently classified into three classes (resistant, tolerant and susceptible) on the basis of symptoms and ELISA tests. Lito fell into the resistant class, whereas Harcot was classified as tolerant.
Resistant seedlings, whose GF 305 rootstock did not show symptoms, were not taken into account as inoculation was considered unsuccessful. In the L × B cross, there were 22 such seedlings amongst those inoculated with PPV-D and four amongst those inoculated with PPV-M but only three in the H × R cross when inoculated with either strain.
The initial assessment considered the maximum degree of susceptibility observed in any of the 3 years of the experiment. This means that seedlings classified in different categories in different years were conservatively assigned to the worst class: 12 and 22% respectively of seedlings of the L × B population inoculated with PPV-M and PPV-D were classified as resistant, 13 and 20% respectively as tolerant and 75 and 58% respectively as susceptible (Fig. 1a, b). The ratio of pooled resistant and tolerant classes to susceptible seedlings was 25:75 for PPV-M and 42:58 for PPV-D, respectively. Both ratios are not consistent with the expected 1:1 ratio for a monogenic dominant resistant trait. These results were confirmed by the analysis of the L × B extended population although it was evaluated for only 2 years. Collectively, these results confirm the data frequently reported in the literature of a polygenic control in apricot and other Prunus species.
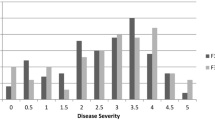
a–f Evaluation of the degree of resistance to PPV (M and D strains) of the individuals derived from the F1 populations Lito × BO81604311 (a–d), Lito × BO81604311 extended (e–f) and Harcot × Reale di Imola (g–h). Data collected during 3 years (two for the L×B extended) have been used to group all the seedlings into three classes: resistant (asymptomatic and ELISA-negative), tolerant (asymptomatic and ELISA-positive) and susceptible (symptoms clearly evident in the leaves). “Max susceptibility” indicates the dataset by considering the maximum degree of susceptibility observed in the 3 years of the tests (a, b, e, f). “With recovery” indicates the dataset that takes into account the recovery in the third year (c, d)
Similar results with an excess of susceptible genotypes were recorded for the H × R progeny, approximately 80% of whose seedlings were recorded as being susceptible to both strains of the virus (Fig. 1g, h).
The PPV-M strain appears to be more virulent at least to the Lito progeny, as 12 seedlings classified as resistant to or tolerant of PPV-D were scored as susceptible to the PPV-M and only two seedlings resistant to or tolerant of PPV-M were susceptible to PPV-D. The same trend was confirmed in the L × B extended progeny in which 36 seedlings resistant or tolerant to PPV-D were susceptible to PPV-M. This difference in susceptibility to either strain was not observed in the H × R progeny, in which only two seedlings differed in their susceptibility to the two PPV strains.
During the third year of phenotyping, 10 seedlings of the L × B progeny susceptible to PPV-M became symptomless and ELISA-negative. At the same time another 12 susceptible seedlings became tolerant and two tolerant seedlings became resistant. A similar trend, but to a lesser extent, was also observed in seedlings inoculated with PPV-D. This observation suggests that several seedlings probably recovered from infection. If recovery is taken into account, the distribution of seedlings in resistant/tolerant and susceptible classes becomes similar for inoculation with the two PPV strains (Fig. 1c, d). Recovery from PPV infection has previously been described in apricot. Karayiannis et al. (2008) reported that seedlings of the SEO × Bebecou progeny recovered the second spring after inoculation. The authors suggested that recovery from PPV symptoms was due to a type of resistance carried by SEO, apparently different from both hypersensitivity reactions and immunity.
In addition, Lambert et al. (2007) observed ten individuals in the Polonais × SEO progeny that showed important changes in virus accumulation over time and eventually recovered. The same phenomenon was also observed in Prunus davidiana (Decroocq et al. 2005).
QTL for sharka resistance in Lito
The genetic map of Lito, upgraded after the first publication (Dondini et al. 2007), currently covers 532 cM with 161 markers at a mean separation of 3.3 cM (data not shown). In particular, the LG1 is covered by 27 evenly spaced markers spanning 91.1 cM with an overall marker density of one marker every 3.4 cM, which reduces to 1.4 cM in the region bearing resistance to sharka (from UDAp-463 to PaCITA5).
The QTL analysis gave similar results using either the Kruskal–Wallis or the Interval Mapping algorithms. A major QTL was observed corresponding to the SSR UDAp-441 with a LOD score of 9.7 (accounting for 32.5% of the variance) when plants were inoculated with the PPV-M strain and scored for the maximum presence of symptoms. An adjacent QTL corresponded to the MA067 marker with a LOD of 16.1 explaining the 53.9% of the variance when the progeny was inoculated with PPV-D (Table 1; Fig. 2). Both results considerably exceeded the LOD threshold of 1.8 calculated by the permutation test. The LOD curve shows a second minor peak corresponding to the SSR PaCITA7 for PPV-M (LOD score 4.5, accounting for 16.7% of the variance) and the SSR aprigms8 (LOD score 3.5, accounting for 15.5% of the variance) for PPV-D (Table 1; Fig. 2). This second QTL should also exceed the LOD threshold of 1.8 calculated by the permutation test. The presence of QTL in addition to the major one in LG1 has been also described in SEO (Lambert et al. 2007) and Harlayne (Marandel et al. 2009b).
QTL for sharka resistance identified on the linkage group 1 of cultivar Lito (Lito from the progeny L×B; Lito extended from the progeny L×B extended). The probability of the association of the markers to the “resistance trait” (after Interval Mapping analysis) is indicated as LOD score (PPV-M in black and PPV-D in grey). The continuous lines indicate the QTL analysis by using the maximum level of susceptibility of the seedlings observed in 3 years of observations. The dotted lines indicate the LOD scores calculated by using the phenotypic data with recovery. Cross-hatched lines indicate the LOD threshold. Arrows indicate the position of QTL
When plant recovery was taken into account, the LOD score of the main QTL in the L × B progeny increased to 14.8 (accounting for 45% of the variability) and the QTL peak shifted slightly from UDAp-441 to the pchcms4 marker (Table 1). LOD score and peak position did not change with PPV-D. For the second QTL the slippage was only for PPV-M and the peak shifted slightly from PaCITA7 to CPPCT26 with a comparable LOD score (Table 1; Fig. 2).
In the L × B extended progeny, the QTL peaked near marker pchcms4 with a LOD of 21.9 and 37.2 respectively for PPV-M and PPV-D strains (LOD threshold of 3.0). In the extended population, the genes controlling resistance appear to be restricted to a 5 cM region. The second QTL peaks in the region of aprigms8 with a LOD score of 8.5 and 8.4 for PPV-M and PPV-D respectively.
No QTL was detected in the susceptible parent BO81604311.
To assess the impact of tolerance on the QTL analysis, we tested the effect of scoring all tolerant seedlings as either susceptible or resistant. In the former case, the main QTL decreased significantly for both strains, while in the latter case it was slightly enhanced (data not shown). This would suggest that tolerance is more correctly associated with resistance rather than being taken as indicating incomplete development of the disease.
The identification of a main QTL on LG1 of Lito is in agreement with Vilanova et al. (2003) and Soriano et al. (2008) who suggested the presence in Lito of a single dominant gene modulated by the activity of other minor genes. A single dominant gene inherited by SEO, Lito’s mother, is also described by Rubio et al. (2007) and Karayiannis et al. (2008) and in the latter paper a 1:1 segregation ratio is reported. In our work, if recovery is considered as a type of resistance to the pathogen, the ratio between the group that includes resistant, tolerant and recovered individuals and the group that is susceptible approaches 1:1.
All the L × B resistant or tolerant seedlings show the UDAp441 allele in coupling with resistance while the 71.1% of the seedlings susceptible to at least one strain are carrying the allele in repulsion (data not shown). Analogously to the L × B extended progeny, 92% of both resistant and tolerant plants shared the UDAp441 “plus allele”, that is the allele of the resistant parental haplotype, while 74% of susceptible plants carried the “minus allele” (in this case the percentages were 93 and 79% respectively using pchcms4 as marker). These results suggest that these markers should be considered for use in MAS to greatly reduce the number of seedlings that have to be phenotyped and, consequently, to save time, space and the costs of the artificial inoculation test.
Other QTL for sharka resistance such as those described in the LGs 3 and 5 of SEO by Lambert et al. (2007) or in the LG3 of Harlayne (Marandel et al. 2009b) were not detected in the present work.
QTL for sharka resistance in Harcot
Two frames of SSR-based maps have been constructed for the parental lines Harcot (478 cM with 69 SSR markers) and Reale di Imola (464 cM with 51 SSRs) in order to cover evenly the eight chromosomes of the apricot haploid genome (data not shown). LG1 in Harcot is 143 cM long and has been oriented and aligned to that of Lito with markers CPPCT16, UDAp-441, MA067, UDP005, CPPCT34, PaCITA7, CPPCT26, BPPCT011 and UDAp-410 (Fig. 2). Several SSRs that mapped in the LG1 of Lito were not polymorphic in H × R progeny and could not be used. In spite of this, probably due to low genetic diversity between the two parents Harcot and Reale di Imola (Geuna et al. 2003), 20 SSRs have been mapped in the Harcot LG1 by testing many new markers isolated from different Prunus species.
QTL analysis revealed a weaker QTL in the same position as in Lito in the LG1 (Table 1; Fig. 3). Kruskal–Wallis analysis identified the SSR CPDCT042 (K index of 12.6 and 13.7 for PPV-M and PPV-D strains respectively) and UDAp-441 (K index of 12.6 and 11.1 for PPV-M and PPV-D strains respectively) as peaks of putative QTLs, but Interval Mapping did not reach the LOD threshold determined by permutation tests. Weak QTL peaks with LOD scores below the threshold were also observed in the LGs 6 and 7 in positions where minor QTL were described in P. davidiana (Decroocq et al. 2005; Marandel et al. 2009a). The low significance of these putative QTL is probably due to the small population size coupled with the very low number of resistant/tolerant plants. The differences observed in the L × B and H × R populations probably reflect the different behaviour of Lito and Harcot in response to PPV infections, the former being resistant and the latter tolerant.
Putative QTL for sharka resistance identified on the linkage group 1 of the apricot cultivar Harcot. The probability of the association of the markers to the “resistance trait” (after Kruskal–Wallis [KW] and Interval Mapping analyses) is indicated as KW index and LOD score respectively (PPV-M in black and PPV-D in grey). Arrows indicate the position of the putative QTL
No QTL was detected in the susceptible parent Reale di Imola.
Conclusions
A genomic region containing genes of resistance to sharka disease has been identified in apricot in the top half of LG1. In that region, a major QTL of resistance to both PPV-M and PPV-D strains was mapped in Lito and probably in Harcot, where a low LOD score was recorded.
A QTL of resistance to PPV-D strain was located in the same region of Goldrich (Hurtado et al. 2002; Soriano et al. 2008), SEO (Lambert et al. 2007; Sicard et al. 2008) and its offspring Lito (Vilanova et al. 2003; Soriano et al. 2008), while the resistance to the PPV strain M, the most aggressive in apricot, was only reported in SEO by Lalli et al. (2008) and in this paper. These results are consistent with those of Marandel et al. (2009b), who used a PPV-rec strain.
The segregation ratios of resistant and susceptible genotypes are not easily calculated because of the complex behaviour of the plant–pathogen interaction. Resistant and susceptible seedlings were easily identified because of the clearcut distinction between these two classes: resistant seedlings were always asymptomatic and negative to the ELISA test, while susceptible trees showed clear symptoms of disease and were positive to the ELISA test. However, there are also tolerant individuals that showed no phenotypic symptoms but were recorded as positive by the ELISA test.
Even if the tolerant and resistant seedlings are considered together, the segregation ratio was not 1:1, as would be expected for the segregation of a single dominant R gene heterozygous in the donor parent. A consistent bias towards an excess of susceptible individuals was always found, ranging from 58 to 80%. For this reason the character was treated as of quantitative nature.
The scenario was complicated by the recovery of some plants, initially classified as susceptible but becoming resistant or tolerant during the third year of scoring. When resistant, tolerant, and such recovered seedlings were pooled together, the ratio of these to susceptible plants approached 1:1. This explains why such a large part of the phenotypic variability is accounted for by a single QTL in the LG1. Other potential QTL either explained a very small part of the variance or were below the LOD threshold. The markers closer to the QTL peaks would already be suitable for marker-assisted breeding. Those plants that recovered should be retained in breeding programs only after careful consideration because such plants, although tolerant or resistant long-term, could be a source of virus infection in the field. However, they could be grown in regions/countries where PPV cannot be eradicated.
From a physiological point of view, resistance of apricots to sharka is based neither on an immunity mechanism (non-host resistance) nor a hypersensitive response (HR) triggered by recognition genes (Dangl and Jones 2001, 2006). According to the literature, it is due either to the absence of initiation factors needed by the virus for its replication (Duprat et al. 2002; Sicard et al. 2008; Marandel et al. 2009a, b) or to the inability of the virus to replicate rapidly, which does not require specific R genes. The first model is based on recessive genes and does not fit our data while the second one is more likely but the genes involved are more difficult to determine in terms of either number or function.
Studies of sharka resistance are easily affected by the small size of populations and the difficulties in scoring the mode of resistance (reviewed in Soriano et al. 2008). We were able to assess at least in Lito × BO81604311 a relatively large population of 349 individuals that allowed the unequivocal identification of a major QTL in the LG1 of several apricot accessions. We also initiated the screening of a random-shear BAC library of Lito with the markers surrounding the QTL region. Preparing a physical map of the region of interest will be the next step in the positional cloning of the gene(s) involved in sharka resistance.
References
Amenduni T, Bazzoni A, Minafra A, Savino V (2004) Evaluation of the susceptibility of seedlings from apricot crosses to the Marcus strain of Plum pox virus. Acta Hort 657:305–308
Aranzana MJ, Pineda A, Cosson P, Dirlewanger E, Ascasibar J, Cipriani G, Ryder CD, Testolin R, Abbott A, King GJ, Iezzoni AF, Arús P (2003) A set of simple-sequence repeat (SSR) markers covering the Prunus genome. Theor Appl Genet 106:819–825
Boscia D, Zeramdini H, Cambra M, Potere O, Gorris MT, Myrta A, Di Terlizzi B, Savino V (1997) Production and characterization of a monoclonal antibody specific to the M serotype of Plum pox potyvirus. Eur J Plant Pathol 103:477–480
Brooks RM, Olmo HP (1997) The Brooks and Olmo register of fruit and nut varieties, 3rd edn. ASHS Press, Alexandria
Cambra M, Capote N, Myrta A, Llácer G (2006) Plum pox virus and the estimated costs associated with Sharka disease. EPPO Bull 36:202–204
Cipriani G, Lot G, Huang WG, Marrazzo MT, Peterlunger E, Testolin R (1999) AC/GT and AG/CT microsatellite repeats in peach [Prunus persica (L.) Batsch]: isolation, characterisation and cross-species amplification in Prunus. Theor Appl Genet 99:65–72
Dangl JL, Jones JD (2001) Plant pathogens and integrated defence responses to infection. Nature 411:826–833
Dangl JL, Jones JD (2006) The plant immune system. Nature 444:323–329
Decroocq V, Foulongne M, Lambert P, Le Gall O, Mantin C, Pascal T, Schurdi-Levraud V, Kervella J (2005) Analogues of virus resistance genes map to QTLs for resistance to Sharka disease in Prunus davidiana. Mol Genet Genomics 272:680–689
Di Terlizzi B, Savino V, Castellano MA, Martelli GP (1988) Presenza della vaiolatura delle drupacee in Puglia e Basilicata. Informatore Fitopatologico 38:53–55
Diaz-Pendon JA, Truniger V, Nieto C, Garcia-Mas J, Bendahmane A, Aranda M (2004) Advances in understanding recessive resistance to plant viruses. Mol Plant Pathol 5:223–233
Dicenta F, Martinez-Gomez P, Burgos L, Egea J (2000) Inheritance of resistance to Plum pox potyvirus (PPV) in apricot, Prunus armeniaca. Plant Breed 119:161–164
Dirlewanger E, Cosson P, Tavaud M, Aranzana MJ, Poizat C, Zanetto A, Arús P, Laigret F (2002) Development of microsatellite markers in peach [Prunus persica (L.) Batsch] and their use in genetic diversity analysis in peach and sweet cherry (Prunus avium L.). Theor Appl Genet 105:127–138
Dirlewanger E, Graziano E, Joobeur T, Garriga-Caldere F, Cosson P, Howad W, Arus P (2004) Comparative mapping and marker-assisted selection in Rosaceae fruit crops. PNAS 101:9891–9896
Dondini L, Lain O, Geuna F, Banfi R, Gaiotti F, Tartarini S, Bassi D, Testolin R (2007) Development of a new SSR-based linkage map in apricot and analysis of synteny with existing Prunus maps. Tree Genet Genomes 3:239–249
Duprat A, Caranta C, Revers F, Menand B, Browning KS, Robaglia C (2002) The Arabidopsis eukaryotic initiation factor (iso)4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J 32:927–934
Geuna F, Toschi M, Bassi D (2003) The use of AFLP markers for cultivar identification in apricot. Plant Breed 122:526–531
Glasa M, Palkovics L, Komınek P, Labonne G, Pittnerova S, Kudela O, Candresse T, Subr Z (2004) Geographically and temporally distant natural recombinant isolates of Plum pox virus (PPV) are genetically very similar and form a unique PPV subgroup. J Gen Virol 85:2671–2681
Guillet-Bellanguer I, Audergon JM (2001) Inheritance of the Stark Early Orange apricot cultivar resistance to Plum pox virus. Acta Hort 550:111–115
Howad W, Yamamoto T, Dirlewanger E, Testolin R, Cosson P, Cipriani G, Monforte AJ, Georgi L, Abbott AG, Arús P (2005) Mapping with a few plants: using selective mapping for microsatellite saturation of the Prunus reference map. Genetics 171:1305–1309
Hurtado MA, Romero C, Vilanova S, Abbott AG, Llacer G, Badenes ML (2002) Genetic linkage map of two apricot cultivars (Prunus armeniaca L.) and mapping of PPV (Sharka) resistance. Theor Appl Genet 105:182–191
Joobeur T, Viruel MA, de Vicente MC, Jauregui B, Ballester J, Dettori MT, Verde I, Truco MJ, Messeguer R, Battle I, Quarta R, Dirlewanger E, Arús P (1998) Construction of a saturated linkage map for Prunus using an almond × peach F2 progeny. Theor Appl Genet 97:1034–1041
Kang BC, Yeam I, Frantz JD, Murphy JF, Jahn MM (2005) The pvr1 locus in Capsicum encodes a translation initiation factor eIF4E that interacts with Tobacco etch virus VPg. Plant J 42:392–405
Karayiannis I, Thomidis T, Tsaftaris A (2008) Inheritance of resistance to Plum pox virus in apricot (Prunus armeniaca L.). Tree Genet Genomes 4:143–148
Kegler H, Fuchs E, Grüntzig M, Schwarz S (1998) Some results of 50 years of research on the resistance to Plum Pox Virus. Acta Virol 42:200–215
Lalli DA, Abbott AG, Zhebentyayeva TN, Badenes ML, Damsteegt V, Polák J, Krška B, Salava J (2008) A genetic linkage map for an apricot (Prunus armeniaca L.) BC1 population mapping Plum pox virus resistance. Tree Genet Genomes 4:481–493
Lambert P, Hagen LS, Arus P, Audergon JM (2004) Genetic linkage maps of two apricot cultivars (Prunus armeniaca L.) compared with the almond Texas × peach Earlygold reference map for Prunus. Theor Appl Genet 108:1120–1130
Lambert P, Dicenta F, Rubio M, Audergon JM (2007) QTL analysis of resistance to Sharka disease in the apricot (Prunus armeniaca L.) ‘Polonais’ × ‘Stark Early Orange’ F1 progeny. Tree Genet Genomes 3:299–309
Lopes MS, Sefc KM, Laimer M, Da Câmara Machado A (2002) Identification of microsatellite loci in apricot. Mol Ecol Notes 2:24–26
Marandel G, Pascal T, Candresse T, Decroocq V (2009a) Quantitative resistance to Plum pox virus in Prunus davidiana P1908 linked to components of the eukaryotic translation initiation complex. Plant Pathol 58:425–435
Marandel G, Salava J, Abbott A, Candresse T, Decroocq V (2009b) Quantitative trait loci meta-analysis of Plum pox virus resistance in apricot (Prunus armeniaca L.): new insights on the organization and the identification of genomic resistance factors. Mol Plant Pathol 10:347–360
Martinez-Gomez P, Dicenta F, Audergon JM (2000) Behaviour of apricot (Prunus armeniaca L.) cultivars in the presence of Sharka (plum pox potyvirus): a review. Agronomie 20:407–422
Mercado JA, El Mansouri I, Jimenez-Bermudez S, Plieco-Alfaro F, Quesada MA (1999) A convenient protocol for extraction and purification of DNA from Fragaria. In Vitro Cell Dev Biol Plant 35:152–153
Messina R, Lain O, Marrazzo MT, Cipriani G, Testolin R (2004) New set of microsatellite loci isolated in apricot. Mol Ecol Notes 4:432–434
Mnejja M, Garcia-Mas J, Howad W, Arús P (2005) Development and transportability across Prunus species of 42 polymorphic almond microsatellites. Mol Ecol Notes 5:531–535
Moustafa TA, Badenes ML, Martínez-Calvo J, Llácer G (2001) Determination of resistance to Sharka (plum pox) virus in apricot. Sci Hort 91:59–70
Nicaise V, German-Retana S, Sanjuan R, Dubrana MP, Mazier M, Maisonneuve B, Candresse T, Caranta C, Le Gall O (2003) The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the potyvirus lettuce mosaic virus. Plant Physiol 132:1272–1282
Nicaise V, Gallois JL, Chafiai F, Allen LC, Schurdi-Levraud V, Browning KS, Candresse T, Caranta C, Le Gall O, German-Retana S (2007) Coordinated and selective recruitment of eIF4E and eIF4G factors for potyvirus infection in Arabidopsis thaliana. FEBS Lett 581:1041–1046
Rubio M, Audergon JM, Martinez-Gomez P, Dicenta F (2007) Testing genetic control hypotheses for Plum pox virus (Sharka) resistance in apricot. Sci Hort 112:361–365
Ruffel S, Dussault MH, Palloix A, Moury B, Bendahmane A, Robaglia C, Caranta C (2002) A natural recessive resistance gene against potato virus Y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E). Plant J 32:1067–1075
Schurdi-Levraud Escalettes V, Hullot C, Wawrzynczak D, Mathieu E, Eyquard JP, Le Gall O, Decroocq V (2006) Plum pox virus induces differential gene expression in the partially resistant stone fruit tree Prunus armeniaca cv. Goldrich. Gene 374:96–103
Sicard O, Marandel G, Soriano JM, Lalli DA, Lambert P, Salava J, Badenes ML, Abbott A, Decroocq V (2008) Flanking the major Plum pox virus resistance locus in apricot with co-dominant markers (SSRs) derived from candidate resistance genes. Tree Genet Genomes 4:359–365
Soriano JM, Vera-Ruiz EM, Vilanova S, Martínez-Calvo J, Llácer G, Badenes ML, Romero C (2008) Identification and mapping of a locus conferring Plum pox virus resistance in two apricot-improved linkage maps. Tree Genet Genomes 4:391–402
Testolin R, Marrazzo MT, Cipriani G, Quarta R, Verde I, Dettori MT, Pancaldi M, Sansavini S (2000) Microsatellite DNA in peach (Prunus persica L. Batsch) and its use in fingerprinting and testing the genetic origin of cultivars. Genome 43:512–520
Testolin R, Messina R, Lain O, Marrazzo MT, Huang WG, Cipriani G (2004) Microsatellites isolated in almond from an AC-repeat enriched library. Mol Ecol Notes 4:459–461
van Ooijen JW, Voorrips RW (2002) Joinmap 3.0. Software for the calculation of genetic linkage maps. Plant Research International, Wageningen
van Ooijen JW, Boer MP, Jansen RC, Maliepaard C (2002) MapQTL 4.0. Software for the calculation of QTL positions on genetic maps. Plant Research International, Wageningen
Vilanova S, Romero C, Abbott AG, Llacer G, Badenes ML (2003) An apricot (Prunus armeniaca L.) F2 progeny linkage map based on SSR and AFLP markers mapping Plum pox virus resistance and self-incompatibility traits. Theor Appl Genet 107:239–247
Vilanova S, Soriano JM, Lalli DA, Romero C, Abbott AG, Llácer G, Badenes ML (2006) Development of SSR markers located in the G1 linkage group of apricot (Prunus armeniaca L.) using a bacterial artificial chromosome library. Mol Ecol Notes 6:789–791
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Yamamoto T, Yamaguchi M, Hayashi T (2005) An integrated genetic linkage map of peach by SSR, STS, AFLP and RAPD. J Jpn Soc Hort Sci 74:204–213
Zhebentyayeva TN, Reighard GL, Lalli DA, Gorina VM, Krška B, Abbott AG (2008) Origin of resistance to Plum pox virus in apricot: what new AFLP and targeted SSR data analyses tell. Tree Genet Genomes 4:403–417
Acknowledgements
This research was supported by the Ministry of Education, University, and Scientific Research (MIUR), National Project PRIN 2006 “Positional cloning of sharka resistance candidate genes in apricot”. We acknowledge Dr Ross Ferguson, Plant & Food Research, New Zealand, for the critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dondini, L., Lain, O., Vendramin, V. et al. Identification of QTL for resistance to plum pox virus strains M and D in Lito and Harcot apricot cultivars. Mol Breeding 27, 289–299 (2011). https://doi.org/10.1007/s11032-010-9431-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-010-9431-3