Abstract
Powdery mildew, caused by Blumeria graminis f. sp. tritici, is one of the most important wheat diseases worldwide. Pyramiding different resistance genes into single cultivar has been proposed as one remedy to provide durable resistance. Powdery mildew resistance genes Pm12 (T6BS-6SS.6SL), transferred from Aegilops speltoides to wheat cv. Wembley, and Pm21 (T6VS.6AL), introduced from Dasypyrum villosum to wheat cv. Yangmai5, conferred broad-spectrum resistance to B. graminis f. sp. tritici. Both Pm12 and Pm21 genes are located on the short arms of homologous group six involved translocated chromosomes 6SS.6BL and 6VS.6AL, respectively. Simple sequence repeat motifs of wheat simple sequence repeat (SSR) and expressed sequence tag (EST) sequences on the short arm of homologous group six chromosomes were analyzed to develop molecular markers for discriminating chromosome arms 6AS, 6BS, 6DS, 6VS, and 6SS. One EST–SSR marker, Xcau127, was polymorphic, and therefore can be used to distinguish the two resistance genes and the respective susceptible alleles. This marker allowed us to develop an efficient “one-marker-for-two-genes” procedure for identifying powdery mildew resistance genes Pm12 and Pm21 for marker-assisted selection and gene pyramiding in wheat breeding programs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat is one of the most important cereal crops worldwide, providing around 35% of food for human consumption. Powdery mildew, caused by Blumeria graminis f. sp. tritici, is a major disease of wheat, particularly in areas with cool or maritime climates. Deployment of diversified powdery mildew resistance genes proved an economical and environmentally sound method to control the disease. Pyramiding resistance genes into a single genotype was proposed as an effective way to achieve durable resistance. However, conventional breeding to combine resistance genes is a tedious process, which can be made more efficient by the use of molecular markers tightly linked to the resistance genes (Tanksley et al. 1989). To date, 39 powdery mildew resistance gene loci have been assigned to specific chromosomes and formally designated. Among them, 24 resistance genes/alleles have been tagged by molecular markers (Huang and Röder 2004; McIntosh et al. 2008).
Pm12, present in a wheat—Aegilops speltoides translocation (T6BS-6SS.6SL) (Miller et al. 1988) and Pm21 in a series of wheat—Dasypyrum villosum translocation (T6VS.6AL) lines (Chen et al. 1995) confer resistance to all isolates of B. graminis f. sp. tritici tested in China (Duan et al. 1998), Europe (Huang and Röder 2004), and the USA (Niewoehner and Leath 1998). The translocation line carrying Pm12 was tagged with a restriction fragment length polymorphism (RFLP) marker (Jia et al. 1996) and then with SSR and EST–SSR markers (Song et al. 2007), whereas Pm21 has been tagged with RFLP (Li et al. 1995, 2005), random amplification of polymorphic DNA (RAPD) (Qi et al. 1996), sequence-characterized amplified region (SCAR) (Liu et al. 1999), and resistance gene analog (RGA) (Cao et al. 2006) markers. All these markers are translocation chromosome (arm) specific and therefore identify only Pm12 or Pm21.
SSRs (or microsatellites), simple sequence length polymorphisms containing tandem repeats of basic motifs <6 bp, are abundant and highly polymorphic in eukaryotic genomes. High-density wheat SSR maps have been published (Röder et al. 1998; Somers et al. 2004; Song et al. 2005). Comparative genetic maps reveal close relationships between homoeologous chromosomes within hexaploid wheat and with other Triticeae species. Study of homoeologous group six RFLP markers indicated homoeologous relationships between D. villosum 6V and wheat chromosomes 6A, 6B, and 6D (Qi et al. 1999). A. speltoides chromosome 6S is very closely related to wheat chromosome 6B even though the species is longer considered to be the B genome donor of polyploid wheat (Kimber and Athwal 1972; Dvorak and Zhang 1990; Huang et al. 2002). The transferability of wheat genomic and EST SSR data to related cereals and grasses has been confirmed by various workers (Gupta et al. 2003; Kuleung et al. 2004; Zhang et al. 2005). Compared with genomic SSRs, EST–SSRs show a very high level of transferability across closely related species because they originate from transcribed regions that are conserved between genomes (Yu et al. 2004). In this paper we describe the results of a comparative sequence analysis of SSR repeat motifs between chromosomes 6AS, 6BS, 6DS, 6SS, and 6VS leading to development of a “one-marker-for-two-genes” assay, thus providing an efficient means to identify and combine Pm12 and Pm21 located in different alien segments.
Materials and methods
Plant materials
The Pm12 and Pm21 powdery mildew resistance gene donors Line #31 (6BS-6SS.6SL) and R149 (6VS.6AL) were kindly provided by Dr. X. Y. Duan, Chinese Academy of Agricultural Sciences, Beijing, and Prof. D. J. Liu, Nanjing Agricultural University. Common wheat cv. Jing 411 was used as the susceptible recurrent parent in backcrosses with Line #31 and R149 to develop near-isogenic lines (NILs), Line#31/8*Jing 411 with Pm12 and R149/8*Jing 411 carrying Pm21. The BC8F2 segregating populations consisting of 190 and 72 individuals and their BC8F3 progenies were used for marker analysis of Pm12 and Pm21, respectively. A F2 population consisting of 85 individuals derived from intercross Line#31/8*Jing 411//R149/8*Jing 411 was selected for marker validation and gene pyramiding. Pm21 cosegregated SCAR1400 (Liu et al. 1999) and Pm12-linked SSR markers (Song et al. 2007) were used to identify the presence of 6VS and 6SS chromosome arms in each F2 individuals. Polymorphic marker was also tested in diversified lines/cultivars containing different powdery mildew genes for allelic diversity (Table 2).
Chinese Spring (CS) ditelosomic lines (Dt6AS, Dt6AL, Dt6BS, Dt6BL, Dt6DS, and Dt6DL) of homoeologous group six (kindly provided by Drs. W. J. Raupp and B. S. Gill, Wheat Genetics Resource Centre, Kansas State University, USA) were used for chromosomal arm assignments of polymorphic markers.
Powdery mildew disease evaluation
Powdery mildew seedling reactions were determined as described by Liu et al. (2002). B. graminis f. sp. tritici race E09 was used for inoculations. Reactions were scored on a 0, 0;, and 1–4 infection type (IT) scale, with 0 representing no visible symptoms, 0; representing necrotic flecks, and 1, 2, 3, 4 for highly resistant, resistant, susceptible, and highly susceptible reactions, respectively. Resistant reaction of 20 seedlings of each F3 progenies were tested to classify the F2 individuals into three types, namely homozygous resistant (RR), heterozygous resistant (Rr), and homozygous susceptible (rr). Due to the complete resistance of Pm12 and Pm21, the reactions of tested seedlings were almost all 0; (resistant) or 4 (susceptible) on this scale.
Genomic DNA isolation and PCR analysis
Genomic DNA was extracted from uninfected leaves by the cetyl trimethylammonium bromide (CTAB) method (Saghai-Maroof et al. 1984). Wheat SSR and EST markers physically mapped on homologous group six (http://wheat.pw.usda.gov, Randhawa et al. 2004) were screened to find polymorphisms between the NILs (Line#31/8*Jing 411 and R149/8*Jing 411) and recurrent parent Jing 411 DNA samples. Each polymerase chain reaction (PCR) was performed in a total volume of 10 μl containing 10 mM Tris–HCl, pH 8.3, 50 mM KCl, 1.5 mM MgCl2, 0.2 mM of dNTP, 25 ng of each primer, 50–100 ng genomic DNA, and 0.75 U Taq DNA polymerase. Amplifications were performed at 94°C for 3 min, followed by 45 cycles at 94°C for 1 min, 50–60°C for 1 min, and 72°C for 2 min, with a final extension at 72°C for 10 min. PCR products were separated on 8% nondenaturing polyacrylamide gels. Gels were silver-stained and photographed.
Chromosomal arm assignments of the polymorphic marker loci
Polymorphic SSR loci between the NILs and Jing 411 were assigned to chromosome arms using Chinese Spring homoeologous group six nullisomic–tetrasomics and ditelosomics.
Results
Polymorphic marker between Pm12, Pm21, and Jing 411
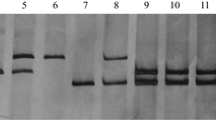
Among 180 genomic and EST–SSR primer pairs screened, EST–SSR marker Xcau127 detected stable polymorphic DNA fragments differentiating the three genotypes, Pm12 (Line#31/8*Jing 411), Pm21 (R149/8*Jing 411), and Jing 411. The CAU127 primer pair amplified 6AS-, 6DS-, and 6SS-specific DNA bands in Line#31 and Line#31/8*Jing 411, but 6BS-, 6DS-, and 6VS-specific DNA bands in R149 and R149/8*Jing 411 (Fig. 1). After testing in two BC8F2 segregating populations, Xcau127 was found to be cosegregated with the powdery mildew resistance genes, indicating the presence of Pm12 and Pm21 genes in the resistant plants, respectively.
Xcau127 was developed from wheat EST sequence CA640857 containing a trinucleotide simple sequence repeat motif (CAG)8. Sequence comparison of CAU127 amplicons on Pm12, Pm21, and Jing 411 revealed simple sequence repeat motif (CAG) n variations between chromosome arms 6AS, 6BS, 6DS, 6SS, and 6VS (Fig. 2). CAU127 amplified 147-, 153-, 159-, 159-, and 165-bp DNA fragments in 6VS, 6BS, 6AS, 6SS, and 6DS, respectively. Both fragments amplified in 6AS and 6SS arms sized 159 bp which could not be distinguished by gel analysis. However, another polymorphic DNA band between 6SS and other chromosome arms could be detected on nondenaturing polyacrylamide gel (Fig. 1).
“One-marker-for-two-genes” approach for molecular discrimination of Pm12 and Pm21 genes
To test if SSR primer pair CAU127 could be used as molecular marker for simultaneously selection of Pm12 and Pm21, the primer was used to amplify the F2 individuals from Line#31/8*Jing 411//R149/8*Jing 411. Xcau127 was able to identify both resistant alleles of Pm12 (AAbb) and Pm21 (aaBB) as well as the susceptible allele of Jing 411 (aabb) in the F2 population. Among the nine possible genotypes derived from intercross Pm12 (AAbb)/Pm21 (aaBB), seven genotypes (AAB_, AaB_, AAbb, Aabb, aaBB, aaBb, and aabb) could be distinguished by amplification patterns of marker Xcau127 in a single gel (Fig. 3; Table 1). The observed individuals number of genotypes A_B_, A_bb, aaB_, and aabb were 50, 14, 17, and 4, which is in agreement with two different genes segregating ratio of 9:3:3:1 (x 2 = 0.59). Presence of 6VS (Pm21) and 6SS (Pm12) chromosome arms in each F2 plants was conformed by Pm21 cosegregated SCAR1400 and Pm12-linked SSR markers, respectively. Powdery mildew testing was also conducted on the F3 progenies to identify the homozygous and heterozygous genotypes of each F2 individual. Both phenotypic test and molecular markers results are in agreement with the Xcau127 screening pattern.
Amplification patterns of EST–SSR CAU127 on Line#31/8*Jing 411 (P1, AAbb, Pm12), R149/8*Jing 411 (P2, aaBB, Pm21), Line#31/8*Jing 411//R149/8*Jing 411 (F1, AaBb, Pm12 + Pm21) and their F2 different genotypes. AA, Aa, bb, BB, Bb, bb represent the homozygous resistant, heterozygous resistant, and homozygous susceptible genotypes of Pm12 and Pm21 alleles, respectively
Marker Xcau127 was further validated in diversified wheat lines containing different powdery mildew resistance genes (Table 2). The A. speltoides 6SS- and D. villosum 6VS-specific DNA fragments were found only on lines carrying genes Pm12, Pm21 or both. No 6SS- or 6VS-specific DNA fragments were detected in any wheat cultivars or lines without Pm12 or Pm21 in their pedigrees in our breeding program, indicating a very low allelic diversity for Xcau127 loci in wheat genome.
Discussion
While RFLP markers linked to Pm12 (Jia et al. 1996) and Pm21 (Li et al. 1995) have been reported, their use for marker-assisted selection is impractical in breeding program. PCR-based markers are more efficient as well as being safe and relatively inexpensive. RAPD, SCAR, and RGA markers linked to Pm21 (Qi et al. 1996; Liu et al. 1999; Cao et al. 2006) have been developed and used extensively in marker-assisted selection of Pm21 in wheat breeding programs. SSR markers linked to Pm12 were also identified recently and used to select a series of introgression lines (Song et al. 2007). While being acceptable for selection of Pm12 or Pm21 alone, these markers are source specific and must be used separately for selection each gene. The marker Xcau127 identified here has the advantage of enabling selection of both genes in a single reaction. Pm12 and Pm21 are highly effective powdery mildew resistance genes against all isolates of B. graminis f. sp. tritici tested in China (Duan et al. 1998), Europe (Huang and Röder 2004), and the USA (Niewoehner and Leath 1998). This “one-marker-for-two-genes” approach provides a highly efficient system for identifying and combining Pm12 and Pm21. The low allelic variations and highly specific Pm12 and Pm21 polymorphic marker Xcau127 provide potential applicability of this system for a wide variety of genetic backgrounds in breeding programs.
References
Cao A, Wang X, Chen Y et al (2006) A sequence-specific PCR marker linked with Pm21 distinguishes chromosomes 6AS, 6BS, 6DS of Triticum aestivum and 6VS of Haynaldia villosa. Plant Breed 125:201–205. doi:10.1111/j.1439-0523.2006.01222.x
Chen P, Qi L, Zhou P et al (1995) Development and molecular cytogenetic analysis of wheat-H. villosa 6VS/6AL translocation lines specifying resistance to powdery mildew. Theor Appl Genet 91:1125–1128. doi:10.1007/BF00223930
Duan X, Sheng B, Zhou Y et al (1998) Monitoring of the virulence population of Erysiphe graminis f. sp. tritici. Acta Phytophylac Sin 25:31–36
Dvorak J, Zhang H (1990) Variation in repeated nucleotide sequences sheds light on the phylogeny of the wheat B and G genomes. Proc Natl Acad Sci USA 87:9640–9644. doi:10.1073/pnas.87.24.9640
Gupta P, Rustgi S, Sharma S et al (2003) Transferable EST–SSR markers for the study of polymorphism and genetic diversity in bread wheat. Mol Genet Genomics 270:315–323. doi:10.1007/s00438-003-0921-4
Huang X, Röder M (2004) Molecular mapping of powdery mildew resistance genes in wheat: a review. Euphytica 137:203–223. doi:10.1023/B:EUPH.0000041576.74566.d7
Huang S, Sirikhachornkit A, Su X et al (2002) Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc Natl Acad Sci USA 99:8133–8138. doi:10.1073/pnas.072223799
Jia J, Devos K, Chao S et al (1996) RFLP-based maps of the homoeologous group–6 chromosomes of wheat and their application in the tagging of Pm12, a powdery mildew resistance gene transferred from Aegilops speltoides to wheat. Theor Appl Genet 92:559–565
Kimber G, Athwal R (1972) A reassessment of the course of evolution of wheat. Proc Natl Acad Sci USA 69:912–915. doi:10.1073/pnas.69.4.912
Kuleung C, Baenziger P, Dweikat I (2004) Transferability of SSR markers among wheat, rye, and triticale. Theor Appl Genet 108:1147–1150. doi:10.1007/s00122-003-1532-5
Li W, Chen P, Qi L et al (1995) Isolation, characterization and application of a species-specific repeated sequence from Haynaldia villosa. Theor Appl Genet 90:526–533. doi:10.1007/BF00221999
Li H, Chen X, Xin et al (2005) Development and identification of wheat-Haynaldia villosa T6DL.6VS chromosome translocation lines conferring resistance to powdery mildew. Plant Breed 124:203–205. doi:10.1111/j.1439-0523.2004.01062.x
Liu Z, Sun Q, Ni Z et al (1999) Development of SCAR markers linked to the Pm21 gene conferring resistance to powdery mildew in common wheat. Plant Breed 118:215–219. doi:10.1046/j.1439-0523.1999.118003215.x
Liu Z, Sun Q, Ni Z et al (2002) Molecular characterization of a novel powdery mildew resistance gene Pm30 in wheat originating from wild emmer. Euphytica 123:21–29. doi:10.1023/A:1014471113511
McIntosh RA, Yamazaki Y, Dubcovsky J et al (2008) Catalogue of gene symbols for wheat. In: Appels R, Eastwood R, Lagudah E et al (eds) Proceedings of the 11th international wheat genetics symposium, Sydney University Press, Sydney, pp 114–121
Miller T, Reader S, Ainsworth C et al (1988) The introduction of a major gene for resistance to powdery mildew of wheat, Erysiphe graminis f.sp. tritici, from Aegilops speltoides into wheat, Triticum aestivum. In: Jorna ML, Slootmaker LAJ et al (eds) Cereal breeding related to integrated cereal production. Pudoc, The Netherlands, pp 179–183
Niewoehner A, Leath S (1998) Virulence of Blumeria graminis f. sp. tritici on winter wheat in the eastern United States. Plant Dis 82:64–68. doi:10.1094/PDIS.1998.82.1.64
Qi L, Cao M, Chen P et al (1996) Identification, mapping, and application of polymorphic DNA associated with resistance gene Pm21 of wheat. Genome 39:191–197. doi:10.1139/g96-025
Qi L, Chen P, Liu D et al (1999) Homoeologous relationships of Haynaldia villosa chromosomes with those of Triticum aestivum as revealed by RFLP analysis. Genes Genet Syst 74:77–82. doi:10.1266/ggs.74.77
Randhawa H, Dilbirligi S, Sidhu D et al (2004) Deletion mapping of homologous group 6-specific wheat expressed sequence tags. Genetics 168:677–686. doi:10.1534/genetics.104.034843
Röder M, Korzun V, Wendehake K et al (1998) A microsatellite map of wheat. Genetics 149:2007–2023
Saghai-Maroof M, Soliman K, Jorgensen R et al (1984) Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal locations and population dynamics. Proc Natl Acad Sci USA 81:8014–8018. doi:10.1073/pnas.81.24.8014
Somers D, Isaac P, Edwards K (2004) A high density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet 109:1105–1114. doi:10.1007/s00122-004-1740-7
Song Q, Shi J, Singh S et al (2005) Development and mapping of microsatellite (SSR) markers in wheat. Theor Appl Genet 110:550–560. doi:10.1007/s00122-004-1871-x
Song W, Xie H, Liu Q et al (2007) Molecular identification of Pm12-carrying introgression lines in wheat using genomic and EST–SSR markers. Euphytica 158:95–102. doi:10.1007/s10681-007-9432-4
Tanksley S, Young N, Paterson A et al (1989) RFLP mapping in plant breeding: new tools for an old science. Biotechnology 7:257–264. doi:10.1038/nbt0389-257
Yu J, La Rota M, Kantety R et al (2004) EST derived SSR markers for comparative mapping in wheat and rice. Mol Genet Genomics 271:742–751. doi:10.1007/s00438-004-1027-3
Zhang L, Bernard M, Leroy P et al (2005) High transferability of bread wheat EST-derived SSRs to other. Theor Appl Genet 111:677–687. doi:10.1007/s00122-005-2041-5
Acknowledgements
The authors are grateful to Drs. X. Y. Duan, D. J. Liu, W. J. Raupp, and B. S. Gill for providing wheat seed stocks and to Dr. R. A. McIntosh for improving the manuscript. This work was financially supported by the National Fund for Distinguished Young Scholars (30425039), the National Natural Science Foundation of China (30571151, 30771341), the Beijing Natural Science Foundation (6061003), the State High Tech Programs (2006AA100102, 2006AA10Z1E9, 2006AA10Z1C4 and 2006BAD01A02), the Programme of Introducing Talents of Discipline to Universities (111-2-03), and the Program for Changjiang Scholars and Innovative Research Team in University.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Wei Song and Chaojie Xie contributed equally to this work
Rights and permissions
About this article
Cite this article
Song, W., Xie, C., Du, J. et al. A “one-marker-for-two-genes” approach for efficient molecular discrimination of Pm12 and Pm21 conferring resistance to powdery mildew in wheat. Mol Breeding 23, 357–363 (2009). https://doi.org/10.1007/s11032-008-9235-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-008-9235-x