Abstract
Human manganese superoxide dismutase (hMnSOD) is a new type of cancer suppressor. Nonamer of arginine (R9) is an efficient protein transduction domain (PTD). The aim of the study was to improve the transduction efficiency of hMnSOD and investigate its activity in vitro. In this study, we designed, constructed, expressed, and purified a novel fusion protein containing the hMnSOD domain and R9 PTD (hMnSOD–R9). The DNA damaged by Fenton’s reagent was found to be significantly reduced when treated with hMnSOD–R9. hMnSOD–R9 fusion protein was successfully delivered into HeLa cells. The MTT assay showed that proliferation of various cancer cell lines were inhibited by hMnSOD–R9 in a dose-dependent manner. In addition, the cell cycle of HeLa cells was arrested at the sub-G0 phase by hMnSOD–R9. hMnSOD–R9 induced apoptosis of HeLa cells in a dose-dependent manner. With hMnSOD–R9 treatment, Bax, JNK, TBK1 gene expression was increased and STAT3 gene expression was gradually down-regulated in HeLa cells. We also found that apoptosis was induced by hMnSOD–R9 in HeLa cells via up-regulation of cleaved caspase-3 and down-regulation phospho-STAT3 pathway. These results indicated that hMnSOD–R9 may provide benefits to cervical cancer treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In human tissues, there are three known isoforms of SOD: the mitochondrial manganese SOD (MnSOD), the cytosolic copper–zinc SOD (Cu/ZnSOD), and the extracellular SOD (EcSOD) [1]. Mainly located in mitochondrial matrix, MnSOD is well known as one of the major antioxidant enzymes against superoxide free radicals and catalyzes dismutation of superoxide radical anion into hydrogen peroxide [2]. Furthermore, MnSOD has been proposed to be a new type of tumor suppressor gene [3]. Most types of cancer cells have reduced the expression of MnSOD compared with their normal cell counterparts. The region of chromosome 6q25.3, where MnSOD is located, is deleted in many cancers cells. Numerous studies have demonstrated that transfection of MnSOD cDNA into various cancer cells resulted in decreasing their tumorigenicity [4–6].
Although MnSOD offers great potential as a therapeutic molecule in many cancers, therapeutic use of MnSOD is limited by poor penetration in tissues and inability to cross cellular membrane. Protein transduction domains (PTDs) are capable of transporting cargo across the plasma membrane into cells [7]. Prior research has shown that peptides containing a higher percentage of arginine will be more effective for transduction. In related studies, it was found that nonamer of arginine (R9: RRRRRRRRR) is an efficient PTD [8].
To improve transduction efficiency of human manganese superoxide dismutase (hMnSOD) while preserving its biological activity, hMnSOD domain and R9 PTD were fused through gene recombination technology, producing a recombinant hMnSOD–R9.
Materials and methods
Materials
Mouse anti-His monoclonal antibody was from TIANGEN (Beijing, China). Horseradish peroxidase (HRP) labeled goat anti-mouse IgG was purchased from Proteintech (Chicago, USA). Ni–NTA resin was from QIAGEN (Hilden, Germany). FITC-labeled rabbit-anti-mouse monoclonal antibody was from BELLANCOM (New Jersey, USA). SYBR Green PCR kit was obtained from TAKARA (Osaka, Japan). Cell cycle and apoptosis analysis kit was obtained from Beyotime Institute of Biotechnology (Haimen, China). An Annexin-V-FLUOS staining kit was from Roche (Basel, Switzerland). Antibodies against phospho-STAT3, cleaved caspase-3 and GAPDH were from Cell Signaling Technology (Danvers, MA, USA).
All other reagents were analytical grade or better and commercially available.
Construction of expression vector
The gene fragment coding for hMnSOD–R9 chimeric peptide was constructed by PCR amplification [9]. Primers (forward: 5′-AAGCACAGCCTCCCCGACCT-3′, reverse: 5′-TTACTTTTTGCAAGCCATGTATCTT-3′) were designed based on the sequence of hMnSOD (Genbank accession NM_000636.2). The hMnSOD–R9 gene was amplified using forward primer 5′-AAGCACAGCCTCCCCGACCT-3′ and reverse primer 5′-TTAACGACGACGACGACGACGACGACGACGCTTTTTGCAAGCCATGTATCTTTC-3′, using template pET15b–hMnSOD. The amplified PCR product was ligated into a modified pET15b (Sma I) expression vector [10], resulting in the recombinant vector pET15b–hMnSOD–R9, then transformed into Escherichia coli DH5α cells [11]. The recombinant plasmid pET15b–hMnSOD–R9 was extracted and confirmed by direct sequencing (Yinjun, Shanghai, China). The pET15b–hMnSOD–R9 plasmid was transformed into Rosetta-gami strain for expression.
Protein expression and purification
The selected colony which contained the pET15b–hMnSOD–R9 recombinant plasmid was cultured in 200 mL LB medium containing 100 μg/mL ampicillin at 37 °C. Protein expression was induced by 0.5 mM IPTG at 30 °C for 8 h. Cells were harvested by centrifugation and disrupted by sonication on ice. The fusion protein was purified by a Ni–NTA–His bind column according to the QIAGEN handbook. The fusion protein was eluted through increasing the imidazole concentration from 10 to 250 mM [12]. The purified fusion protein was dialyzed overnight against a buffer containing 20 mM sodium phosphate, 500 mM NaCl (pH 7.4) to remove imidazole [10]. The concentration of fusion protein was assayed by bicinchoninic acid (BCA) method. SDS-PAGE was used to analyze the expression, purity and content of the fusion protein. The purified fusion protein was confirmed by Western blot. The percentage of fusion protein was analyzed by scanning a coomassie-stained gel using Quantity One-4.4.1 Bio-Rad software. SOD activity assay kit (Jiancheng, Nanjing, China) was used to detect the SOD activity of hMnSOD–R9.
DNA oxidation damage protective assay
DNA damage protective activity of hMnSOD–R9 was performed utilizing supercoiled pUC-19 DNA. The hMnSOD–R9 (5–60 μg/mL) was added to 6 μL DNA (0.5 μg) in an ultimate volume of 10 μL. Experiment was conducted at room temperature for 10 min. Then, 10 μL Fenton’s reagent was added to the mixture of the hMnSOD–R9 and pUC-19 DNA. The mixture was incubated for 30 min at 37 °C. The pUC-19 DNA was analyzed on 1 % agarose gel using ethidium bromide staining [13].
Confocal laser scanning microscope assay
HeLa cells were grown in plates and treated with hMnSOD or hMnSOD–R9 (40 μg/mL) for 90 min, then incubated with primary antibody (mouse anti-His, 1:500) and conjugated secondary antibody (FITC-labeled rabbit-anti-mouse, 1:1,000). The control hMnSOD without R9 PTD was expressed by our earlier research [10]. After nuclear staining with DAPI (0.5 mg/mL), the HeLa cells were observed by confocal laser scanning microscope (TCS SP2 AOBS LEICA, Germany) [14, 15].
MTT assay
The cancer cells (HeLa, HepG-2, SGC-7901, A549) were incubated with the hMnSOD–R9 at the appropriate concentration (0, 5, 10, 20, and 40 μg/mL) for 48 h. Subsequently, the growth inhibitory effect of hMnSOD–R9 toward the cancer cells was evaluated by means of MTT assay [16].
Cell cycle analysis
Cell cycle and apoptosis analysis kit was applied to analyze the cell cycle of HeLa cells. HeLa cells were treated with (0, 10, 20, and 40 μg/mL) hMnSOD–R9 for 48 h. After being fixed in 70 % ice-cold ethanol, the HeLa cells were incubated with 0.5 ml propidium iodide (PI) staining buffer (200 mg/mL RNaseA, 50 μg/mL PI) at 37 °C for 30 min in the dark. The cell cycles were analyzed by a flow cytometer (FACS Aria BD, USA) at 488 nm [17].
Annexin V/PI assay
HeLa cells were treated with (0, 10, 20, and 40 μg/mL) hMnSOD–R9 for 48 h. Apoptosis analysis was performed using an Annexin-V-FLUOS staining kit according to the manufacturer’s instruction. Apoptotic cells were defined as Annexin-V-position, PI-negative cells. Analysis was performed under a flow cytometer (FACS Aria BD, USA) [17].
Q-PCR assay
The HeLa cells were treated with various concentrations (0, 10, 20, and 40 μg/mL) of hMnSOD–R9 for 48 h. The expression of some apoptosis-related genes (STAT3, Bak, Bax, JNK, TBK1, p53) were detected by Real-time PCR. Total RNA were extracted from HeLa cells. The mRNA level of GAPDH was used as an internal control. The expression of apoptosis-related genes were measured using a SYBR Green PCR kit in Bio Rad CFX Manager Version PCR machine [18].
Western blot analysis
Western blot was performed to evaluate the protein expression of p-STAT3 and cleaved caspase-3. The HeLa cells were treated with various concentrations (0, 10, 20, and 40 μg/mL) of hMnSOD–R9 for 48 h. The total proteins of HeLa cells were extracted. The total protein concentrations were determined by the BCA method. Total proteins were separated by SDS-PAGE and blotted onto a PVDF membrane. The PVDF membrane was blocked with 5 % non-fat dry milk and incubated with a diluted primary antibody (rabbit anti-mouse cleaved caspase-3, rabbit anti-mouse phospho-STAT3, 1:1,000) overnight at 4 °C. The membrane was washed and incubated with a diluted secondary antibody (goat anti-rabbit IgG, 1:3,000). After extensive washing, proteins were visualized by an ECL-chemiluminescent kit (ECL-plus, Thermo Scientific). Blots were visualized using autoradiography on X-ray film (Kodak, Rochester, NY, USA) [19].
Statistical analysis
Data were processed using Statistical Package for Sciences and Society (SPSS 18.0) (SPSS Inc, Chicago, IL, USA). The data were subjected to one-way analysis of variance (ANOVA). The results were expressed as mean ± SD from at least three independent experiments. P < 0.05 was considered statistically significant, P < 0.01 was considered statistically highly significant.
Results
Construction of expression vector
The size of hMnSOD–R9 gene fragment was 624 bp. The hMnSOD–R9 gene fragment was inserted into pET15b (Sma I) vector, achieving recombinant plasmid pET15b–hMnSOD–R9. The size of recombinant expression plasmid pET15b–hMnSOD–R9 was about 6,300 bp [20]. The recombinant plasmid was confirmed by automated sequencing. Sequence analysis indicated that it encoded hMnSOD–R9 fusion protein with an estimated molecular mass of 26 kDa.
Expression and purification of recombinant hMnSOD–R9
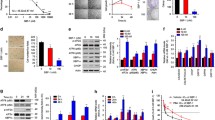
Cells were grown under several conditions of temperature and IPTG concentration to optimize protein expression. The fusion protein reached its peak expression level after induction with 0.5 mM IPTG at 30 °C for 8 h. As shown in Fig. 1a, clear differences can be observed on the SDS-PAGE gel between induced and uninduced cells. hMnSOD–R9 fusion protein (26 kDa) was the major product of the expression while it could not be detected without the addition of IPTG. Cells were lysed by sonication in the presence of lysozyme.
Expression, purification, characterization, and antioxidant ability of hMnSOD–R9. a Expression and purification: lane 1 extract from the uninduced cells, lane 2 extract from cells after IPTG induction, lanes 3, 4 supernatant and cell debris, respectively, obtained after centrifugation of the sonicated cells, M Protein marker, lane 5 flow-through, lane 6 elution fractions. b Western blot detected fusion protein with anti-His mAb: lane 1 cell lysate prior to induction, lane 2 the sample after Ni–NTA affinity chromatography. c DNA damage protective activity: lane 1 native pUC-19 DNA, lane 2 DNA + Fenton’s reagent, lane 3–6 DNA + Fenton’s reagent + hMnSOD–R9 (60, 40, 20, and 5 μg/mL, respectively)
The hMnSOD–R9 was virtually in the supernatant of the lysis. Availability of the expressed hMnSOD–R9 in the soluble fraction greatly facilitated its isolation and purification. Most of the impurities were in the flow-through while His–hMnSOD–R9 fusion protein was retained on the column. His–hMnSOD–R9 fusion protein appeared as an almost unique band after Ni–NTA purification from SDS-PAGE gel.
Furthermore, the hMnSOD–R9 protein was identified by Western blot with anti-His antibody (Fig. 1b). These results indicated that soluble hMnSOD–R9 protein was successfully expressed and purified.
The expression quantity of hMnSOD–R9 was approximate 57.1 mg/L of cultures. Purity of eluted His-tagged hMnSOD–R9 was >95 %. Table 1 summarized the yield of hMnSOD–R9 during purification. SOD activity assay showed that the activity of the hMnSOD–R9 was 1,800.2 ± 86.5 U/mg.
DNA damage protective activity of hMnSOD–R9
As shown in Fig 1c, the supercoiled plasmid pUC-19 DNA without pre-incubation with hMnSOD–R9 was almost completely damaged by Fenton’s reagent. While, the nicked DNA bands damaged by Fenton’s reagent were weakened and the supercoiled DNA bands were enhanced with hMnSOD–R9 concentration increasing. The pUC-19 DNA was protected by hMnSOD–R9 from damage by Fenton’s reagent in a dose-dependent manner. At concentration 60 μg/mL hMnSOD–R9 protection was more effective. The result suggested that hMnSOD–R9 has a DNA damage protective activity.
Trans-membrane effect observed by confocal laser scanning microscope assay
The intracellular delivery of hMnSOD–R9 into HeLa cells was confirmed by direct fluorescence analysis. As shown in Fig 2, all of the cells within the field contained green fluorescence from the FITC-labeled hMnSOD–R9. In contrast, cells treated with PBS and FITC-labeled hMnSOD exhibited no green fluorescence. These results suggested that the hMnSOD–R9 accumulated in the nucleus and cytoplasm, while, hMnSOD protein could not penetrate into cells without R9 transduction at the same concentration [13, 21]. R9 tag is a useful tool which could deliver hMnSOD into HeLa cells.
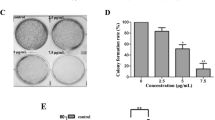
Growth inhibition of hMnSOD–R9 in different cancer cell lines
5, 10, and 20 μg/mL hMnSOD–R9 showed little or no cytotoxicity in four cell lines, while, at a dose of 40 μg/mL, the cell viability decreased significantly (Fig. 3). Cell viability of A549, HeLa, HepG-2, and SGC-7901 cells respectively, treated with 40 μg/ml of hMnSOD–R9 for 48 h, was approximately 77.9 ± 2.0, 55.9 ± 2.7, 70.1 ± 1.1, and 74.7 ± 1.1 %. In general, among the tested human cancer cell lines, HeLa cell line was more sharply inhibited by 40 μg/mL hMnSOD–R9 than A549, HepG-2 and SGC-7901 cell lines. These data suggested hMnSOD–R9 inhibited the proliferation of the four human cancer cell lines in a dose-dependent manner.
Effect of hMnSOD–R9 on cancer cells proliferation: several human cancer cell lines were treated with various concentrations of hMnSOD–R9 for 48 h and the inhibition of cell growth was measured by MTT assay. Results are expressed as mean ± SD of three independent experiments. Statistically significant and highly significant differences from the control in each group are indicated as *P < 0.05 and **P < 0.01 respectively
Cell cycle and apoptosis
HeLa cells were analyzed by flow cytometry after treatment with 40 μg/mL hMnSOD–R9 for 48 h. Figure 4a showed that the percentage of HeLa cells in sub-G0 phase increased from 4.6 ± 0.9 to 13.1 ± 0.4 % with concentration increasing of hMnSOD–R9. The hMnSOD–R9 treatment arrested the cell cycle of HeLa cells at the sub-G0 phase in a dose-dependent manner. The appearance of a sub-G0 peak which is also called apoptosis peak indicated apoptosis occurrence [22].
Effect of hMnSOD–R9 on apoptosis in HeLa cells. HeLa cells were treated with various concentrations (0, 10, 20, and 40 μg/mL) of hMnSOD–R9 for 48 h. HeLa cells treated with 0 μg/mL hMnSOD–R9 served as a control group. Results are expressed as mean ± SD of three independent experiments. Statistically significant differences from the control in each group are indicated as *P < 0.05, **P < 0.01 is considered highly significant. a The relative numbers of cells within each cell cycle were determined by flow cytometry. b The percentage of apoptosis cells were analyzed by flow cytometry. c The expression of apoptosis-related genes were analyzed by Real-time PCR. d The expression of apoptosis-related proteins were analyzed by Western blot
The percentage of total apoptotic cells (Annexin V+) increased from 6.5 ± 0.8 to 19.0 ± 0.6 % with concentration of hMnSOD–R9 increasing (0–40 μg/mL) (Fig. 4b). The result suggested that hMnSOD–R9 induced apoptosis of HeLa cells in a dose-dependent manner.
Q-PCR assay apoptosis-related genes
To investigate the mechanism by which hMnSOD–R9 induced apoptosis, we analyzed the expressions of apoptotic genes in HeLa cells, using Q-PCR. As shown in Fig 4c, with various concentrations of hMnSOD–R9 treatment for 48 h, expression of Bax, JNK, TBK1 gene were increased while STAT3 gene was decreased in HeLa cells. Expression of Bak, p53 gene were virtually unchanged in HeLa cells.
Western blot analysis apoptosis-related proteins
The HeLa cells were treated with various concentrations (0–40 μg/mL) of hMnSOD–R9 for 48 h. As shown in Fig. 4d, the expression of phospho-STAT3 (p-STAT3) was significantly decreased while the expression of cleaved caspase-3 was markedly increased in a concentration-dependent fashion. The result suggested that the apoptosis caused by hMnSOD–R9 was mediated by p-STAT3 suppression and cleaved caspase-3 activation pathway.
Discussion
Molecular cloning and expression of MnSOD have been reported in various prokaryotic and eukaryotic species, for example, Lactobacillus casei [23], Clonorchis sinensis [24], and pistachio nut [25]. However, human immune rejection has restricted the therapeutic use of MnSOD from other species. In previous studies, we expressed and purified human MnSOD [10]. Application of MnSOD was limited by the unfortunate ability to cross cellular membrane. We solved the issues of human immune rejection and poor ability to cross cellular membrane through constructed hMnSOD–R9 fuse protein. We also found that hMnSOD–R9 protected DNA from oxidation damage in vitro, compare to previous studies played in vivo.
In this study, we found that hMnSOD–R9 fusion protein was efficiently transduced into HeLa cells. Fluorescence signals of hMnSOD–R9 were found to be distributed uniformly in HeLa cells, suggesting that hMnSOD–R9 were translocated into both the cytoplasm and the nucleus.
Growth suppression was observed in human breast carcinoma cells [26], human glioma cells [27], human oral squamous carcinoma cells [28], and human prostatic carcinoma cells [29], via overexpression of MnSOD by cDNA transfection. We proved that MnSOD suppressed the proliferation of various cancer cell lines using hMnSOD–R9 trans-membrane protein, dissimilar from most studies using MnSOD cDNA transfection [30, 31]. Many anticancer agents regulate the cell cycle. We tested whether hMnSOD–R9 could also inhibit cell cycle progression in HeLa cells. The result suggested that hMnSOD–R9 arrested the cell cycle at sub-G0 phase in a dose-dependent manner. The appearance of sub-G0 peak (apoptosis peak) usually indicated apoptosis occurrence.
We used Annexin V/PI analysis to confirm apoptosis occurrence. Our result showed that hMnSOD–R9 induced HeLa cells apoptosis in a dose-dependent manner. To gain insight into the molecular mechanism involved in HeLa cells apoptosis caused by hMnSOD–R9, expression of the apoptosis-related genes and proteins were examined. STAT3, Bak, Bax, JNK, TBK1, p53 are some genes which are closely associated with apoptosis. With hMnSOD–R9 treatment, Bax, JNK, TBK1 gene expression were increased and STAT3 gene expression was gradually down-regulated in HeLa cells. c-Jun N-terminal kinases (JNKs) is a stress-activated member of the mitogen-activated protein kinase (MAPK) family [32]. It is now known that JNK is one of the molecules activated by oxidative stress and JNK signaling pathway contributes to the regulation of cell proliferation and apoptosis [33]. JNK activates the Bax which is a critical effector of apoptosis. During apoptosis, Bax translocates from the cytosol to insert into the mitochondrial outer membrane (OM) and releases cytochrome c. Cytosolic cytochrome c leads to caspase-9 activation, subsequent caspase-3 activation and final apoptosis [34].
TANK-binding kinase 1 (TBK1) which is one of IκB kinases negatively regulates noncanonical NF-κB signaling. TBK1 indirectly facilitates the activation of apoptosis protein caspase-3 [35]. Caspase-3 is a final apoptosis effector of Bax, JNK, and TBK1. Numerous studies with cancer cell lines and patient samples provided that signal transducer and activator of transcription 3 (STAT3) is one of the decisive players in human cancer formation [36]. Direct inhibition of STAT3 induces cell growth inhibition and apoptosis of breast and prostate cancer cells [37, 38]. Previous studies demonstrated that the expression of STAT3 was elevated in human cervical cancer cells [39]. Cleaved caspase-3 is the activated form of caspase-3. p-STAT3 is the activated form of STAT3. We found that the expression of p-STAT3 was significantly decreased, while the expression of cleaved caspase-3 was markedly increased in hMnSOD–R9 treatment HeLa cells. We conjecture that the hMnSOD–R9 crossed the cellular membrane into HeLa cells and activated JNK and subsequently Bax. The activated Bax and TBK1 caused caspase-3 activation. The caspase-3 activation and STAT3 suppression finally resulted in apoptosis.
References
Michiels C, Raes M, Toussaint O, Remacle J (1994) Importance of Se-glutathione peroxidase, catalase, and Cu/Zn-SOD for cell survival against oxidative stress. Free Radic Biol Med 17:235–248
Pani G, Colavitti R, Bedogni B, Fusco S, Ferraro D, Borrello S, Galeotti T (2004) Mitochondrial superoxide dismutase: a promising target for new anticancer therapies. Curr Med Chem 11:1299–1308
Bravard A, Sabatier L, Hoffschir F, Ricoul M, Luccioni C, Dutrillaux B (1992) SOD2: a new type of tumor-suppressor gene? Int J Cancer 51:476–480
Zhong W, Oberley LW, Oberley TD, Yan T, Domann FE, St Clair DK (1996) Inhibition of cell growth and sensitization to oxidative damage by overexpression of manganese superoxide dismutase in rat glioma cells. Cell Growth Differ 7:1175–1186
Oberley TD (2004) Mitochondria, manganese superoxide dismutase, and cancer. Antioxid Redox Signal 6:483–487
Li Z, Khaletskiy A, Wang J, Wong JY, Oberley LW, Li JJ (2001) Genes regulated in human breast cancer cells overexpressing manganese-containing superoxide dismutase. Free Radic Biol Med 30:260–267
van den Berg A, Dowdy SF (2011) Protein transduction domain delivery of therapeutic macromolecules. Curr Opin Biotechnol 22:888–893
Mitchell DJ, Kim DT, Steinman L, Fathman CG, Rothbard JB (2000) Polyarginine enters cells more efficiently than other polycationic homopolymers. J Pept Res 56:318–325
Zhou HH, Zhang AX, Zhang Y, Zhu DY (2012) Cloning, expression, and purification of a recombinant Tat-HA-NR2B9c peptide. Protein Expr Purif 85:239–245
Feng W, Mei S, Wenjie Y, Luyuan H (2011) High-level soluble expression of recombinant human manganese superoxide dismutase in Escherichia coli, and its effects on proliferation of the leukemia cell. Protein Expr Purif 77:46–52
Li CL, Xu TT, Chen RB, Huang XX, Zhao YC, Bao YY, Zhao WD, Zheng ZY (2013) Cloning, expression and characterization of antimicrobial porcine beta defensin 1 in Escherichia coli. Protein Expr Purif 88:47–53
Liu D, Zhang R, Yang X, Xu Y, Tang Z, Tian W, Shen Q (2011) Expression, purification and characterization of two thermostable endoglucanases cloned from a lignocellulosic decomposing fungi Aspergillus fumigatus Z5 isolated from compost. Protein Expr Purif 79:176–186
Yamada S, Kanno H, Kawahara N (2012) Trans-membrane peptide therapy for malignant glioma by use of a peptide derived from the MDM2 binding site of p53. J Neurooncol 109:7–14
Verma AR, Vijayakumar M, Rao CV, MatHeLa CS (2010) In vitro and in vivo antioxidant properties and DNA damage protective activity of green fruit of Ficus glomerata. Food Chem Toxicol 48:704–709
Kim YG, Park HY (2004) The effects of Pycnogenol on DNA damage in vitro and expression of superoxide dismutase and HP1 in Escherichia coli SOD and catalase deficient mutant cells. Phytother Res 18:900–905
Chen W, Shen X, Xia X, Xu G, Ma T, Bai X, Liang T (2012) NSC 74859-mediated inhibition of STAT3 enhances the anti-proliferative activity of cetuximab in hepatocellular carcinoma. Liver Int 32:70–77
He Y, Huang C, Sun X, Long XR, Lv XW, Li J (2012) MicroRNA-146a modulates TGF-beta1-induced hepatic stellate cell proliferation by targeting SMAD4. Cell Signal 24:1923–1930
De Preter K, Speleman F, Combaret V, Lunec J, Laureys G, Eussen BHJ, Francotte N, Board J, Pearson ADJ, De Paepe A, Van Roy N, Vandesompele J (2002) Quantification of MYCN, DDX1, and NAG gene copy number in neuroblastoma using a real-time quantitative PCR assay. Mod Pathol 15:159–166
Yao Y, Zhang YW, Sun LG, Liu B, Bao YL, Lin H, Zhang Y, Zheng LH, Sun Y, Yu CL, Wu Y, Wang GN, Li YX (2012) Juglanthraquinone C, a novel natural compound derived from Juglans mandshurica Maxim, induces S phase arrest and apoptosis in HepG2 cells. Apoptosis 17:832–841
Chumanov RS, Burgess RR (2010) Expression, purification, and refolding of active Nrf2 transcription factor fused to protein transduction TAT tag. Protein Expr Purif 74:280–288
Choi HS, An JJ, Kim SY, Lee SH, Kim DW, Yoo KY, Won MH, Kang TC, Kwon HJ, Kang JH, Cho SW, Kwon OS, Park J, Eum WS, Choi SY (2006) PEP-1-SOD fusion protein efficiently protects against paraquat-induced dopaminergic neuron damage in a Parkinson disease mouse model. Free Radic Biol Med 41:1058–1068
Clarke RG, Lund EK, Johnson IT, Pinder AC (2000) Apoptosis can be detected in attached colonic adenocarcinoma HT29 cells using annexin V binding, but not by TUNEL assay or sub-G0 DNA content. Cytometry 39:141–150
Liu QL, Hang XM, Liu XL, Tan J, Li DT, Yang H (2012) Cloning and heterologous expression of the manganese superoxide dismutase gene from Lactobacillus casei Lc18. Ann Microbiol 62:129–137
Li AH, Kong Y, Cho SH, Lee HW, Na BK, Pak JK, Kim TS (2005) Molecular cloning and characterization of the copper/zinc and manganese superoxide dismutase genes from the human parasite Clonorchis sinensis. Parasitology 130:687–697
Noorbakhsh R, Mortazavi SA, Sankian M, Shahidi F, Assarehzadegan MA, Varasteh A (2010) Cloning, expression, characterization, and computational approach for cross-reactivity prediction of manganese superoxide dismutase allergen from pistachio nut. Allergol Int 59:295–304
Li JJ, Oberley LW, St Clair DK, Ridnour LA, Oberley TD (1995) Phenotypic changes induced in human breast cancer cells by overexpression of manganese-containing superoxide dismutase. Oncogene 10:1989–2000
Zhong W, Oberley LW, Oberley TD, St Clair DK (1997) Suppression of the malignant phenotype of human glioma cells by overexpression of manganese superoxide dismutase. Oncogene 14:481–490
Liu R, Oberley TD, Oberley LW (1997) Transfection and expression of MnSOD cDNA decreases tumor malignancy of human oral squamous carcinoma SCC-25 cells. Hum Gene Ther 8:585–595
Li N, Oberley TD, Oberley LW, Zhong W (1998) Overexpression of manganese superoxide dismutase in DU145 human prostate carcinoma cells has multiple effects on cell phenotype. Prostate 35:221–233
Mansouri A, Tarhuni A, Larosche I, Reyl-Desmars F, Demeilliers C, Degoul F, Nahon P, Sutton A, Moreau R, Fromenty B, Pessayre D (2010) MnSOD overexpression prevents liver mitochondrial DNA depletion after an alcohol binge but worsens this effect after prolonged alcohol consumption in mice. Dig Dis 28:756–775
H-l Guo, Zhao H-w Xu, Z-f Ma H, Song X, Guan J, Z-j Li, J-m Yu (2005) Manganese superoxide dismutase gene transfection of mouse small intestinal epithelial cells protects them from radiation injury. Zhonghua Zhongliu Zazhi 27:672–675
Bogoyevitch MA, Ngoei KRW, Zhao TT, Yeap YYC, Ng DCH (2010) c-Jun N-terminal kinase (JNK) signaling: recent advances and challenges. Biochim Biophys Acta 1804:463–475
Coffey ET, Smiciene G, Hongisto V, Cao J, Brecht S, Herdegen T, Courtney MJ (2002) c-Jun N-terminal protein kinase (JNK) 2/3 is specifically activated by stress, mediating c-Jun activation, in the presence of constitutive JNK1 activity in cerebellar neurons. J Neurosci 22:4335–4345
Westphal D, Dewson G, Czabotar PE, Kluck RM (2011) Molecular biology of Bax and Bak activation and action. Biochim Biophys Acta 1813:521–531
Jin J, Xiao YC, Chang JH, Yu JY, Hu HB, Starr R, Brittain GC, Chang M, Cheng XH, Sun SC (2012) The kinase TBK1 controls IgA class switching by negatively regulating noncanonical NF-kappa B signaling. Nat Immunol 13:1101–1109
Siddiquee KAZ, Turkson J (2008) STAT3 as a target for inducing apoptosis in solid and hematological tumors. Cell Res 18:254–267
Mora LB, Buettner R, Seigne J, Diaz J, Ahmad N, Garcia R, Bowman T, Falcone R, Fairclough R, Cantor A, Muro-Cacho C, Livingston S, Karras J, Pow-Sang J, Jove R (2002) Constitutive activation of Stat3 in human prostate tumors and cell lines: direct inhibition of Stat3 signaling induces apoptosis of prostate cancer cells. Cancer Res 62:6659–6666
Garcia R, Bowman TL, Niu GL, Yu H, Minton S, Muro-Cacho CA, Cox CE, Falcone R, Fairclough R, Parsons S, Laudano A, Gazit A, Levitzki A, Kraker A, Jove R (2001) Constitutive activation of Stat3 by the Src and JAK tyrosine kinases participates in growth regulation of human breast carcinoma cells. Oncogene 20:2499–2513
Chen CL, Hsieh FC, Lieblein JC, Brown J, Chan C, Wallace JA, Cheng G, Hall BM, Lin J (2007) Stat3 activation in human endometrial and cervical cancers. Br J Cancer 96:591–599
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (30700704), Science and Technology Innovation Project from Department of Education of Guangdong Province (2012KJCX0017) and the Key Laboratory of Regenerative Biology, Guangzhou Institute of Biomedicine and Health, Chinese Academy of Sciences (KLRB201105).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, Z., Huang, L., Wu, Q. et al. A recombinant trans-membrane protein hMnSOD–R9 inhibits the proliferation of cervical cancer cells in vitro. Mol Cell Biochem 385, 79–86 (2014). https://doi.org/10.1007/s11010-013-1816-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-013-1816-2