Abstract
Reintroduction projects represent viable options for animal conservation. They allow the establishment of new local populations and may contribute to recreating functional networks within a metapopulation. In the latter case, landscape connectivity may be a major determinant of the phase of spread of the reintroduced populations. Here, we deal with an example of a red deer (Cervus elaphus) translocation planned to enable the connection among existing isolated populations of the species in the Italian Alps. Our aim was to assess whether the analysis of landscape suitability and the simulation of dispersal of released individuals could shed light on the actual process of population spread. For these purposes, we adopted a modelling approach using radiotracking data to develop a habitat suitability map. On the basis of this map, we simulated the dispersal of the animals after release and we then compared the simulation results with the outcome of null models and with the observed population redistribution. The results suggest that the spread of the subpopulation was easier north-westward than southward. Taking into account landscape suitability, our simulations produced a reliable estimate of the ease of colonization of the valleys neighbouring the release-site and they allowed the identification and validation of a potential pathway for animal dispersal. The suitability model based on the monitoring of individuals in the earliest phase of establishment shed light on the spread of the population and on its potential connections with other deer subpopulations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The spatial configuration of habitat can have an important influence on populations and biological communities. The habitat spatial pattern determines the persistence of natural populations (Hanski 1999) and in conservation biology the effects of habitat loss and fragmentation have been regarded as major threats to biodiversity (Fahrig 2003).
However, the biological consequences of landscape structure have rarely been considered in reintroduction biology. Most papers on reintroduction projects focus on the explicit assessment of the status and viability of released populations, on captive management and on genetics. When habitat is taken into account, attention is concentrated on the release-site suitability, on habitat preparation and restoration, or on habitat selection by released animals (Seddon et al. 2007). Focusing mainly on habitat availability, this approach does not contextualize reintroduction biology in the more complete framework of metapopulation ecology and it does not provide adequate tools to tackle management problems related to the redistribution of reintroduced populations.
Recent studies have adopted new approaches, considering the dispersal of the reintroduced individuals in the landscape and reporting that limited landscape connectivity could significantly affect the results of reintroduction projects, making them questionable in the long run if connections among local populations are not ensured (Kramer-Schadt et al. 2005). Taking into account landscape structure and in particular landscape connectivity (Taylor et al. 1993) would enable the assessment of the usefulness of reintroductions to re-establish a network of (sub) populations, rather than isolated ones.
Here, we provide an example of how spatial modelling and dispersal simulations can be applied to investigate the potential spread of a reintroduced species. In particular, our aim was to assess whether the simulation of dispersal by released animals could shed light on the dynamics of spread of a population towards areas neighbouring the release-site and to outline a procedure for the identification of ecological corridors potentially used by animals during the recolonization process. As a case study, we used a recent reintroduction of red deer (Cervus elaphus) in Chisone valley, Western Italian Alps. In this area, the red deer was present until the 17th century, but its populations then declined and finally disappeared because of unrestricted hunting and human-induced habitat degradation. Reintroduction programs carried out in the second half of the 1900s in neighbouring valleys attempted to rebuild the red deer subpopulations. While they were locally successful, they were not able to recreate a continuous distribution of the species in the Western Italian Alps. A new translocation project was planned in 1999–2000 to re-establish a deme of red deer in the lower Chisone valley. Animals were then expected to disperse from the release-site to the north and south, thus connecting existing isolated populations.
Study area
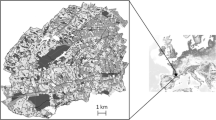
The reintroduction of red deer was carried out in the Wildlife Management Unit of Chisone, Pellice and Germanasca valleys (Fig. 1a), Western Italian Alps (Piedmont, Italy; Fig. 1b). The Wildlife Management Unit consists of 716 km2; altitude ranges between 360 and 3171 m asl and vegetation consists mainly of woodlands (51%), grasslands and alpine meadows (26%), while shrubs account for 10% of the total area. The climate is continental, with a mean annual precipitation of 900 mm.
The following ungulates are present in the Wildlife Management Unit: roe deer (Capreolus capreolus), chamois (Rupicapra rupicapra), ibex (Capra ibex), mouflon (Ovis orientalis) and wild boar (Sus scrofa). In 2000, the red deer presence was limited to the upper Chisone valley. Thanks to reintroduction projects carried out in 1962 and 1964, red deer became established in the neighbouring Susa valley and then spread to the upper Chisone valley. In 2000 and 2002, other translocations carried out in the Po, Varaita and Maira valleys led to the establishment of new subpopulations south of our study area. With the exception of ibex and the newly founded red deer subpopulations, ungulates are hunted in winter by stalking according to conservative shooting plans.
The release-site for the reintroduction of red deer was located in the Pramollo area, in the lower Chisone valley (Fig. 1a), at 1010 m asl. The lateral Pramollo valley is oriented NW-SE, it is characterised by limited human disturbance and the dominance of woodlands, including beech (Fagus sylvatica), chestnut (Castanea sativa) and larch (Larix decidua) woods. Shrubs form a broad transition zone between woodlands and meadows located at high altitudes (approximately 1600–2300 m). Shrubs include Rhododendron sp. and Vaccinium sp., but the green alder (Alnus viridis) is the most widespread species in this transition zone, since it rapidly colonized surfaces that had been deforested for human use, especially livestock grazing. At present, there is only one livestock pasture in the boundary land between Germanasca and Chisone valley.
Materials and methods
Release and monitoring of red deer
Data collection for connectivity modelling
On February 27th 2002, 25 red deer (15 does and 10 bucks) were shipped from Carinthia (Austria) to Pramollo valley. All animals were fitted with ear-tags before release, and 10 females were also fitted with radio-collar transmitters (650 g). From February 2002 to May 2003 operators located the marked animals via triangulation using mobile receiving stations (receiver TRX 64S, Wildlife Materials Inc.), adopting the same sampling scheme for all individuals, with a time distance between fixes of about 5 days, and ensuring the collection of an adequate sample of locations (Seaman et al. 1999).
We only analysed summer radiotracking data (collected in the snow-free months—from March 10th, 2002, to October 10th, 2002) to estimate home ranges and derive a habitat suitability map (see further below), since we expected animal dispersal and population spread to be limited in winter because of snow cover and temperature constraints (Pépin et al. 2009).
Data collection for the validation of the dispersal corridor
From 2002 to 2006, technicians performed observations of the animals from vantage points or along transects uniformly distributed in the Chisone, Germanasca and Pellice valleys, and they collected data on field signs detected along the transects or in sampling plots. We referred all these data to a grid of 250 × 250 m laid on the map of the Wildlife Management Unit using GIS.
We exploited information provided by direct observations and field signs collected in 2002 and 2003 to validate the dispersal corridors, while we used locations of the animals collected in 2004–2006 to verify the population redistribution.
Analysis of the potential spread of the released animals
Landscape structure may affect individual dispersal abilities, thus directing the process of population redistribution (Gardner and Gustafson 2004), since the individual ability to colonise habitat patches may be determined by the biophysical nature of the routes between suitable patches and by the biology and behaviour of the animals (Henein and Merriam 1990). The degree to which the landscape facilitates or impedes movement among habitat patches is referred to as landscape connectivity (Taylor et al. 1993), and connectivity values can be used to estimate the landscape resistance to movement (Gonzales and Gergel 2007). When connectivity maps can not be developed, habitat maps may be coupled with behaviour rules to model dispersal throughout the landscape (e.g. Kramer-Schadt et al. 2004).
Indeed, to evaluate the ease of spread of the population towards the neighbouring valleys, we used suitability maps to compare two dispersal scenarios: (1) deer spread is affected by landscape structure; (2) deer dispersal is modelled as a random process, i.e., the spread of the released animals is not affected by habitat features (we also refer to this model as the null model). For scenario no. 1, we estimated values for each cell (250 × 250 m) of the grid of the study area according to the observed spatial distribution of the released animals. We first developed the habitat suitability map, and we later used it as a basis for dispersal simulations. In the scenario no. 2, we assigned all cells of the grid a fixed suitability value.
Habitat suitability model
To develop the habitat suitability map for scenario no. 1, different statistical approaches can be adopted. We tested several of them, including the development of Resource Selection Probability Functions (RSPF, Lele and Keim 2006), and we finally adopted the procedure described by Marzluff et al. (2004) and Rittenhouse et al. (2008) for the estimation of Resource Utilization Functions (RUFs), since it was best suited for our dataset and it produced the best results concerning the corridor validation (see further below and the discussion for a comparison).
We based the habitat suitability map on the radiotracking data collected in summer 2002 and their analysis involved four steps: (1) for each animal, estimate the Utilization Distribution (UD) as a probabilistic measure of animal space use and measure the density estimate at each cell within the UD; (2) determine the values of associated environmental variables at the same cells; (3) hypothesize different candidate models and use multiple regression to estimate the cell-by-cell relationship between the density estimates (heights of the UD) and the values of environmental variables for each cell; (4) select the most supported model for each individual from the candidate set and estimate an overall RUF for the reintroduced animals via model averaging.
First, we estimated the seasonal home ranges of the marked animals via a UD technique, adopting a fixed kernel method (Worton 1989) and selecting the smoothing parameter via Least Squares Cross Validation (LSCV) (Seaman et al. 1999). We defined the spatial extent of space use as the 99% fixed-kernel home range boundary and we then took the height of the UD for each cell (250 × 250 m) of the study area as a measure of its relative use (Marzluff et al. 2004).
Second, we created a separate raster image in GRASS 6.2.2 (GRASS Development Team 2008) for each of the following variables, adopted as environmental predictors of red deer distribution: (1) distance from villages; (2) distance from main roads; (3) distance from rivers; (4) altitude; (5) aspect; (6) slope; (7) percentage of shrublands in each cell; (8) percentage of open areas; (9) distance from woodlands. We considered vegetation and topography because they represent cover for many ungulate species and affect important aspects of ungulate biology and behaviour (Coughenour 1991; Mysterud and Ostbye 1999). Human disturbance also plays an important role because it may affect red deer movements (Conner et al. 2001), behaviour (Hodgetts et al. 1998) and distribution (Rowland et al. 2000), and we modelled it with derived variables such as distance from roads and villages. We considered also the variable distance from rivers, because suitable corridors for red deer may be localized near rivers (Patthey 2003). As the explanatory variables were not normally distributed, we performed a correlation analysis based on Spearman’s coefficient and we analysed the paired plots of explanatory variables to exclude highly correlated (rs < 0.5; Binzenhöfer et al. 2005) predictors from the subsequent modelling procedure.
Taking into account the observed correlations, we developed several candidate models (Burnham and Anderson 2002; Rittenhouse et al. 2008) relating the UD height to environmental predictors. Each model included different subsets of explanatory variables. We also tested the models including the squared terms of the predictors to account for potential unimodal relationships with the dependent variable (Binzenhöfer et al. 2005).
For each model, we regressed resources on use according to the Marzluff et al. (2004) procedure, performing a Box-Cox transformation of the dependent variable to achieve normality and adopting a Matern correlation function (Handcock and Stein 1993) to remove the spatial autocorrelation (SA) induced by the kernel analysis. This model adjusts SA by fitting a regression model to the UD with SA as a function of the Euclidean distance between the cells. For each animal the regression model taking into account SA was (Marzluff et al. 2004):
where V(x, y) is the vector of resource attributes at the location (x, y) within the region R of positive density for the animal and β values are the corresponging RUF coefficients. The term Z(x, y) approximates the correlation among values of the estimated UD function at different locations within the range determined by the bandwidth used in each individual animal’s kernel density estimate.
We fitted all candidate models separately for each animal and we then determined the best approximating model (with lowest AIC) from the model set, we ranked candidate models using ΔAIC, and we determined the relative likelihood of each of them using Akaike weights (Burnham and Anderson 2002).
Finally, we estimated an overall RUF for the reintroduced animals by taking the average of the RUF coefficients from the most supported model for each individual (Rittenhouse et al. 2008) and for each coefficient we obtained a conservative estimate of variation (see Marzluff et al. 2004 and details therein).
We used the unstandardized RUF coefficients to develop the habitat suitability map, while we analysed the standardized ones to compare the relative influences of explanatory variables on the UD height. Their sign indicates whether use increases or decreases with increase in the quantity of the resource, while their magnitude indicates the change in UD for a unit change in the quantity of the resource.
Since the procedure of model averaging relies on the assumption of independence among animals, we used the association coefficient (Cole 1949; Warrens 2008) to measure the relationship between each pair of radio-collared females. This coefficient measures the degree to which the observed proportion of joint occurrences of two animals exceeds or falls short of the proportion of joint occurrences expected on the basis of chance alone. In particular, we considered the number of times animals were located in the same or different cells of the grid. To avoid bias in the association coefficient due to the large amount of cells without deer locations, we defined the study area for the coefficient estimate including only cells located within the 99% home range of the animals.
We represented the outcome of the RUF analysis via a habitat suitability map (Fig. 2), estimating suitability for each cell (250 × 250 m) as a function of the explanatory variables included in the average RUF model and using a linear stretch to scale the predicted values between 0 and 1 (Johnson et al. 2004). The latter was applied for representation purposes since the RUF analyses produce relative estimates of suitability but they did not provide absolute presence probabilities. Statistical analyses were carried out using R 2.11.0 (R Development Core Team 2010).
Dispersal simulations
To assess the ease of deer spread from the release-site, we performed simulations of the dispersal of animals based on the previously developed model. Coupling the habitat suitability map with movement rules, each simulated step was performed taking into account the habitat suitability values of the grid cells of the map, thus making simulations as realistic as possible. We considered that both the perceptual range and the dispersal distance are important parameters in modelling animal dispersal (Vuilleumier and Metzger 2006). Taking into account deer biology and body size, we assumed that coupling our scale of resolution (250 × 250 m) with a perceptual distance of the eight nearest-neighbouring cells could be adequate for the analyses. Concerning the dispersal ability, according to available literature data, movement paths of about 1000 steps (250 m/step) would be consistent with the distance walked in a 6-months summer period by a free-ranging adult red deer (Pépin et al. 2008, 2009—average walking distance per day recorded in summer by these authors ranged from 1173 to 2075 m, while according to our simulations it was about 1400–1900 m), but we simulated dispersal paths of 2000 steps in order to take into account wider movements or a longer dispersal period.
In scenario no. 1, we simulated the interaction between landscape structure and animal behaviour by allowing the animals to choose one of the eight nearest-neighbouring cells according to their suitability values. Thus, the probability of selecting a cell at a simulation step was not based on probabilities of deer presence, but rather on the relative suitability in comparison to neighbouring ones.
In deterministic (Det) simulations, we allowed an optimal selection of neighbouring cells resulting from always choosing one of the cells with the highest value. The moves were not directionally biased, except that at each step animals could not go back to the previously occupied cell (first-order self-avoiding walk—Gustafson and Gardner 1996). In the probabilistic (Pr) simulations, for each neighbourhood we estimated the empirical cumulative distribution function (ecdf) of suitability values and we then sampled one of the cells according to this function (probability of selection based on suitability values; Gustafson and Gardner 1996). Finally, we coupled probabilistic selection with first-order correlated random walks, producing correlated probabilistic (CPr) simulations (Gardner and Gustafson 2004). A probability of selection of each cell due to the correlation of turning angles was derived from a normal distribution with 40, 60 or 80° SDA (standard deviation of turning angles, reflecting the magnitude of the search area of the individuals; Byers 2001) centred on the angle of the previous step. The code used for simulations at first allowed continuous values of the movement angles, but we then discretized the random angles thus producing a grid-based random walk. The final probability of selecting a cell was obtained by multiplying the probability due to correlation of successive moves for the selection based on suitability values and at each step the movement direction was determined by random sampling according to this final distribution (Gardner and Gustafson 2004).
For the null model (scenario no. 2), we simulated individual dispersal as uncorrelated or correlated simple random walks (Table 1). In uncorrelated random walks (RW), at each step we allowed the animals to randomly select one of the eight nearest-neighbouring cells (completely random selection), while in correlated random walks (CRW) the animals randomly chose the movement direction at the first step, but we then allowed the previous direction to influence the direction of the next step (directionally biased, first order correlated random walks), with a frequency distribution of turning angles described by a normal distribution with 40, 60 or 80° SDA.
For both scenarios and for each movement rule, we simulated 999 exploratory paths, made up of 2000 steps starting from the release-site, and we calculated: (a) the arrival rate in the neighbouring valleys (Schippers et al. 1996); (b) the number of times each grid cell was included in the exploratory paths. We graphically compared the arrival rates in the three valleys with the frequency of actual deer locations collected in 2004–2006. Taking into account the differences concerning the directional bias and movement rules (Table 1), we considered the frequency of inclusion of each cell in the dispersal paths under the suitability scenario and the corresponding null model. For each comparison (Det vs. RW, Pr vs. RW, CPr SDA 40° vs. CRW, CPr SDA 60° vs. CRW, CPr SDA 80° vs. CRW), we then performed a G-test to compare the frequencies recorded under the two scenarios. When we observed an overall significant difference, we identified grid cells with higher than expected use and we mapped them to plot the potential corridor used by the animals during the process of range expansion. Dispersal simulations were carried out in R 2.11.0.
To assess whether deer effectively used the corridors identified via the dispersal simulations based on scenario no. 1, we used independent data on deer presence collected in 2002 and 2003. We calculated the number of deer locations inside and outside each corridor and we repeated this classification procedure for a set of pseudo-absence (random) points generated in the study area. We used these data to produce a confusion matrix that classified our results according to the above mentioned binary rules and to calculate the True Skill Statistics (TSS, Allouche et al. 2006) as a measure of predictive performance.
Results
Habitat suitability model
In summer 2002, the kernel home ranges of the radio-collared animals laid within the lower Chisone valley, with a median extent of 28.7 km2 (99% home ranges, interquartile range = 6.6 km2).
We detected a linear correlation (rs > 0.5) between altitude and the distance from villages, main roads and rivers and among these three latter variables. We also observed a relationship between altitude and slope (rs = 0.73) and between altitude and the distance from woodlands (rs = 0.62). These results were partly expected, since in Alpine valleys villages are mainly located in the lowlands, along main rivers, and woodlands usually extend at low altitudes as well. On the contrary, aspect did not show a clear trend of correlation with other variables. Via the inspection of paired plots, we also observed a relationship between open areas and shrublands, with the latter declining steadily (although non-linearly) with increasing open areas.
To avoid multicollinearity, we kept only four explanatory variables (altitude, aspect, percentage of open areas, distance from woodlands) in the subsequent analyses and we combined them to test three hypothesis (Table 2).
According to Akaike weights, the most supported model varied among deer. Indeed, the Akaike weights suggested that a single model could not adequately explain the spatial distribution of all animals and only models excluding squared terms of habitat type variables received support from the data, suggesting that linear rather than unimodal relationships occurred among the deer presence and these habitat variables (Table 2).
Since we did not detect a strong affiliation between pairs of radio-collared females (average association coefficient = 0.06, SD = 0.05), we averaged the coefficients of the models. We observed a positive relationship of deer presence with the percentage of open areas and distance from woodlands, while an unimodal relationship was found for aspect and altitude (Table 3).
Finally, we represented the outcome of the analysis via a habitat suitability map (250 × 250 m, Fig. 2) based on the averaged unstandardized RUF coefficients.
Dispersal simulations and corridor validation
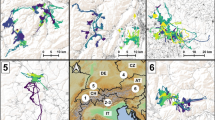
Our simulations showed that the arrival rate of the animals in the valleys neighbouring the release-site differed according to the landscape model. We observed that most of the uncorrelated dispersal paths (Fig. 3: RW, Det and Pr) ended in the low Chisone valley, without reaching the neighbouring sites. Anyway, the simulations based on landscape connectivity and in particular on optimal selection (deterministic simulations, Det) predicted a higher arrival rate in the Germanasca than in the Pellice valley. We also observed that almost all the probabilistic paths (Pr) ended in the low Chisone valley: the few paths reaching the neighbouring areas ended in the Germasca valley and no path reached the Pellice valley. On the contrary, via uncorrelated random simulations (Fig. 3: RW) we could not detected a difference between the arrival rates in the neighbouring valleys.
Frequency of actual deer locations collected in 2004–2006 (FIELD DATA) in the three valleys of the wildlife management unit compared with the arrival rates obtained via dispersal simulations based on the null model [scenario no. 2: uncorrelated random walks (RW) and uncorrelated random walks (CRW)] and on the suitability map [scenario no. 1: deterministic (Det), probabilistic (Pr) and correlated probabilistic(Cpr)]. Results of correlated simulations are differentiated according to the Standard Deviation on turning Angles (SDA)
Concerning correlated walks, we observed that random paths (Fig. 4: CRW SDA 40, 60 and 80°) allowed the animals to reach the Germanasca valley, but that a very limited number of paths ended in the low Chisone valley. In contrast, according to correlated walks taking into account landscape suitability (Fig. 3: CPr SDA 40, 60 and 80°) most of the paths ended in the low Chisone valley and some of them reached the Germanasca valley, while the frequency of arrival in the Pellice valley was very limited.
When we compared this results with the actual population distribution (Fig. 3: FIELD DATA), we observed that the 434 deer locations were concentrated in the low Chisone valley (312) and that a few animals were detected in the Pellice valley. This distribution pattern was consistent with simulations based on the connectivity model, even if they overestimated the settlement in the low Chisone valley.
Via the G-tests, we verified that the frequencies of use of the cells under the scenarios nos.1 and 2 differed. The log likelihood ratio statistic (G), was highly significant (df = 3968, P < 0.0001) for all comparisons between the null models and the corresponding dispersal models. We plotted the cells with a higher than expected use rate to identify the ecological corridor corresponding to each set of simulations and we observed that they were mainly located in the low Chisone valley (Fig. 4).
In 2002 and 2003, we recorded 179 deer locations in the release area and neighbouring valleys. Together with 179 pseudo-absence points, these data allowed us to validate the dispersal corridors. The most supported was the one obtained via the uncorrelated probabilistic simulations (Fig. 3: uncorr. probabilistic, Fig. 4b, TSS = 0.61, Table 1), while the TSS for the corridors obtained via the correlated probabilistic walks (Fig. 4c, d and e) was about 0.55. The corridor obtained via the deterministic simulations (Fig. 4a) was less supported by the field data (TSS = 0.20).
Discussion
Reintroduction projects can represent viable options for animal conservation. They allow the establishment of new local populations and they may contribute to recreating functional networks within a metapopulation. While establishment in the release-site depends upon several factors, including primarily habitat suitability, landscape connectivity may be a major determinant of the subsequent phase of spread of the reintroduced populations.
In our study, we considered a reintroduction of red deer planned to connect existing and isolated populations of the species in the Italian Alps. To assess the role of landscape connectivity in determining the population redistribution we analysed radio-tracking data to obtain a habitat suitabilty map, and we then simulated dispersal of the released individuals.
The first step of our procedure involved the analysis of the relationship between deer space use and environmental variables and we assessed the relative probability of using resources within the home ranges of the animals according to the UD (Marzluff et al. 2004).
Since the suitability map is the basis for the following simulations, the modelling approach adopted to estimate the suitability values is a key step. In particular, it may be argued that a map of absolute presence probabilities could be more adequate to perform the dispersal simulations. To tackle this issue, we also performed statistical analyses with the approach suggested by Lele and Keim (2006), which allows one to obtain absolute probabilities of deer presence by comparing presence and pseudo-absence points (Appendix 1). Anyway, in our case study the number of radio-tracking locations for each animal was adequate to derive the individual home ranges, but fitting distinct Resource Selection Probability Functions (RSPFs) for each animal using presence/pseudo-absence data was not straightforward. Adopting the RSPF approach hence required pooling of data across animals (which is generally not recommended, Gillies et al. 2006). The map of presence probabilities obtained by fitting RSPFs (one for each hypothesised model) was also used to run the simulations, but the resulting corridors were not highly supported by the validation dataset (with maximum TSS value = 0.21). We thus decided to base our simulation on the relative suitability map obtained via the RUF analysis. Although it does not provide absolute presence probabilities, it quantifies use with a probabilistic and continuous metric (Marzluff et al. 2004) and the resulting dispersal corridor was highly supported by the data (maximum TSS = 0.61). Moreover, examples of dispersal simulation based on habitat maps rather than on maps of absolute presence probabilities are documented in the literature (Gardner and Gustafson 2004, Walters 2007) and suitability maps are generally regarded as a sensible tool to identify wildlife corridors (Chetkiewicz and Boyce 2009).
Via the RUF procedure, different environmental predictors were selected to explain the spatial distribution of individual animals. The topography seemed to be the major determinant of deer distribution, as observed also by other authors (Pompilio and Meriggi 2001). In alpine areas topography is expected to affect other environmental variables (Pfeffer et al. 2003) and even when habitat types are not explicitly included in the regression models, the unimodal relationship occurring between the topographic features and the deer UD (Topography model 2b, Table 2) suggests that open areas are important for deer: during summer deer negatively select intermediate altitudes, where open pastures are less abundant than woodlands. These findings agree with the results of models explicitly including open areas or distance from woodlands as independent variables (Table 2). The standardized coefficients obtained by model averaging (Table 3) suggest that a linear relationship occurs between the deer UD and the percentage of open areas, as well as between the UD and the distance from woodlands. Indeed, the red deer is considered ubiquitous and may live in various type of habitats, but originally it was probably an open habitat animal (Clutton-Brock et al. 1982), which may actually select forest rather than open areas mainly to avoid human disturbance (Gonzales and Pépin 1996).
In the second phase of our research, we performed dispersal simulations on the map obtained via the RUF analysis. Due to the difficulty in gathering data on animal dispersal, simulation models have become a cost-effective approach to understand dispersal dynamics (Vuilleumier and Metzger 2006). They can be based on the assumption of diffusive movement (Okubo 1980) and they can employ simple random walks, if simulations are preferred to analytical solutions. Both diffusion models and discrete simulations not considering the variability in landscape attributes can adequately describe animal movement only under the assumption that landscape connectivity does not affect dispersal (i.e., under the assumption of environmental homogeneity). In contrast, animal species can perceive the landscape structure and have complex strategies for foraging and dispersal (Gustafson and Gardner 1996; Vuilleumier and Metzger 2006). To tacke this and other issues, basic diffusion models have been widely extended, being actually able to incorporate information on biased/correlated movement directions, on interaction among individuals and in particular on habitat heterogeneity (Turchin 1998; Ovaskainen 2004). Recently, Ovaskainen (2008) facilitated the use of diffusion-based models for the analysis of movement in heterogeneous landscapes. In spite of this, simulation of correlated random walks have often been preferred for empirically oriented research because they are flexible and allow the inclusion of many realistic parameters. As Ovaskainen (2008) pointed out, the drawback of increased realism is a high specify with the cases under study and the difficult generalization of simulation results. While this may be a disadvantage to generalize the effects of landscape structure on animal dispersal, on the opposite a high specificity is desirable when the aim of the research is the identification of actual corridors for animal dispersal, as in our study.
We hypothesised that dispersing red deer reacts to landscape features and to support this hypothesis we compared the outcome of random walks with simulations based on the connectivity map. We adopted a “habitat selection” approach for dispersal simulations (Gonzales and Gergel 2007), assuming that positively selected habitat are correlated with ease of movement and resource availability.
The results of simulations based on the null model were not consistent with the observed distribution of the animals in 2004–2006. The field data suggested that 4 years after the release most of deer locations were still detected in the release area, but also that the Germanasca valley was actually colonized, while only a few direct observations of deer were recorded for the Pellice valley. In contrast, under a completely random dispersal scenario, the number of simulated paths ending in the Germanasca valley was almost equal to the number of those ending in the Pellice valley, while dispersal simulations based on correlated random walks correctly predicted a higher arrival rate in the Germanasca than in the Pellice valley, but the number of paths ending in the release area was clearly underestimated if compared to field data (Fig. 4).
On the contrary, the ranking of the valleys according to the simulations based on the suitability map (low Chisone valley > Germanasca valley > Pellice valley) was consistent with the observed population redistribution, although the arrival rates were partially different. This pattern of spread suggests that in the future the new deer population will connect with existing deer populations located on the north-west of the release-site, while connection with southern populations seems more difficult because of the low connectivity characterizing the watershed between the release area and the Pellice valley. In general, these findings suggest that misleading results may be obtained when ignoring the effects of landscape spatial structure on the spread of red deer populations.
An important outcome of our simulations was the identification of a potential corridor for animal spread in the low Chisone valley. From the release-site, the corridor runs eastward and then north-westward, providing a pathway for dispersal toward the Germanasca valley (Fig. 4b). This result was achieved by comparing the frequencies of use of the cells according to the null models versus the frequencies under the connectivity scenario, and then further comparing the outlined corridors with field data on deer presence collected in 2002–2003. The corridor most supported by the field data was the one obtained via uncorrelated probabilistic simulations of deer dispersal, although the True Skill Statistic was quite satisfactory also for the other corridors based on probabilistic movement rules. Similar algorithms have been suggested by other authors (Hargrove et al. 2005), but dispersal pathways have been seldom validated. Alternative approaches for finding potential corridors include the analysis of least-cost paths (LaRue and Nielsen 2008), but they assume that animals will follow an optimum route between patches due to a complete knowledge of their environment, which is not achievable by released animals.
Our results thus suggest that the spatial pattern of habitat suitability plays a major role in determining the redistribution of released animals, since dispersal simulations ignoring this spatial pattern produced misleading predictions of colonization of the areas neighbouring the release-site. On the contrary, both deterministic and probabilistic simulations based on connectivity maps may be a valuable tool to predict the population redistribution and to identify ecological corridors used by released animals, respectively.
Identification of dispersal pathways for reintroduced species and assessment of the connectivity among populations are topics of major concern for reintroduction biology, since reintroduction projects should not be isolated efforts. We think our procedure could be useful to tackle issues related to translocations of more selective species of conservation concern in terrestrial ecosystems. Since the procedure was of value in predicting the spread of a rather generalist species, it should work even better when applied to habitat specialists. Finally, the procedure is calibrated only on data collected in the first months after release and this feature should enable its application in most reintroduction projects, which should always include a post-release monitoring phase, providing the data needed for statistical analysis.
References
Allouche O, Tsoar A, Kadmon R (2006) Assessing the accuracy of species distribution models: prevalence, kappa and the true skill statistic (TSS). J Appl Ecol 43:1223–1232
Binzenhöfer B, Schröder B, Strauss B, Biedermann R, Settele J (2005) Habitat models and habitat connectivity analysis for butterflies and burnet moths: the example of Zygaena carniolica and Coenonympha arcania. Biol Cons 126:247–259
Burnham KP, Anderson DR (2002) Model selection and multimodel inference A practical information-theoretic approach, 2nd edn. Springer, New York
Byers JA (2001) Correlated random walk equations of animal dispersal resolved by simulation. Ecology 82:1680–1690
Chetkiewicz C-LB, Boyce MS (2009) Use of resource selection functions to identify conservation corridors. J Appl Ecol 46:1036–1047
Clutton-Brock TH, Guinness FE, Albon SD (1982) Red deer: behavior and ecology of two sexes. The University of Chicago Press, Chicago
Cole LC (1949) The measurement of interspecific association. Ecology 30:411–424
Conner MM, White GC, Freddy DJ (2001) Elk movement in response to early-season hunting in Northwest Colorado. J Wildl Manage 65:926–940
Coughenour MB (1991) Spatial components of plant-herbivore interactions in pastoral, ranching, and native ungulate ecosystems. J Range Manage 44:530–542
Fahrig L (2003) Effects of habitat fragmentation on biodiversity. Annu Rev Ecol Evol Syst 34:487–515
Gardner RH, Gustafson EJ (2004) Simulating dispersal of reintroduced species within heterogeneous landscapes. Ecol Model 171:339–358
Gillies CS, Hebblewhite M, Nielsen SE, Krawchuk MA, Alldridge CL, Frair JL, Saher DJ, Stevens CE, Jerde CL (2006) Application of random effects to the study of resource selection by animals. J Anim Ecol 75:887–898
Gonzales E, Gergel SE (2007) Testing assumptions of cost surface analysis as a tool for invasive species management. Landscape Ecol 22:1155–1168
Gonzales G, Pépin D (1996) Le cerf (Cervus elaphus) en milieu montagnard et nordique. I. Paléontologie, occupation de l’espace, utilisation du temps et des ressources. Revue bibliographique. Gibier Faune Sauvage 13:27–52
GRASS Development Team 2008. Geographic resources analysis support system (GRASS) Software. Open source geospatial foundation project. Available from http://grass.osgeo.org. Accessed July 2009
Gustafson EJ, Gardner RH (1996) The effect of landscape heterogeneity on the probability of patch colonization. Ecology 77:94–107
Handcock MS, Stein ML (1993) A Bayesian analysis of kriging. Technometrics 35:403–410
Hanski IA (1999) Metapopulation ecology. Oxford University Press, Oxford
Hargrove W, Hoffman F, Efroymson R (2005) A practical map-analysis tool for detecting potential dispersal corridors. Landscape Ecol 20:361–373
Henein K, Merriam G (1990) The element of connectivity where corridor is variable. Landscape Ecol 4:157–170
Hodgetts BV, Wass JR, Matthews LR (1998) The effects of visual and auditory disturbance on the behaviour of red deer (Cervus elaphus) at pasture with and without shelter. Appl Anim Behav Sci 55:337–351
Johnson CJ, Seip DR, Boyce MS (2004) A quantitative approach to conservation planning: using resource selection functions to map the distribution of mountain caribou at multiple spatial scales. J Appl Ecol 41:238–251
Kramer-Schadt S, Revilla E, Wiegand T, Breitenmoser U (2004) Fragmented landscapes, road mortality and patch connectivity: modelling influences on the dispersal of Eurasian lynx. J Appl Ecol 41:711–723
Kramer-Schadt S, Revilla E, Wiegand T (2005) Lynx reintroductions in fragmented landscapes of Germany: projects with a future or misunderstood wildlife conservation? Biol Cons 125:169–182
LaRue MA, Nielsen CK (2008) Modelling potential dispersal corridors for cougars in midwestern North America using least-cost path methods. Ecol Model 212:372–381
Lele SR, Keim JL (2006) Weighted distributions and estimation of resource selection probability functions. Ecology 87:3021–3028
Marzluff JM, Millspaugh JJ, Hurvitz P, Handcock MS (2004) Relating resources to a probabilistic measure of space use: forest fragments and steller’s jay. Ecology 85:1411–1427
Mysterud A, Ostbye E (1999) Cover as a habitat element for temperate ungulates: effects on habitat selection and demography. Wildlife Soc B 27:385–394
Okubo A (1980) Diffusion and ecological problems: mathematical models. Springer, Verlag
Ovaskainen O (2004) Habitat-specific movement parameters estimated using mark-recapture data and a diffusion model. Ecology 85:242–257
Ovaskainen O (2008) Analytical and numerical tools for diffusion-based movement models. Theor Popul Biol 73:198–211
Patthey P (2003) Habitat and corridor selection of an expanding red deer (Cervus elaphus) population. Université de Lausanne, Faculté des Sciences, Thèse de doctorat
Pépin D, Adrados C, Janeau G, Joachim J, Mann C (2008) Individual variation in migratory and exploratory movements and habitat use by adult red deer (Cervus elaphus L.) in a mountainous temperate forest. Ecol Res 23:1005–1013
Pépin D, Morellet N, Goulard M (2009) Seasonal and daily walking activity patterns of free-ranging adult red deer (Cervus elaphus) at the individual level. Eur J Wildl Res 55(5):478–486
Pfeffer K, Pebesma EJ, Burrough PA (2003) Mapping alpine vegetation using vegetation observations and topographic attributes. Landscape Ecol 18:759–776
Pompilio L, Meriggi A (2001) Modelling wild ungulate distribution in Alpine habitat: a case study. Ital J Zool 68:281–289
R Development Core Team (2010) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available from http://www.R-project.org. Accessed May 2010
Rittenhouse CD, Millspaugh JJ, Hubbard MW, Sheriff SL, Dijak WD (2008) Resource selection by translocated three-toed box turtles in Missouri. J Wildl Manage 72:268–275
Rowland MM, Wisdom MJ, Johnson BK, Kie JG (2000) Elk distribution and modeling in relation to roads. J Wildl Manage 64:672–684
Schippers P, Verboom J, Knaapen JP, Apeldoorn RC (1996) Dispersal and habitat connectivity in complex heterogeneous landscapes: an analysis with a GIS-based random walk model. Ecography 19:97–106
Seaman DE, Millspaugh JJ, Kernohan BJ, Brundige GC, Raedeke KJ, Gitzen RA (1999) Effects of sample size on kernel home range estimates. J Wildl Manage 63:739–747
Seddon PJ, Armstrong DP, Maloney RF (2007) Developing the science of reintroduction biology. Conserv Biol 21:303–312
Taylor PD, Fahrig L, Henein K, Merriam G (1993) Connectivity is a vital element of landscape structure. Oikos 68:571–573
Turchin P (1998) Quantitative analysis of movement: measuring and modeling population redistribution in animals and plants. Sinauer associates, Sunderland
Vuilleumier S, Metzger R (2006) Animal dispersal modelling: handling landscape features and related animal choices. Ecol Model 190:159–170
Walters S (2007) Modeling scale-dependent landscape pattern, dispersal, and connectivity from the perspective of the organism. Landscape Ecol 22:867–881
Warrens MJ (2008) On association coefficients for 2 × 2 tables and properties that do not depend on the marginal distributions. Psychometrika 73:777–789
Worton BJ (1989) Kernel methods for estimating the utilization distribution in home-range studies. Ecology 70:164–168
Acknowledgments
Thanks are due to the Wildlife Managers Marco Giovo and Federica Gaydou for providing radiotracking data, to Olivier Friard for his technical support, and to Stefano Focardi for a review of an early draft of the paper.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
La Morgia, V., Malenotti, E., Badino, G. et al. Where do we go from here? Dispersal simulations shed light on the role of landscape structure in determining animal redistribution after reintroduction. Landscape Ecol 26, 969–981 (2011). https://doi.org/10.1007/s10980-011-9621-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10980-011-9621-3