Abstract
High levels of attrition characterize undergraduate science courses in the USA. Predictive analytics research seeks to build models that identify at-risk students and suggest interventions that enhance student success. This study examines whether incorporating a novel assessment type (concept inventories [CI]) and using machine learning (ML) methods (1) improves prediction quality, (2) reduces the time point of successful prediction, and (3) suggests more actionable course-level interventions. A corpus of university and course-level assessment and non-assessment variables (53 variables in total) from 3225 students (over six semesters) was gathered. Five ML methods were employed (two individuals, three ensembles) at three time points (pre-course, week 3, week 6) to quantify predictive efficacy. Inclusion of course-specific CI data along with university-specific corpora significantly improved prediction performance. Ensemble ML methods, in particular the generalized linear model with elastic net (GLMNET), yielded significantly higher area under the curve (AUC) values compared with non-ensemble techniques. Logistic regression achieved the poorest prediction performance and consistently underperformed. Surprisingly, increasing corpus size (i.e., amount of historical data) did not meaningfully impact prediction success. We discuss the roles that novel assessment types and ML techniques may play in advancing predictive learning analytics and addressing attrition in undergraduate science education.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction: Machine Learning and Predictive Learning Analytics
Predictive learning analytics uses educational data to develop mathematical frameworks suitable for modeling student outcomes (Brooks and Thompson 2017). In recent years, machine learning (ML) has altered the ways in which these predictions are generated (Rovira et al. 2017). ML techniques are capable of extracting complex patterns from vast stores of data and, unlike many traditional statistical models (such as regression), they can integrate and assess independent sources of educational records that do not follow a prespecified statistical distribution (Tekin 2014; Rovira et al. 2017). Multiple ML algorithms (e.g., a combination of decision trees) that individually would perform less accurately are often combined to bolster prediction performance (Rokach 2010). These ML algorithms, called ensemble methods, have the potential to yield more accurate and timely predictions of student performance (Kotsiantis et al. 2010; Ade and Deshmukh 2014; Amrieh et al. 2016).
ML has been used to develop assessments (e.g., standardized aptitude tests, computer-based formative assessments) in order to (1) evaluate complex constructs in science, (2) enhance inferences about student comprehension, and (3) advance automaticity and accuracy of scoring (Zhai et al. 2020). These tools have also used assessment data to predict student success and identify at-risk students in undergraduate science classrooms (Baker 2010; Silva and Fonseca 2017; PCAST 2012; Chang et al. 2014). Our study follows this latter research tradition and explores whether novel assessment types from different university sources and ML-based prediction models (1) enhance prediction quality, (2) reduce the time point of successful predictions, and (3) suggest more actionable course-level interventions. This work has relevance for several national initiatives focused on enhancing student success and diversifying the life science professions (e.g., American Association for the Advancement of Science [AAAS] 2011; PCAST 2012).
Early Warning Systems and Assessment Sources
A primary application of using ML techniques is the development of Early Warning Systems (EWS), which are computational systems that track and report at-risk students to faculty or other university stakeholders (Macfadyen and Dawson 2010; Neild et al. 2007). Traditionally, these systems have used (1) interactive dashboards, (2) email/instructor notifications, and (3) placement schemes to suggest educational interventions (see Table 1) (Daniel 2019). EWSs rely on prediction algorithms developed using ML and related statistical techniques.
Using predictive analytics to develop EWSs requires examining how the amalgamation of independent assessment data sources can enhance current prediction algorithms and identifying which methods are best suited to model the corpora being examined. Few studies have assessed the efficacy of ML techniques in predicting final course outcomes in introductory science, technology, education, and mathematics (STEM) classes using both university and course-specific assessment data. The focus in past STEM retention studies has been on using university assessment data for these efforts, which typically contain information about prior student academic performance and achievement. Predictors often included (but were not limited to) standardized test scores (e.g., SAT) and student background variables (Rath et al. 2007; Dobson 2008; Orr and Foster 2013; Eddy et al. 2014). More recently, research has been examining how assessment information gathered during students’ current academic experiences may be leveraged in predictive analytics research (e.g., Minaei-Bidgoli et al. 2003; Lykourentzou et al. 2009; Lee et al. 2015). Concept inventories (CI) are increasingly used assessment tools designed for use in large undergraduate STEM classes. They may be valuable sources for enhancing predictions and suggesting interventions.
Concept Inventories in Undergraduate Biology Education
Over the last three decades, a significant body of work in science education has focused on the development of assessment instruments for rigorously measuring undergraduate understanding of disciplinary core ideas (Libarkin 2008; Haudek et al. 2011; Nehm 2019). CI assessments are designed to measure both normative understanding and common misconceptions in introductory science settings (Hake 1998). CIs have also been used to diagnose learning barriers, thereby enabling instructors to modify instruction and improve educational outcomes (Haudek et al. 2011; Bennett 2011; National Research Council 2012). These assessments now cover an array of disciplinary core ideas, ranging from evolution to force and motion (see Sayre and Heckler 2009; Nehm 2019 for examples).
In undergraduate biology education, evolution is widely recognized as a core idea unifying the curriculum (AAAS 2011) and, therefore, a large body of work has focused on developing and using CIs to study student knowledge and misunderstandings of this topic (e.g., Kalinowski et al. 2016; Nehm et al. 2012; Furrow and Hsu 2019). Research has used CIs to document that students often struggle with an array of naïve ideas that are differentially evoked depending upon biological contexts (Nehm and Reilly 2007; Nehm 2019). CIs have also been used to examine the co-occurrence of naïve and scientific ideas and to study how they change throughout a semester (Opfer et al. 2012; Colton et al. 2018). Overall, the centrality of evolution to introductory biology and the well-documented challenges of naïve ideas suggest that CIs may be a useful starting point for studying whether they are able to enhance the quality, timeliness, and usability of predictions.
Literature Review
Research in predictive learning analytics and ML continues to grow. Table 2 includes recent work in this area and highlights features relevant to our study, notably assessment types, ML algorithms utilized, and corpus sizes.
Predictive analytics work has primarily relied upon data from traditional assessment types to build ML prediction models for in-person, hybrid, and online learning environments; no work to our knowledge has utilized CIs. Traditional assessment sources have included (1) standardized tests [e.g., SAT] (Adekitan & Noma-Osaghae (2019); Alexandro (2018); Aulck et al. (2017); Beemer et al. (2018); Getachew (2017); Kumar and Singh (2017)), (2) classroom/virtual assignments and examinations (Al-Shabandar et al. 2017; Lisitsyna and Oreshin 2019), and (3) collaborative group and participation activities (Bucos and Drăgulescu 2018).
In addition to these assessments, prior work suggests that non-academic characteristics should also be incorporated along with academic factors from different university databases (Lee et al. 2015; Xue 2018). Examples include (1) student financial aid data (Adekitan and Noma-Osaghae 2019), (2) demographic characteristics (Al-Shabandar et al. 2017), and (3) learning management system (LMS) variables (Conijn et al. 2016). LMS data, in conjunction with academic characteristics (e.g., grade point average) and personal data records have been shown to improve prediction success (Vovides et al. 2017; Zhai et al. 2020).
Predictions need to be generated early enough in the semester for timely interventions (Feng et al. 2009; PCAST 2012; Lee et al. 2015; Beemer et al. 2018). The differential predictive success of ML techniques at distinct time points has been examined in prior work (Waterhouse et al. 1993; Aulck et al. 2017; Costa et al. 2017). While many studies have used ML for predicting failure, they have often been limited by the timeliness of the predictions (Bayer et al. 2012; Watson et al. 2013; Márquez-Vera et al. 2013; Khobragade and Mahadik 2015). Moreover, these studies did not utilize course-specific assessments in their analyses and only focused on demographic, non-assessment academic endeavors (e.g., number of credits taken), and course activities. Timely predictions incorporating both non-assessment and assessment features may provide greater predictive power in generating accurate forecasts of student attrition and retention.
Although many studies in learning analytics have investigated different approaches for analyzing educational data and predicting learning outcomes (see Lang et al. 2017 for a review), much less work has rigorously explored how increasingly used CI assessments may differentially inform predictions suitable for instructor actions in undergraduate science classrooms. None of the papers we reviewed used CI data to predict student performance. To that end, our study explores the potential use of CI assessments and ML methods for enhancing the (i) accuracy, (ii) timeliness, and (iii) actionability of learning analytics predictions when combined with assessment and non-assessment features from the university data warehouse.
Research Questions
Our study addressed three research questions:
(RQ 1) Do CI assessments significantly enhance (i) accuracy and (ii) timeliness (e.g., week 3, 6 of a 15-week semester) of predictions when combined with non-assessment predictors?
(RQ 2) Do ensemble ML techniques outperform more traditional, non-ensemble approaches?
(RQ 3) Does corpus size (i.e., number of previous semesters) and testing semester (i.e., fall or spring) significantly impact ML prediction efficacy?
Institutional Context
Our investigation focused on predictive analytics in a gateway in-person, lecture-based biology course at a public research university in the Northeastern USA. The university enrolls a large number of first-generation (~ 40%) and underrepresented minorities (23%), many of whom are pursuing STEM-related majors and careers. This institutional context is well suited for our work in predictive analytics and ML given national and institutional calls for addressing persistence in STEM degrees among first-generation and underrepresented minorities (PCAST 2012; Seymour and Hunter 2019). At our institution, student success (i.e., C or above) in gateway science courses is moderate (~ 70–80%), and STEM attrition rates are high (approximately 38% of students pursuing a STEM-related career switch to non-STEM majors at our university). Both of these factors motivated the use of institutional and course-specific CI data for knowledge generation, prediction, and action.
The course studied is a large (n > 1000/year) undergraduate biology class required for completion of a baccalaureate degree in biology and many STEM-related professions (e.g., nursing, pre-medical, general science). Prerequisites are a high school biology course and a college-ready mathematics background. This three-credit course aligns with five core concepts of biological literacy that are detailed in the AAAS Vision and Change policy document: (1) evolution, (2) information flow, (3) nature and practice of science, (4) structure and function, and (5) systems (AAAS 2011). Emphasis is placed on evolution in this course.
University and Course Corpora
Records for 3225 students were obtained for six academic semesters: fall 2014, spring 2015, fall 2015, spring 2016, fall 2016, and spring 2017 (Table 3). The target variable was the student’s transcript grade for the class divided into a binary category: a failing course grade included the grades D, D+, F, I (incomplete), I/F (incomplete course mark whose final grade was not reported by the university-specified deadline), NC (no credit), and W (withdrawal), and was denoted with a ‘0’. A passing course grade included the grades A, A−, B+, B, B−, C, C+, and C− and was denoted with a ‘1’. The overall failing percentage was 11.7% (n = 378) over six semesters. Fall semester failure rates ranged between 14.5 and 22.7%, which was three times as high as the spring failure rates ranging between 4.2 and 7.5%. Fall and spring semesters differ in several variables that are likely to contribute to failure rate disparities. The fall semester enrolls students with reduced high school GPAs (91.8 vs. 93.0), fewer arts and sciences majors (76.2% vs. 84.5%), more transfer students (8.7% vs. 4.7%), more commuters (15.3% vs. 11.5%), and more students in their first term at the university (18.1% vs. 4.7%).
A total of 53 university and course CI-specific predictors were used in this study. Two IRBs were obtained to gather data from these two sources—one for the university-specific corpus and one for the CI data. For the university corpus, we were permitted to use data from all students enrolled in the course during the six semesters examined (the study was considered to be non-human subjects research by the IRB). However, for the course-specific CIs, data could only be used for consenting students (2,311 [71.7%] in total). These individuals consented to participate in this observational study by completing an online survey pre- and post-course. All data were de-identified to protect student privacy and confidentiality in line with IRB requirements (#504271-3).
Only variables with less than 40% missing data were considered, irrespective of pass/failure status, based on the recommendation of two prior simulation studies (Dong and Peng 2013; Jakobsen et al. 2017). On average, all students were missing six predictors (standard deviation = 8). Students who failed the course exhibited a slightly larger number of missing predictors (mean = 10, standard deviation = 9) compared with those who passed the course (mean = 6, standard deviation = 8). Table S5 and Table S6 detail summary statistics and the percentages of missing data for each variable prior to imputation. From these tables, it is noticeable that there is a disproportionate amount of missing entries for failing students compared with passing students pertaining to pre-collegiate academic characteristics and CI assessments. This raises an important issue in quantitative higher education research that missing data, subsequently imputed, can bias ML prediction results (Peugh and Enders 2004; Croninger and Douglas 2005). We present analyses in the supplementary materials (Table S9 and Table S10) examining additional and more restrictive cutoffs for missing data in our corpora. Based on this, we found that neither the differential amount of missing data between passing and failing students nor our original cutoff substantially biased predictive efficacy.
Missing data were imputed by the MICE (Multivariate Imputation via Chained Equations) package in R using 50 iterations of the predictive mean matching multiple imputation technique (Buuren and Groothuis-Oudshoorn 2010). Multiple imputation is advantageous compared with other imputation techniques (e.g., listwise deletion, maximum likelihood estimation) since it is capable of preserving the underlying data distribution following imputation, and can replicate variability in the corpus by using a pooled estimate of random error terms over multiple corpora (Graham 2009; Cox et al. 2014). While there is no universally accepted imputation technique (Marshall et al. 2010), MICE is one of the “best performing multiple imputation methods with regards to the quality of the predictions” for a binary target outcome (Ambler et al. 2007, p. 277). More specifically, the predictive mean matching technique is also the preferred method in MICE for generating the least biased predictions (Marshall et al. 2010).
The 50 university covariates can be divided into five categories: (1) demographics (e.g., age, sex, ethnicity) [missing data ranged between 0.0 and 1.0%], (2) pre-college characteristics (e.g., high school grade point average (GPA) [missing data ranged between 9.6 and 32.9%], (3) college characteristics (e.g., credits taken, prospective major) [missing data ranged between 0.0 and 14.5%], (4) LMS logins [missing data ranged between 0.1 and 0.2%], and (5) financial aid [missing data ranged between 25.6 and 29.2%] (see Table S3 and Table S4).
Three course assessment variables (see Table S4) were included in the course-level results for consenting students. These predictors came from CI assessments administered at the beginning of the course. Given that evolution is a central focus of the life sciences as well as the introductory biology course that we studied (AAAS 2011), we used scores from two published, validated, and commonly employed CI assessments: the Assessment of Contextual Reasoning about Natural Selection ([ACORNS]; Nehm et al. 2012) and the Conceptual Inventory of Natural Selection ([CINS]; Anderson et al. 2002). These tools are widely used in the evolutionary biology classroom for providing valid inferences pertaining to student’s understanding of evolutionary concepts (Nehm 2019).
The ACORNS is a constructed response assessment that requires a student to generate expository explanations to describe the evolutionary change in various settings presented. The student is graded on inclusion of normative key concepts which include competition, differential survival, differential reproduction, heritability, limited resources, and non-adaptive reasoning, and penalized for evolutionary “misconceptions” or naive ideas (Nehm and Reilly 2007). Students’ responses were scored using EvoGrader’s ML algorithms (Moharreri et al. 2014). This computerized grading system is equivalent to human scoring (Beggrow et al. 2014). The second predictor is the number of key concepts the student used in their responses to the ACORNS assessment. Out of the 2311 consenting students, 193 (8.4%) are missing the ACORNS predictors.
The third variable, CINS, is a multiple-choice assessment that measures 10 evolutionary concepts. Each correct response is given a score of 1 and each incorrect response yields a score of 0. Each question has four answer choices with one correct answer and three incorrect answers. Out of the 2311 consenting students, 217 (9.4%) are missing CINS scores.
Methods
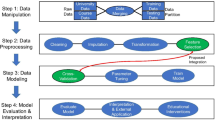
Prediction performance for five widely used ML algorithms in the predictive learning analytics literature was evaluated: (1) logistic regression (LR), (2) support vector machine (SVM), (3) generalized linear model with elastic net (GLMNET), (4) random forest (RF), and (5) stochastic gradient boosting (GBM) (Table 4). Specifically, differences in the area under the receiver operating characteristic curve (AUC) [see Evaluation Metrics] results for models that solely utilized university data and those that combined university and CI data were compared. GLMNET, RF, and GBM are ensemble methods which use a combination of multiple ML techniques to generate predictions. The ML techniques chosen are classified as supervised algorithms because they learn from a set of labeled features in order to generate predictions. These are the most common ML techniques employed in the education literature. Therefore, unsupervised (e.g., latent Dirichlet allocation) and semi-supervised (e.g., various clustering techniques) ML approaches were not explored in our study. A drawback to these techniques is that they require extensive human effort to assemble (Zhai et al. 2020). These ML algorithms were implemented using the caretList function in the R caret package (Kuhn 2015; R Core Team 2017). This statistical package has been used in recent predictive learning analytics studies (Tops et al. 2012; Knowles 2015; Shepherd 2016; Alexandro 2018; Chung and Lee 2019).
Evaluation Metrics
The primary metric calculated to assess the differential efficacy of the ML techniques is the AUC. We also report the accuracy and misclassification rates of each model. Five additional performance metrics are tabulated, but in the interest of space, they are not discussed (see Supplementary Materials C).
The receiver operating characteristic curve graphically depicts the tradeoff between sensitivity and specificity for all possible thresholds to classify observations (Jago et al. 2007) (see Fig. 1). By choosing a different threshold value, \(\tau\), the model will yield different sensitivity and specificity values. Sensitivity is defined as the probability of correctly predicting a student who will pass the course. Specificity is the probability of correctly predicting a student who will fail the course. The closer the AUC is to 1, the more accurate the ML algorithm is. Curves closest to the reference line are less accurate since these predictions are those obtained by chance (Friedman et al. 2001). Unlike other performance metrics, the AUC is independent of the threshold, \(\tau\). Different thresholds were compared to discern the cut-point that optimized model performance. For each ML method, the cut-point \(\tau\)* that maximized Youden’s index (Youden’s J statistic) (Eq. 1) was chosen using the cutpointr package in R (Schisterman et al. 2005).
Youden’s index has been used in a wide variety of ML studies (Bekkar et al. 2013; Mwitondi and Said 2013; Luo et al. 2017; Alexandro 2018). Moreover, \(\tau\)* is the unique optimal cutpoint for maximizing overall correct classifications and minimizing misclassification rates for both positive and negative outcomes (Perkins and Schisterman 2006).
Prediction Methodology
Models were constructed at three time points during the semester: pre-course, week 3, and week 6. Student demographic and pre-collegiate records were included at all time points in the model. Pre-course models did not contain any information about past credit information and any course data, except for the CI assessments. Week 3 and week 6 included LMS logins up to week 3 and week 6, respectively (Table S7). Four sizes of training data (Fig. 2) were examined: the first used two semesters to train the model and the subsequent semester to test the model. The latter three corpora used three, four, and five semesters to predict the following semester, to see whether increasing the amount of historical data was associated with prediction performance.
A comprehensive data pipeline (Fig. 3) was developed in R software for pre-processing and model evaluation. This included (1) imputing missing data, (2) standardizing covariates, and (3) using oversampling to balance the number of passing and failing students in the training corpus. Ten-fold cross-validation was used to optimally tune hyperparameters for each ML method in the pipeline. The values for these hyperparameters can be found in Table S1. A detailed discussion of the data pipeline can be found in section B of the supplementary materials.
Statistical Analyses
To address all research questions, a multiple regression model was used to perform an analysis of the AUC by examining main effects between (1) the number of training semesters, (2) testing semester (fall or spring), (3) time frame, (4) ML technique, and (5) data source (see Table 5). The level of significance was set at 0.01. Furthermore, mean accuracy and mean misclassification rates for passing and failing students were computed for all corpora properties (Table 6). Detailed summary statistics for the evaluation metrics can be found in Supplementary Materials B.
Results
RQ 1—Do CI assessments significantly enhance the (i) accuracy and (ii) timeliness (e.g., week 3, 6 of a 15-week semester) of predictions when combined with non-assessment predictors? A regression model was used to compare the performance of different ML methods at various time points to determine how the addition of course-specific CI data impacted prediction success relative to university data alone. Overall, the regression model explained 72.4% of the variation in the AUC measure, across all prediction time frames, corpora, ML algorithms, testing semesters, and number of training semesters (multiple regression, p value < 0.0001; see Table 5). The university-specific corpus produced significantly lower AUC results by 0.025 points, compared with models run using university and CI data together (t value = − 3.944, p < 0.0001). Both corpora achieved similar prediction accuracy (university: 0.670; composite: 0.676) and misclassification rates for passing (university: 0.333, composite: 0.329) and failing students (university: 0.276, composite: 0.241) averaged across all training semesters, DMMs, and time frames.
Given that a major goal of predictive analytics is to generate actionable knowledge, it is important to consider the time point at which robust predictions can be made. Our study therefore examined prediction success using different ML techniques at pre-course, week three, and week six. Compared with pre-course models, ML methods run at week 3 and week 6 resulted in a significant increase in the mean AUC (week 3: by 0.093, t value = 11.805, p value < 0.0001; week 6: by 0.105, t value = 13.364, p value < 0.001). As the semester progressed, the number of misclassifications decreased. Compared with pre-course, the average number of misclassified passing students decreased by 30, while for failing students, this number decreased by five.
RQ 2—Do ensemble ML techniques outperform more traditional, non-ensemble approaches? Given that few studies have focused on datasets from introductory biology courses, we compared the differential efficacy of three common ensemble ML techniques (GLMNET, RF, GBM) relative to two non-ensemble methods (LR and SVM). Figure 4 depicts the AUC results for each of the five ML techniques, averaged across all training data sizes and time frames. The ensemble methods of GLMNET, RF, and GBM had higher mean AUC values for all three time points than SVM and LR, with GLMNET having the highest mean AUC at week 6 (0.87) using university and course-specific CI assessment data. Based on the regression model, SVM and LR yielded AUC values that were 0.071 (t value = − 6.990, p value < 0.0001) and 0.224 (t value = − 22.017, p value < 0.0001) points lower than GLMNET. The mean AUC values for LR were substantially lower than those obtained for the other models. Overall, ensemble methods outperformed non-ensemble methods.
The model with the highest AUC point estimate was achieved using the following conditions: spring 2017 testing semester using the university corpus, ML algorithm GLMNET, week 6 with five training semesters (AUC: 0.94). Three students (11.1%) out of 27 students were misclassified as passing, while 76 students (13.8%) out of 549 students were misclassified as failing. The specificity rate is 0.86, while the sensitivity rate is 0.89 (Table S14). GLMNET achieved the highest mean accuracy (0.762 with a standard deviation of 0.074). Compared with GLMNET, GBM, SVM, RF, and LR lowered the accuracy by 0.068, 0.073, 0.111, and 0.192 points, respectively.
RQ 3—Does corpus size (i.e., number of previous semesters) and testing corpus (i.e., fall or spring) significantly impact ML prediction efficacy? In the social and applied sciences, using larger data sets (“big data”) has been shown to yield more accurate prediction models (Patel and Sharma 2014; Marr 2015; Prinsloo et al. 2015). Three and four semesters of training data, on average, increased the AUC by 0.009 (t value = 1.137, p value = 0.256) and 0.005 (t value = 0.576, p value = 0.565) points, respectively (compared with two semesters). Five training semesters negatively impacted the AUC metric. Compared with models using two semesters of training data, the use of five semesters of training data significantly lowered the AUC value by 0.025 points (t value = − 2.746, p value = 0.006). While the p value for five training semesters is significant at a 1% level of significance, only one replicate was performed (see Fig. 2). Moreover, as additional training corpora were aggregated, the mean accuracy of prediction models decreased. Compared with five training corpora (0.630), the mean accuracy of models using two training corpora was 0.680. While the average number of misclassified passing students increased when using two to five training semesters (156 to 207), the number of misclassified failing students decreased (17 to 7). Therefore, increasing corpus size neither increased the AUC nor improved predictive success by a meaningful amount.
Since fall semesters had higher failure rates than spring semesters, we also investigated whether testing semester (either fall or spring) impacted the AUC metric. While fall testing corpora yielded AUC values that were 0.016 points lower than spring corpora, this difference was not significant at the 0.01 level (t value = − 2.243, p value = 0.026) and did not improve overall prediction performance. Fall semesters were more accurate at predicting passing students than failing students. On average, the misclassification rates for passing students in the fall and spring were 0.323 and 0.336, respectively. For failure, the misclassification rates were 0.398 and 0.233 for fall and spring semesters, respectively.
Discussion
Our study explored three research questions relevant to predictive analytics in an introductory biology class at a public university. This research context was chosen because student success in gateway courses at the institution is moderate, STEM attrition rates are high, and a large corpus of university and course-specific assessments has been gathered for use in predictive research for knowledge generation, prediction, and action.
The answer to RQ 1 is that the addition of course-specific CI assessments significantly increases the AUC for each of the ML techniques that we examined (p values < 0.0001). Student characteristics and pre-course CI metrics were highly predictive of final course grades. This differs from some literature which found that adding academic assessment predictors in conjunction with student demographic information did little to appreciably enhance predictive accuracy (Allensworth and Easton 2005; National Research Council and National Academy of Education 2011). A likely (and expected) reason for this result is that the CI assessments we used directly tested students on the content that was covered in the course. Therefore, these predictors had significantly greater power in modeling course performance. Utilizing both assessment data sources—university and course CI data—generated the most robust predictions, and both can be considered pragmatic inputs for building predictive models and developing EWSs.
We had anticipated that the course-specific CI assessment data would have a much greater effect on predictive performance. Given that evolution was a core concept emphasized throughout the course, and that the pre-course CIs rigorously measured competencies in this domain, it was surprising that their additive contribution to the overall AUC was moderate. Inclusion of course assessment data increased the AUC and accuracy by 0.025 and 0.006, respectively. Bias from the large number of non-consenting students may have also impacted prediction results. Our expectation was that CI assessments would be a valuable data source for helping to guide instructors in the development and deployment of responses to learning analytics predictions (e.g., unsuccessful performance). For example, while many STEM instructors may be unsure how to address general predictions of student failure developed using university-specific indicators, they may be more confident in their ability to address conceptual difficulties (e.g., misconceptions or low levels of understanding in prerequisite concepts relevant to course themes). Our work shows that while domain-specific knowledge was a significant predictor, its contribution in our study was less than expected. Despite this, the role of domain-specific knowledge remains a crucial but understudied aspect of predictive analytics.
While pre-course predictions are accurate and can be used to implement targeted interventions prior to course commencement, week 3 is the earliest time point during the semester at which robust predictions of successful course performance can be made. This finding reinforces the potential utility of predictive analytics research. At week 3, instructors have time to reformulate student groups, provide supplemental instruction, direct students to tutoring, and engage in psychosocial support structures (Table 1). For the university corpus, the addition of aggregated LMS logins at week 3 and week 6 bolstered predictive performance, reducing the misclassification rate for failing students by 0.066 and 0.094, respectively, compared with pre-course. Moreover, the mean AUC values at week 3 and week 6 for composite corpora were 0.770 and 0.786, respectively, across all ML methods and prediction designs. These positive results indicate the benefit of using LMS data when designing ML-driven tools. Although we found that week 6 models have significantly higher predictive power than models run at all other time frames, it may be more difficult for interventions introduced at this time point to be as effective for struggling students during a standard 15-week semester.
RQ 2 addressed the question of whether ensemble ML algorithms are more effective at predicting performance in an introductory biology course, compared with traditional non-ensemble prediction models. The three ensemble ML techniques achieved the highest mean AUC values, with GLMNET performance the best, compared with the non-ensemble methods SVM and LR. GLMNET achieved the highest mean accuracy across all testing semesters, corpora, DMM, and time frames (0.762 with a standard deviation of 0.074; Table 6). It is important to note that the superior performance of this method using our corpora may not generalize to other educational contexts. Research outside of education has shown that GLMNET’s high predictive performance can be attributed to this method’s approach of selecting or excluding correlated independent variables (Lu and Petkova 2014; Kirpich et al. 2018; Jiménez et al. 2019). However, it must be noted that a few papers have reported that GLMNET performs worse than other ensemble ML algorithms such as RF (Ransom et al. 2019). Additional techniques (i.e., the remaining ML methods in Table 2 not examined in this work) could be applied to determine whether they produce more accurate predictions than the ones obtained.
Our mean AUC results for week 3 using university and course CI assessment data with the GLMNET algorithm (0.85) are lower than Aulck et al. (2017) top three ML methods, but are higher than those of Beemer et al. (2018) (see Table 2). These papers did not use GLMNET. Aulck et al. (2017) achieved a higher mean AUC using GBM (0.89) compared with our model’s mean AUC at week 3 using university and course data (0.80). However, our RF method for this same period was slightly better than the results of Beemer et al. (2018) (our research: 0.81; Beemer et al.: 0.79). While this demonstrates predictive variability among the same algorithms when applied to independent target populations, it shows that these methods have the potential to make evidence-based instructional decisions promoting student success (e.g., Table 1). Until more findings are published for introductory STEM courses, it will be difficult to make confident ML recommendations. Researchers in predictive learning analytics are encouraged to try multiple ML algorithms in order to achieve optimal performance predictions when constructing data-driven tools for their respective disciplines.
The answer to RQ 3 is that increasing the size of the training database did not significantly improve prediction performance. This finding differs from prior work which found that utilizing large data sets improve predictions when examining educational outcomes (Epling et al. 2003; Boyd and Crawford 2011). Although the instructors, course content, and course difficulty were kept relatively constant throughout the semesters that we studied, our surprising results may be attributed to variability in student performance based on (1) heterogeneity in the aptitude of the student body over different academic years, (2) variability of student engagement and retention, (3) administrative changes such as transfer student percentages, (4) lack of sufficient statistical power, and (5) evaluating model adequacy on a single semester corpus (either fall or spring). Overall, our results suggest that larger (and longer-term) corpora may not always appreciably improve predictive success.
Testing semester (fall or spring) also did not significantly enhance prediction performance. During pre-processing, we employed a common oversampling technique (SMOTE: see section B of the supplementary materials) to balance the number of passing and failing students in the training corpus. In predictive analytics literature, oversampling aids ML algorithms in learning data with different prevalence of class levels (i.e., passing and failing rates). Our result is consistent with prior education studies which found that balanced training corpora yield more accurate predictions of student retention and attrition than unbalanced education corpora (Kotsiantis 2009; Márquez-Vera et al. 2010; Radwan and Cataltepe 2017; Alexandro 2018).
A total of 300 prediction models were run exploring the behavior between different data properties and ML algorithms that researchers in predictive learning analytics commonly encounter in their corpora (see Literature Review). The regression analysis and p values presented in Table 5 provide measurements of statistical significance, indicating whether the results obtained can be attributed to sampling variability or by chance. It is important to distinguish between statistical and practical significance in evaluating the usefulness of these results for educational domains. Conflation of these distinctions is common across many disciplines (not only education), leading to unreplicable and impractical predictions (see Ioannidis 2005; Baker 2016; Wasserstein and Lazar 2016). Therefore, to help provide actionable guidance for instructors and stakeholders, we specified model accuracy and misclassification rates for all corpora properties (Table 6) so readers can make use of general predictions of course failure. Our primary goal was to correctly identify students who failed the course (i.e., minimize the amount of failure misclassifications). By week 3, on average, only 14 [3.7%] failing students (out of 378 in total) were misclassified. While the number of misclassified passing students is greater than the number of misclassified failing students, it is more detrimental if struggling students are predicted incorrectly and therefore, do not receive psychosocial support structures and/or supplementary instruction (Table 1). Students on track to succeed in the course are likely to benefit from additional resources, but for failing students, timely identification is critical to reduce attrition. Our prediction results, tools, and pragmatic performance metrics can be applied in educational domains not exclusive to biology in order to develop timely prediction models and EWSs using ML algorithms to improve educational outcomes within their respective discipline. These findings are a small but important first step for improving learning outcomes in gateway science courses. The models developed in this study will be implemented in future semesters in an attempt to reduce failure rates.
Limitations
Our promising results are data-dependent and may not be replicable because of (1) CI choice, (2) content coverage within introductory courses, (3) institution type (e.g., public or private), and (4) class size. Our study also used CIs focused on the core concepts of evolution; it is possible that CIs focused on other topics (e.g., genetics, meiosis, photosynthesis) might not have comparable predictive power. It is likely that courses that are much shorter in duration (e.g., a quarter-long course) have a different time point at which accurate course performance can be generated. Furthermore, we were only able to analyze CI assessment data from consenting students in this study. This percentage of non-participants (28.3%) may have minimized the predictive power of the CI assessments on the AUC metric. While consent rates for the CIs differed between failing and passing students, future studies should attempt to collect CI data from all students. In cases where this is not possible, separate prediction models could be developed for passing students, failing students, and non-consenting students (for whom CI scores are lacking).
For university data, we used a rather exhaustive list of student attributes that encompassed both traditional and non-academic assessments. Even though we suspect that these variables are sufficient to measure a student’s academic performance, our study does not include data that directly measure students’ social behaviors, motivation magnitudes, nor study habits (Gundlach et al. 2015). These measures would need to be collected via surveys and standardized instruments. Prior work has incorporated survey data with amalgamated university data sources to develop analytical models (Beck and Davidson 2001; Yukselturk et al. 2014). However, there is evidence supporting the claim that students are overconfident on survey data and cannot evaluate themselves accurately (Grimes 2002). Survey data, combined with university and CI data, may increase predictive accuracy but could simultaneously introduce bias into the results and cloud intervention planning.
LMS logins for the course were obtained through the university data warehouse, and therefore, these records were incorporated with institution-specific data despite the fact that they may be classified as course-specific. While this may have minimized the impact of the course-specific data corpus on ML results, we still maintain our overall recommendation to combine university and classroom assessment data. Future research should examine the utility of other information extracted from LMSs (e.g., student access to course deliverables), aside from login data, in order to assess how digital tools impact student performance predictions.
Attendance was not utilized in this study because of insufficient variability (attendance was required). Clicker scores were also not used, although the percentage of variation explained by the AUC regression model (72.4%) exceeded findings incorporating clickers in prior work (60.1%; Lee et al. 2015). That paper also suggested week 3 as a reasonable time point for intervention.
Main effects were only examined in our regression analysis. We did not consider interactions between different corpora properties (e.g., corpus size, time point) and ML algorithms. A comprehensive study of these interactions should be examined in future studies.
Conclusion
The special issue Applying Machine Learning in Science Assessment: Opportunities and Challenges highlights many conceptual and methodological advances in the development of science assessments. However, there are additional ways in which ML and science assessments may be used to transform the use of evidence to infer student progress and outcomes in science classrooms. The use of CIs and ML in predictive analytics research has been the focus of remarkably few studies in science education. Our study seeks to advance work in this area by illustrating the potential of ML methods and high-quality science assessments to improve predictions of student success (and associated degree outcomes) in undergraduate science classes. Robust predictions could be linked to inclusive pedagogies (Table 1) in order to address concerning levels of attrition for underrepresented minorities in introductory science classes (PCAST 2012; Seymour & Hunter 2019). Science educators should continue to explore the potential of ML methods and CI assessments for addressing this challenge.
References
Ade, R., & Deshmukh, P.R. (2014, October). Classification of students by using an incremental ensemble of classifiers. In Proceedings of the 3rd International Conference on Reliability, Infocom Technologies and Optimization (pp. 1-5). IEEE.
Adekitan, A. I., & Noma-Osaghae, E. (2019). Data mining approach to predicting the performance of first year student in a university using the admissions requirement. Education and Information Technologies, 24(2), 1527–1543.
Alexandro, D. (2018). Aiming for Success: Evaluating Statistical and Machine Learning Methods to Predict High School Student Performance and Improve Early Warning Systems. (Doctoral Dissertation). University of Connecticut, Storrs, Connecticut.
Allensworth, E. M., & Easton, J. Q. (2005). The on-track indicator as a predictor of high school graduation. Chicago, Illinois: Consortium on Chicago School Research.
Al-Shabandar, R., Hussain, A., Laws, A., Keight, R., Lunn, J., & Radi, N. (2017). Machine learning approaches to predict learning outcomes in Massive open online courses. 2017 International Joint Conference on Neural Networks (IJCNN) (pp. 713–720). Anchorage: IEEE.
Ambler, G., Omar, R. Z., & Royston, P. (2007). A comparison of imputation techniques for handling missing predictor values in a risk model with a binary outcome. Statistical methods in medical research, 16(3), 277–298.
American Association for the Advancement of Science (2011). Vision and change in undergraduate biology education. AAAS, Washington D.C.
Amrieh, E. A., Hamtini, T., & Alijarah, I. (2016). Mining educational data to predict student’s academic performance using ensemble methods. International Journal of Database Theory and Application, 9(8), 119–136.
Anderson, D. L., Fisher, K. M., & Norman, G. J. (2002). Development and evaluation of the conceptual inventory of natural selection. Journal of research in science teaching, 39(10), 952–978.
Aulck, L., Aras, R., Li, L., L’Heureux, C., Lu, P., & West, J. (2017). STEM-ming the tide: Predicting STEM attrition using student transcript data. Knowledge Discovery and Data Mining (KDD): Halifax.
Baker, M. (2016). Reproducibility crisis. Nature, 533(26), 353–366.
Baker, R. (2010). Data mining for education. International Encyclopedia of Education, 7(3), 112–118.
Bayer, J., Bydzovská, H., Géryk, J., Obšıvac, T., & Popelinský, L. (2012). Predicting Drop-Out from Social Behaviour of Students. Proceedings of the 5th International Conference on Educational Data Mining - EDM 2012, (pp. 103–109). Chania, Greece.
Beck, H. P., & Davidson, W. D. (2001). Establishing an early warning system: Predicting low grades in college students from survey of academic orientations scores. Research in Higher Education, 42(6), 709–723.
Beemer, J., Spoon, K., He, L., Fan, J., & Levine, R. (2018). Ensemble learning for estimating individualized treatment effects in student success studies. International Journal of Artificial Intelligence in Education, 28(3), 315–335.
Beggrow, E. P., Ha, M., Nehm, R. H., Pearl, D., & Boone, W. J. (2014). Assessing scientific practices using machine-learning methods: How closely do they match clinic interview performance? Journal of Science Education and Technology, 23(1), 160–182.
Bekkar, M., Djemaa, H. K., & Alitouche, T. A. (2013). Evaluation measures for models assessment over imbalanced data sets. Journal of Information Engineering and Applications, 3(10), 27–38.
Bennett, R. E. (2011). Formative assessment: A critical review. Assessment in Education: Principles, Policy, & Practice, 18(1), 5–25.
Boyd, D., & Crawford, K. (2011). Six provocations for big data. A decade in internet time: Symposium on the dynamics of the internet and society (Volume 21). Oxford, UK: Oxford Internet Institute.
Brooks, C., & Thompson, C. (2017). Predictive modelling in teaching and learning. In C. Lang, G. Siemens, A. Wise, & D. Gašević. Handbook of learning analytics (pp. 61–68). SOLAR, Society of Learning Analytics and Research.
Bucos, M., & Drăgulescu, B. (2018). Predicting student success using data generated in traditional educational environments. TEM Journal, 7(3), 617.
Buuren, S. V., & Groothuis-Oudshoorn, K. (2010). mice: Multivariate Imputation by Chained Equations in R. Journal of Statistical Software, 45(3), 1–68.
Chang, M. J., Sharkness, J., Hurtado, S., & Newman, C. B. (2014). What matters in college for retaining aspiring scientists and engineers from underrepresented racial groups. Journal of Research in Science Teaching, 51(5), 555–580.
Chawla, N. V., Bowyer, K. W., Hall, L. O., & Kegelmeyer, W. P. (2002). SMOTE: Synthetic minority over-sampling technique. Journal of artificial intelligence research, 16, 321–357.
Chung, J. Y., & Lee, S. (2019). Dropout early warning systems for high school students using machine learning. Children and Youth Services Review, 96, 346–353.
Cohen, W. (1995). Fast effective rule induction. In Machine Learning Proceedings 1995 (pp. 115–123). Elsevier.
Colton, J., Sbeglia, G., Finch, S. J., & Nehm, R. H. (2018). A quasi-experimental study of short-and long-term learning of evolution in misconception-focused classes. Paper presented at the American Educational Research Association International conference. New York: NY.
Conijn, R., Snijders, C., Kleingeld, A., & Matzat, U. (2016). Predicting student performance from LMS data: A comparison of 17 blended courses using Moodle LMS. IEEE Transactions on Learning Technologies, 10(1), 17–29.
Costa, E. B., Fonseca, B., Santana, M. A., de Araújo, F. F., & Rego, J. (2017). Evaluating the effectiveness of educational data mining techniques for early prediction of students’ academic failure in introductory programming courses. Computers in Human Behavior, 73, 247–256.
Croninger, R. G., & Douglas, K. M. (2005). Missing data and institutional research. New directions for institutional research, 2005(127), 33–49.
Cox, B. E., McIntosh, K., Reason, R. D., & Terenzini, P. T. (2014). Working with missing data in higher education research: A primer and real-world example. The Review of Higher Education, 37(3), 377–402.
Daniel, B.K. (2019). Improving the Pedagogy of Research Methodology through Learning Analytics. Electronics Journal of Business Research Methods, 17(1).
Davidson, A.C. & Hinkley, D.V. (1997). Bootstrap Methods and their Application (Volume 1). Cambridge University Press.
Dobson, J. L. (2008). The use of formative online quizzes to enhance class preparation and scores on summative exams. Advances in Physiology Education, 32(4), 297–302.
Domingos, P. (1999, August). A general method for making classifiers cost-sensitive. In Proceedings of the Fifth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (pp. 155–164).
Dong, Y., & Peng, C. Y. J. (2013). Principled missing data methods for researchers. SpringerPlus, 2(1), 222.
Eddy, S. L., Brownell, S. E., & Wenderoth, M. P. (2014). Gender gaps in achievement and participation in multiple introductory biology classrooms. CBE - Life Sciences Education, 13(3), 478–492.
Epling, M., Timmons, S., & Wharrad, H. (2003). An educational panopticon? New technology, nurse education and surveillance. Nurse Education Today, 23(6), 412–418.
Feng, M., Beck, J.E., & Heffernan, N.T. (2009). Using Learning Decomposition and Bootstrapping with Randomization to Compare the Impact of Different Educational Interventions on Learning. International Working Group on Educational Data Mining.
Fox, J., & Weisberg, S. (2018). An R Companion to Applied Regression. Sage Publications.
Friedman, J., Hastie, T., & Tibshirani, R. (2001). The Elements of Statistical Learning (Volume 1, No. 10). New York: Springer .
Furrow, R.E., & Hsu, J.L. (2019). Concept inventories as a resource for teaching evolution. Evolution: Education and Outreach, 12(1), 2.
Getachew, M. (2017). Students' Placement Prediction Model: A Data Mining Approach. (Doctoral Dissertation). Addis Ababa University, Arada, Ethiopia.
Graham, J. W. (2009). Missing data analysis: Making it work in the real world. Annual Review of Psychology, 60(1), 549–576.
Grimes, P. (2002). The overconfident principles of economics student: An examination of a metacognitive skill. Journal of Economic Education, 33(1), 15–30.
Gundlach, E., Richards, K., Nelson, D., & Levesque-Bristol, C. (2015). A comparison of student attitudes, statistical reasoning, performance, and perceptions for web-augmented traditional, fully online, and flipped sections of a statistical literacy class. Journal of Statistics Education, 23(1), 1.
Hake, R. R. (1998). Interactive-engagement versus traditional methods: A six-thousand-student survey of mechanics test data for introductory physics courses. American Journal of Physics, 66, 64–74.
Haudek, K. C., Kaplan, J. J., Knight, J., Long, T., Merrill, J., Munn, A., et al. (2011). Harnessing technology to improve formative assessment of student conceptions in STEM: Forging a national network. CBE - Life Science Education, 10(2), 149–155.
Ioannidis, J. P. (2005). Why most published research findings are false. PLoS medicine, 2(8), e124.
Jago, R., Zakeri, I., Baranowski, T., & Watson, K. (2007). Decision boundaries and receiver operating characteristic curves: New methods for determining accelerometer cutpoints. Journal of sports sciences, 25(8), 937–944.
Jakobsen, J. C., Gluud, C., Wetterslev, J., & Winkel, P. (2017). When and how should multiple imputation be used for handling missing data in randomised clinical trials – a practical guide with flowcharts. BMC Medical Research Methodology, 17(1), 162.
James, G., Witten, D., Hastie, T., Tibshirani, R. (2013). An Introduction to Statistical Learning (Vol. 112, p. 184). New York: Springer.
Jiménez, S., Angeles-Valdez, D., Villicaña, V., Reyes-Zamorano, E., Alcala-Lozano, R., Gonzalez-Olvera, J.J., & Garza-Villarreal, E.A. (2019). Identifying cognitive deficits in cocaine dependence using standard tests and machine learning. Progress in Neuro-Psychopharmacology and Biological Psychiatry, 109709.
Kalinowski, S. T., Leonard, M. J., & Taper, M. L. (2016). Development and validation of the conceptual assessment of natural selection (CANS). CBE - Life Sciences Education, 15(4), 64.
Khobragade, L. P., & Mahadik, P. (2015). Students’ academic failure prediction using data mining. International Journal of Advanced Research in Computer and Communication Engineering, 4(11), 290–298.
Kirpich, A., Ainsworth, E. A., Wedow, J. M., Newman, J. R., Michailidis, G., & McIntyre, L. M. (2018). Variable selection in omics data: A practical evaluation of small sample sizes. PLoS, 13(6), e0197910.
Knowles, J. E. (2015). Of needles and haystacks: Building an accurate statewide dropout early warning system in Wisconsin. Journal of Educational Data Mining, 7(3), 18–67.
Kotsiantis, S. (2009). Educational data mining: A case study for predicting dropout-prone students. International Journal of Knowledge Engineering and Soft Data Paradigms, 1(2), 101–111.
Kotsiantis, S., Patriarcheas, K., & Xenos, M. (2010). A combinational incremental ensemble of classifiers as a technique for predicting students’ performance in distance education. Knowledge-Based Systems, 23(6), 529–535.
Krstajic, D., Buturovic, L. J., Leahy, D. E., & Thomas, S. (2014). Cross-validation pitfalls when selecting and assessing regression and classification models. Journal of cheminformatics, 6(1), 1–15.
Kuhn, M. (2015). Caret: classification and regression training. Astrophysics Source Code Library.
Kumar, M., & Singh, A. (2017). Evaluation of data mining techniques for predicting student’s performance. International Journal of Modern Education and Computer Science, 9(8), 25–31.
Lang, C., Siemens, G., Wise, A., & Gašević, D. (2017). The Handbook of Learning Analytics. ISBN: 978–0–9952408–0–3. DOI: https://doi.org/10.18608/hla17.
Lavesson, N., & Davidsson, P. (2006, July). Quantifying the impact of learning algorithm parameter tuning. In AAAI (Vol. 6, pp. 395–400).
Lee, U. J., Sbeglia, G. C., Ha, M., Finch, S. J., & Nehm, R. H. (2015). Clicker score trajectories and concept inventory scores as predictors for early warning systems for large STEM classes. Journal of Science Education and Technology, 24(6), 848–860.
Libarkin, J. C. (2008, October 13–14). Concept inventories in higher education science. Prepared for the national research council promising practices in undergraduate STEM education workshop 2. Washington D.C., United States.
Lisitsyna, L., & Oreshin, S. (2019). Machine Learning Approach of Predicting Learning Outcomes of MOOCs to Increase Its Performance. Smart Education and e-Learning 2019 (pp. 107–115). Springer.
Lu, F., & Petkova, E. (2014). A comparative study of variable selection methods in the context of developing psychiatric screening instruments. Statistics in Medicine, 33(3), 401–421.
Lu, W., Benson, R., Glaser, K., Platts, L., Corna, L., Worts, D., et al. (2017). Relationship between employment histories and frailty trajectories in later life: Evidence from the English Longitudinal Study of Ageing. Journal of Epidemiology Community Health, 71(5), 439–445.
Luengo, J., García, S., & Herrera, F. (2012). On the choice of the best imputation methods for missing values considering three groups of classification methods. Knowledge and information systems, 32(1), 77–108.
Luo, Y., Li, Z., Guo, H., Cao, H., Song, C., Guo, X., & Zhang, Y. (2017). Predicting congenital heart defects: A comparison of three data mining methods. PLoS ONE, 12(5), e0177811–e0177811.
Lykourentzou, I., Giannoukos, I., Mpardis, G., Nikolopoulos, V., & Loumos, V. (2009). Early and dynamic student achievement prediction in e-learning courses using neural networks. Journal of the American Society for Information Science and Technology, 60(2), 372–380.
Macfadyen, L. P., & Dawson, S. (2010). Mining LMS data to develop an “early warning system” for educators: A proof of concept. Computers & education, 54(2), 588–599.
Márquez-Vera, C., Morales, C. R., & Soto, S. V. (2013). Predicting school failure and dropout by using data mining techniques. IEEE Revista Iberoamericana de Tecnologias del Aprendizaje, 8(1), 7–14.
Márquez-Vera, C., Romero, C., & Ventura, S. (2010). Predicting School Failure Using Data Mining. 4th International Conference on Educational Data Mining, (p. 271). Eindhoven, Netherlands.
Marr, B. (2015). Big Data: Using SMART big data, analytics and metrics to make better decisions and improve performance. John Wiley & Sons, 2015.
Marshall, A., Altman, D. G., Royston, P., & Holder, R. L. (2010). Comparison of techniques for handling missing covariate data within prognostic modelling studies: A simulation study. BMC medical research methodology, 10(1), 7.
Minaei-Bidgoli, B., Kashy, D. A., Kortemeyer, G., & Punch, W. F. (2003, November). Predicting student performance: An application of data mining methods with an education web-based system. 33rd Annual Frontiers in Education, 2003. FIE 2003. (Vol. 1, pp.T2A-13). IEEE.
Moharreri, K., Ha, M., & Nehm, R. H. (2014). EvoGrader: an online formative assessment tool for automatically evaluating written evolutionary explanations. Evolution: Education and Outreach, 7(1), 15.
Mwitondi, K. S., & Said, R. A. (2013). A data-based method for harmonising heterogeneous data modelling techniques across data mining applications. Journal of statistics applications and probability, 2(3), 157–162.
National Research Council. (2012). Thinking evolutionarily: evolution education across the life sciences. Washington D.C: National Academic Press.
National Research Council and National Academy of Education. (2011). High school dropout, graduation, and completion rates: better data, better measures, better decisions. Washington D.C.: The National Academics Press.
Nehm, R. H. (2019). Biology education research: Building integrative frameworks for teaching and learning about living systems. Disciplinary and Interdisciplinary Science Education Research, 1(1), 15.
Nehm, R. H., & Reilly, L. (2007). Biology majors’ knowledge and misconceptions of natural selection. BioScience, 57(3), 263–272.
Nehm, R. H., Beggrow, E. P., Opfer, E. P., & Ha, M. (2012). Reasoning about natural selection: diagnosing contextual competency using the ACORNS instrument. The American Biology Teacher, 74(2), 92–98.
Neild, R. C., Balfanz, R., & Herzog, L. (2007). An early warning system. Educational leadership, 65(2), 28–33.
Opfer, J. E., Nehm, R. H., & Ha, M. (2012). Cognitive foundations for science assessment design: Knowing what students know about evolution. Journal of Research in Science Teaching, 49(6), 744–777.
Orr, R., & Foster, S. (2013). Increasing student success using online quizzing in introductory (majors) biology. CBE - Life Sciences Education, 12(3), 509–514.
Patel, J.A., & Sharma, P. (2014, August). Big data for better health planning. In 2014 International Conference on Advances in Engineering & Technology Research (ICAETR-2014). (pp. 1–5). IEEE.
PCAST, PsCoSaT. . (2012). Engage to excel: Producing one million additional college graduates with degrees in science, technology, engineering, and mathematics. Washington DC: Executive Office of the President.
Perkins, N. J., & Schisterman, E. F. (2006). The inconsistency of “optimal” cutpoints obtained using two criteria based on the receiver operating characteristic curve. American Journal of Epidemiology, 163(7), 670–675.
Peugh, J. L., & Enders, C. K. (2004). Missing data in educational research: A review of reporting practices and suggestions for improvement. Review of Educational Research, 74(4), 525–556.
Prinsloo, P., Archer, E., Barnes, G., Chetty, Y., & Van Zyl, D. (2015). Big(ger) data as better data in open distance learning. International Review of Research in Open and Distributed Learning, 16(1), 284–306.
R Core Team. (2017). R: A Language for Statistical Computing. Vienna Austria: R Foundation for Statistical Computing. https://www.R-project.org.
Radwan, A., & Cataltepe, Z. (2017). Improving performance prediction on education data with noise and class imbalance. Intelligent Automation & Soft Computing, 1–8.
Ransom, C. J., Kitchen, N. R., Camberato, J. J., Carter, P. R., Ferguson, R. B., et al. (2019). Statistical and machine learning methods evaluated for incorporating soil and weather into corn nitrogen recommendations. Computers and Electronics in Agriculture, 164, 104872.
Rath, K., Peterfreund, A., Xenos, S., Bayliss, F., & Carnal, N. (2007). Supplemental instruction in introductory biology I: Enhancing the performance and retention of underrepresented minority students. CBE- Life Science Education, 6(3), 203–216.
Rebok, G. W., Ball, K., Guey, L. T., Jones, R. N., Kim, H. Y., Kim, H. Y., et al. (2014). Ten-year effects of the advanced cognitive training for independent and vital elderly cognitive training trial on cognition and everyday functioning in older adults. Journal of the American Geriatrics Society, 62(1), 16–24.
Rokach, L. (2010). Ensemble-based classifiers. Artificial Intelligence Review, 33(1–2), 1–39.
Rovira, S., Puertas, E., & Igual, L. (2017). Data-driven system to predict academic grades and dropout. PLoS, 12(2), e0171207.
Sayre, E. C., & Heckler, A. F. (2009). Peaks and decays of student knowledge in an introductory E&M course. Physical Review Special Topics-Physics Education Research, 5(1), 1–5.
Schisterman, E. F., Perkins, N. J., Liu, A., & Bondell, H. (2005). Optimal cut-points and its corresponding Youden index to discriminate individuals using pooled blood samples. Epidemiology, 16(1), 73–81.
Seymour, E. & Hunter, A.B. (Eds.) (2019). Talking about Leaving Revisited. Springer. Nature: Switzerland.
Shepherd, D. L., (2016). The open door of learning - Access restricted: School effectiveness and efficiency across the South African education system. (Doctoral Dissertation). Stellenbosch University, Stellenbosch, South Africa .
Silva, C., & Fonseca, J. (2017). Educational Data Mining: A Literature Review. Europe and MENA Cooperation Advances in Information and Communication Technologies: Advances in Intelligent Systems and Computing, vol 520 (pp. 87–94). Springer, Cham.
Tekin, A. (2014). Early prediction of students’ grade point averages at graduation: A data mining approach. Eurasian Journal of Educational Research, 54, 207–226.
Thai-Nghe, N., Gantner, Z., & Schmidt-Thieme, L. (2010). Cost-sensitive learning methods for imbalanced data. In The 2010 International Joint Conference on Neural Networks (IJCNN) (pp. 1–8). Barcelona, Spain, 2010.
Tops, W., Callens, M., Lammertyn, J., Van Hees, V., & Brysbaert, M. (2012). Identifying students with dyslexia in higher education. Annals of Dyslexia, 62(3), 186–203.
Vovides, Y., Sanchez-Alonso, S., Mitropoulou, V., & Nickmans, G. (2007). The use of e-learning course management systems to support learning strategies and to improve self-regulated learning. Educational Research Review, 2(1), 64–74.
Wasserstein, R. L., & Lazar, N. A. (2016). The ASA statement on p-values: Context, process, and purpose. The American Statistician, 70(2), 129–133.
Waterhouse, J. K., Carroll, M. C., & Beeman, P. B. (1993). National council licensure examination success: Accurate prediction of student performance on the post-1988 examination. Journal of Professional Nursing, 9(5), 278–283.
Watson, C., Li, F., & Godwin, J. (2013). Predicting performance in an introductory programming course by logging and analyzing student programming behavior. 2013 IEEE 13th International Conference on Advanced Learning Technologies (pp. 319–323). Beijing: IEEE.
Xue, Y. (2018, June). Testing the differential efficacy of Data Mining Techniques to predicting student outcomes in higher education. (Doctoral Dissertation). Stony Brook University, Stony Brook, New York.
Yang, Q., & Wu, X. (2006). 10 challenging problems in data mining research. International Journal of Information Technology & Decision Making, 5, 597–604.
Yukselturk, E., Ozekes, S., & Turel, Y. K. (2014). Predicting dropout student: An application of data mining methods in an online education program. European Journal of Open, Distance, and e-learning, 17(1), 118–133.
Zhai, X., Yin, Y., Pellegrino, J. W., Haudek, K. C., & Shi, L. (2020). Applying machine learning in science assessments: A systematic review. Studies in Science Education, 56(1), 111–151.
Acknowledgements
We sincerely thank Drs. Yaqi Xue and Nora Galambos for assembling the databases that we analyzed in this study. We thank the guest editor and anonymous reviewers for providing thoughtful and helpful comments to improve this manuscript.
Funding
The Howard Hughes Medical Institute Science Education Program provided funding. The views in this contribution reflect those of the authors and not necessarily those of HHMI.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This article does not contain any studies with animals performed by any of the authors.
Informed Consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bertolini, R., Finch, S.J. & Nehm, R.H. Testing the Impact of Novel Assessment Sources and Machine Learning Methods on Predictive Outcome Modeling in Undergraduate Biology. J Sci Educ Technol 30, 193–209 (2021). https://doi.org/10.1007/s10956-020-09888-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10956-020-09888-8