Abstract
Cervical cancer is the fourth most communal malignant disease amongst women worldwide. In maximum circumstances, cervical cancer indications are not perceptible at its initial stages. There are a proportion of features that intensify the threat of emerging cervical cancer like human papilloma virus, sexual transmitted diseases, and smoking. Ascertaining those features and constructing a classification model to categorize, if the cases are cervical cancer or not is an existing challenging research. This learning intentions at using cervical cancer risk features to build classification model using Random Forest (RF) classification technique with the synthetic minority oversampling technique (SMOTE) and two feature reduction techniques recursive feature elimination and principle component analysis (PCA). Utmost medical data sets are frequently imbalanced since the number of patients is considerably fewer than the number of non-patients. For the imbalance of the used data set, SMOTE is cast-off to solve this problem. The data set comprises of 32 risk factors and four objective variables: Hinselmann, Schiller, Cytology and Biopsy. Accuracy, Sensitivity, Specificity, PPA and NPA of the four variables remains accurate after SMOTE when compared with values obtained before SMOTE. An RSOnto ontology has been created to visualize the progress in classification performance.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
In order to replace the dead and damaged cells a normal human body produces 50 to 70 billion cells every day. At times the growth of cells remain uncontrolled which results in benign or malignant. Malignant tumors are referred as a cancer case. This paper emphasis on a specific type of cancer called cervical cancer. Two main factors those are responsible for cervical cancer is
-
1.
Modifiable factors like sexual intercourse
-
2.
Non-modifiable factors like mutational hormones [1].
One of the serious health issue faced by women nowadays is cervical cancer [2]. 80% of cervical cancer cases prevail in developing countries [3]. The United States estimate 13.240 new cervical cancer cases in 2018 and about 4.170 estimated death [4] which means that the death ratio is nearly 31.5%. Cervical cancer affects the reproductive system of women by attacking women’s cervix. At the early stages it develops without any symptoms and these symptoms starts appearing only at later stage after spreading to all other organs. So it is very important to diagnose the infection at the early stage and increase the survival ratio.
Since the ratio of infected widely increases Machine learning techniques are used to resolve these problems in medical and disease diagnosis. In thispaper we apply Random Forest (RF) algorithm to deal withunbalanced data sets, to increase the performance [5, 6]. It remains better than simple neural networks technique.
Synthetic Minority Oversampling Technique (SMOTE) algorithm used balances the dataset classes there by quantitatively increasing the minority class. The increase of minority classes is based on k-nearest neighbors to nearly equal classes. In order to lessen the processing time and remove unimportant features in the classification Recursive Feature Elimination (RFE) and Principle Component Analysis (PCA) are used. Then Random Forest classification technique classifies the cases into 2 categories, cervical cancer and non-cervical. The completed performance is measured before and after SMOTE for further results.
The paper is structured as follows.
-
Section II - Related work of cervical cancer classification.
-
Section III - Methods of machine learning, oversampling, features reduction techniques used.
-
Section IV - Experimental results discussed. Analysis and comparison shown.
-
Section V - Ontological Representation
-
Section VI - Conclusion and Future work presented.
Related Work
Researchers have made many researches in the field of cervical cancer. Researchers used various approachesto detect and diagnose their presence. Various classification and segmentation methods are used at various time periods to enhance the research in this area. The enhanced versions are used so that they help to identify various risk factors in cervical cancer. Game theory model [7, 8], dynamic genetic algorithms [9] and Artificial Bee Colony based clustering approach [10] are play the vital role to develop a medical system model and ontological representation. This papers presents an ontological representation RSOnto for the enhanced study with SMOTE to enrich the research in this area.
In 2013, Tseng et al. [11] obtained the highest results in accuracy by using three classification models
-
1.
C5.0
-
2.
Support vector machine
-
3.
Extreme machine learning in cervical cancer.
The dataset collected form the Medical University Hospital, Chung Shan was with 12 features for 168 cases where two risk factors were identified. The results proved C5.0 obtained the highest classification.
In 2014, Hu et al. [12] using artificial neural networks obtained the highest classification accuracy by back substitution in cervical cancer.
In 2016, Sharma [13] obtained accurate results using naïve bayes which outperforms logistics regression.
In 2016 Sobar et al. [14] used the theory of behavior in social science and obtained accurate results using naïve bayes which outperforms logistics regression [15].
In 2017 Wu and Zhou [16] experimented a classification model based on Support Vector Machine (SVM) and obtained the highest accuracy ratio. Four target variables Hinselmann, Schiller, Cytology and Biopsy were determined by the relevant factors available. RFE and PCA techniques were used to reduce the processing time.
In 2019 3rd April KwandaNgwenduna (www.colloquium2019.org.za/wp.../2019/04/kwanda_sydwell_ngwenduna_10h45.pdf) stated that there remains class imbalance still and SMOTE can be combined with under sampling and remains comprehensive to regression and time series.
Proposed Methods
Random Forest (RF)
A renowned classification technique used in diverse classification areas is Random Forest (RF) [17, 18]. RF is also recognized as bagged decision trees [19, 20]. This algorithm [21] works on using group of weak learners to formulate strong learner. RF customizes 2 techniques
-
1.
Classification technique
-
2.
Regression Tree (CART) technique [22].
These techniques progresses uncorrelated combination or multiple decision trees centered on bootstrap aggregation (bagging) technique [23].CART technique enables us to learn the correct classification amongst some dependent variables (y) and some independent variables (x) and relationbetween them. Random Forest technique selects a subset randomly to build an independent decision tree. It is a repetition process which splits the selected random subset from the root node to a child node [24]. This splitting continues till each tree reaches a leaf node without cropping. Each tree makes the classification of the features and the objective variable independently and votes for the final tree class [25]. The overall classification is based on the majority acquired trees voting.
Steps for RF construction:
-
N - Quantitative amount of bootstrap samples
-
M - Total number of attributes
-
m - Sample size
-
k - Next node
-
Step1.
Creates N bootstrap samples from the dataset.
-
Step2.
Every node (sample) takes attributes randomly of size m where m < M.
-
Step3.
Builds a split for the m attributes selected in Step 2 and identifies the k node by utilizing the best split point.
-
Step4.
Split the tree repeatedly till one leaf node is reached and now the tree remains completed.
-
Step5.
The algorithm is trained on each bootstrapped separately.
-
Step6.
Using trees classification voting predicted data is collected from the trained trees (n).
-
Step7.
The final RF model is build using the highest voted features.
The proposed flow of activities has been diagrammatically represented in Fig. 1.
Features Selection Techniques
Two feature selection methods are used.
-
1.
Principle Component Analysis (PCA)
-
2.
Recursive Feature Elimination (RFE).
These selection techniques reduce the features remaining without degrading the model performance. The remaining features remain in the full features dataset.
Principle Component Analysis (PCA)
A statistical mathematical procedure that uses eigenvector to describe the feature orientation is PCA. This analysis maps the n-dimension feature space into k-dimension where k < n, known as principle component. The covariance matrix is calculated. The calculated result is used for defining eigenvectors and Eigen values [26]. Principle component is the Eigen vector with the highest Eigen value. This principle component is chosen from the cervical cancer dataset since it reveals the most important relationship amongst the data set attributes [7]. The Eigen values are arranged in ascending order where the most significant data is chosen and the least significant data is discarded. The highest dimension data is reduced to a lower dimension data [27].
In order to define the deviation of data in the data set calculate the variance (1) which depicts the spread of data.
Then covariance is determined to identify the relation of the dataset features. The high values express the high relation amongst features and zero values identifies that there is no relation amongst features. The covariance is calculated using eq. (2).
The Eigenvalues and Eigenvectors for the covariance matrix are determined. The determined eigenvalues are then transformed (varimax orthogonal rotation) using eq. (3).
Recursive Feature Elimination (RFE)
RFE algorithm is also used with random forest for variable importance grouping [28]. RFE is proposed by Guyon et al. [29]. It was used in gene microarray where the number of features was thousands. Díaz-Uriarte and Alvarez de AndrØs [30] used RFE-RF for gene selection and class prediction; they used a back-word selection method in linear support vector machine. It also works with other linear classification methods. Figure 2 shows the pseudo-code for the algorithm.
Synthetic Minority Oversampling Technique (SMOTE)
Machine learning techniques facing troubles when one class dominates the dataset which means that the number of records in one class highly exceeds the number of the other classes. Dataset in this case is called imbalanced dataset and this kind of dataset misleads the classification and affects the results. SMOTE is used to solve this problem. SMOTE is one of the oversampling techniques that was introduced by Chawla et al. [31]. It is used synthetically to increase the minority class based on k-nearest neighbors [31], to balance the dataset. The SMOTE algorithm is used in different fields to solve the unbalanced problem like network intrusion detection systems [32], breast cancer detection [33] and sentence boundary in speech [34]. SMOTE technique synthetically increase the minority class.
SMOTE can be described by the following steps.
-
Step1.
Identifies the feature vector xi and identify the K-nearest neighborsxknn.
-
Step2.
Calculates the difference between the feature vector and k-nearest neighbor.
-
Step3.
Multiplies the difference by a random number between 0 and 1.
-
Step4.
Adds the output number to feature vector to identify a new point on the line segment.
-
Step5.
Repeats the process from 1 to 4 for identifying the feature vectors.
Cervical Cancer Dataset
The cervical cancer dataset consisted of medical record history, routines and demographic information for 858 cases with 32 features for each and every case [35]. Dataset may have missing values because there are cases which decide not to answer all questions for confidential concern. The information predicts the
-
1.
Dataset features
-
2.
Total number of entries and
-
3.
The missing value for each feature.
The dataset is from https://www.kaggle.com/c/intel-mobileodt-cervical-cancer-screening.
Missing values are visible in dataset. We can find a lot of missing values, which are removed and the number of features are decreased to 30. We use the mean equation to handle the missing values. Figure 3 represents the features in the dataset. Figure 4A and 4B illustrate the values related to the features.
Schiller’s test is used to diagnose cervical cancer by applying iodine solution in the cervix [36]. Hinselmann is used to examine the cervix, vulva and vagina [35]. Cytology is the test which checks for cancer, precancerous conditions, and urinary tract infection. Biopsy, a piece of tissue from the body used to examine and suggest if it is normal or not. These four tests are examined and the results are compared before and after SMOTE.
For each case of the 858 cases labeled with Hinselmann, Schiller, Cytology and Biopsy, an ontological representation for the same is provided in this paper. The objective variables articulate a form of cervical cancer examination.
Quantitatively the number of examinations of the patients are compared to the non-patients before and after SMOTE.
-
1.
Before SMOTE the data remains imbalanced.
-
2.
After the implementation of SMOTE algorithm the dataset remains balanced.
Evaluation Metrics
Using unbalanced dataset the accuracy, sensitivity, specificity, positive predicted accuracy (PPA) and negative predicted accuracy (NPA) are measured which predicts the performance of the classification. Using random forest with SMOTE and two feature reduction techniques the cervical cancer prediction is performed. In the pre-processing stage the unbalanced dataset with missing values and lack of information are deleted. Apply SMOTE to balance the unbalanced dataset. Apply the feature selection techniques PCA and RFE which reduces the number of features and decrease the processing time of the dataset. The second phase signifies the classification phase in which training is performed using random forest. The next phase emphasizes on 10-fold cross validation technique for validation and testing purpose. The concluding phase of the model compares the results with and without SMOTE algorithms and the obtained result with methodology is applied in ontology [37].
Simulation Experiment
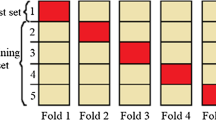
The cost of misdiagnose of a cervical cancer case or vice versa is high. The used dataset is unbalanced as the number of malignant records is fewer than the number of normal records so SMOTE algorithm is used to balance the number of classes. In this section three RF-based approaches were used to classify cervical cancer cases to identify the patient and the non-patient ones. For validating our model performance, 10-fold cross validations were used. The experiments were done before and after SMOTE with and without feature selection. Each experiment was executed separately to ensure the highest accuracy and avoid classification mislead due to the nature of the dataset. The experiments will be conferred in the imminent sections with reference count as in Table 1.
Objective Variable: Hinselmann
In Hinselmann examination test, the RF before SMOTE was achieved with total accuracy of 95.91% with 35 patient records and 823 non-patient records. After using SMOTE algorithm RF achieved a total accuracy of 97.91% with number of patients 805 and non-patients 823. SMOTE algorithm increased the accuracy ratio with sensitivity ratio, PPA and as shown in Table 2 and Table 3.
Objective Varaible: Schiller
In Schiller examination test, the RF before SMOTE was achieved with total accuracy of 91.48 with 35 patient records and 823 non-patient records. After using SMOTE algorithm RF achieved a total accuracy of 95.02% with number of patients 805 and non-patients 823. SMOTE algorithm increased the accuracy ratio with sensitivity ratio, PPA and NPA as shown in Table 4 and Table 5.
Objective Varaible: Cytology
In Cytology examination test, the RF before SMOTE was achieved with total accuracy of 94.58% with 35 patient records and 823 non-patient records. After using SMOTE algorithm RF achieved a total accuracy of 95.02% with number of patients 805 and non-patients 823. SMOTE algorithm increased the accuracy ratio with sensitivity ratio, PPA and NPA as shown in Table 6 and Table 7.
Objective Varaible: Biopsy
In Biopsy examination test, the RF before SMOTE was achieved with total accuracy of 93.48% with 35 patient records and 823 non-patient records. After using SMOTE algorithm RF achieved a total accuracy of 94.02% with number of patients 805 and non-patients 823. SMOTE algorithm increased the accuracy ratio with sensitivity ratio, PPA and NPA as shown in Table 8 and Table 9.
Analysis and Comparison
The results has proved the practice of Random Forest technique to categorize the biased dataset to get a better accuracy ratio in classifying cervical cancer data has been graphically represented using Figs. 5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32.
A comparative table using SVM and SMOTE has been tabulated by using values given in reference 16. Accuracy, sensitivity, specificity, PPA and NPA are the features calculated for 805 patients among 835 non-patients and given using Tables 10, 11, 12 and 13.
Ontological Representation
Knowledge representation is ontology. Knowledge is in the form of vocabulary of concepts which are explicitlydefined with relationships amongst the concepts. Ontologies is also a structured view of the domain with rich semanticmeaning. Since the size and diversityof datasets semantically represened is growing dramatically,the computational load have been increased significantly.
A knowledge based graph on ontologytake an advantage of exihibiting relevant information visually which helps us to effectively and efficiently analyze the crucial need to find computationload without losing any data. The aforementionedrequirements and explanations stimulate us to educate how to recognizeand proceed these inherent semantic structures and hierarchies todetermine new perceptions and elevate prevailing services.
Figure 33 represents the ontographical representation of RSOnto which depict the relation amongst various classes. Figure 34 illustrates the classes and sub-classes in RSOnto ontology. Figure 35 and 36 illustrates the comparitive study with SVM and SMOTE. This ontology graphically represents the comparitive study of Hinselmann, Schiller, Biopsy and Cytology tests before and after SMOTE. The study relates the tests using the objective variable.
RSOnto depicts the accuracy, sensitivity, specificity, PPA and NPA for Hinselmann, Schiller, Biopsy and Cytology comparitively before and after SMOTE proving the efficiency of SMOTE. Figure 35 and Fig. 36 represents the graphical representation of tests before and after SMOTE. This framework is based on RDF/OWL which captures the dependencies amongst low level domain and complex activities. This defines the tests to capture the knowledge for detecting complex activities. This ontology-based semantic fusion aids as a baseline to recognize events in a universal view where complete multimodal is recorded.
Limitations
SMOTE is used only for 2 dimensional data here. When moving to higher dimensions smote is not very effective, since it does not consider adjacent nodes which results in overlapping, resulting in inaccuracy. In further study a higher version of SMOTE can be implemented for higher dimensions.
Conclusion and Future Work
The services and systems, provided for cervical cancer requires accurate and reliable considerations for the degree of expectation. Measuring the evaluation metrics of features is not much easier, since they remain with various uncertainties. It is a difficult and ambiguous task. In order to balance the imbalanced data set SMOTE is applied which is visualized using RSOnto ontology which increases the quality of metrics.
We presented the brief evaluation of metrics which in future work can be proved more efficient and accurate with several algorithms and various case studies.
References
Cancer Facts & Figures, American Cancer Society, Atlanta, GA, USA, 2018.
Saha, A., Chaudhury, A. N., Bhowmik, P., and Chatterjee, R., Awareness of cervical cancer among female students of premier colleges in Kolkata, India. Asian Paci c J. Cancer Prevention 11(4):1085 1090, 2010.
El-Moselhy, E. A., Borg, H. M., and Atlam, S. A., Cervical cancer: Sociode-mographic and clinical risk factors among adult Egyptian females. J. Oncol. Res. Treat. 1(1):7, 2016.
Siegel, R. L., Miller, K. D., and Jemal, A., Cancer statistics, 2018. CA,Cancer J. Clin. 68(1):7 30, Jan. 2018.
Vimal, S., Kalaivani, L., and Kaliappan, M., Collaborative approach on mitigating spectrum sensing data hijack attack and dynamic spectrum allocation based on CASG modeling in wireless cognitive radio networks. Cluster Computing, 2017. https://doi.org/10.1007/s10586-017-1092-0.
Mariappan. E, Kaliappan. M, Vimal S, “Energy Efficient Routing protocol using Grover’s searching algorithm using MANET”, Asian Journal of Information Technology, Vol: 15, no.24, 2016.
Kaliappan, M., and Paramasivan, B., Enhancing secure routing in Mobile Ad Hoc Networks using a Dynamic Bayesian Signalling Game model. Journal of Computers & Electrical Engineering 41:301–313, 2015.
B. Paramasivan, M.J VijuPrakash, M. Kaliappan, 2015 Development of a Secure Routing Protocol usingGame Theory Model in Mobile Ad Hoc Networks, Journal of Communications and Networks, Vol. 17, No. 1
Kaliappan, M., Augustine, S., and Paramasivan, B., Enhancing energy efficiency and load balancing in mobile ad hoc network using dynamic genetic algorithms. Journal of Network and Computer Applications 73:35–43, 2016.
SudhakarIlango, S., Vimal, S., Kaliappan, M., and Subbulakshmi, P., Optimization using Artificial Bee Colony based clustering approach for big data. Cluster Computing. https://doi.org/10.1007/s10586-017-1571-3.
Tseng, C.-J., Lu, C.-J., Chang, C.-C., and Chen, G.-D., Application of machine learning to predict the recurrence-proneness for cervical cancer. Neural Comput. Appl. 24(6):1311 1316, May 2014.
Hu, B. et al., A risk evaluation model of cervical cancer based on etiol-ogy and human leukocyte antigen allele susceptibility. Int. J. InfectionDiseases 28:8 12, 2014.
Sharma, S., Cervical cancer stage prediction using decision tree approach of machine learning. Int. J. Adv. Res. Comput. Commun. Eng. 5(4):345 348, 2016.
Sobar, S., Machmud, R., and Wijaya, A., Behavior determinant based cervical cancer early detection with machine learning algorithm, in Proc.4th Int. Conf. Internet Services Technol. Inf. Eng., vol. 4, pp. 3120 3123, Jun. 2016.
Kannan, N., Sivasubramanian, S., Kaliappan, M., Vimal, S., and Suresh, A., Predictive big data analytic on demonetization data using support vector machine. Cluster Comput, 2018. https://doi.org/10.1007/s10586-018-2384-8 March 2018.
Wu, W., and Zhou, H., Data-driven diagnosis of cervical cancer with support vector machine-based approaches. IEEE Access 5:25189 25195, 2017.
Lin, W.-Z., Fang, J.-A., Xiao, X., and Chou, K.-C., iDNA-Prot: Identica-tion of DNA binding proteins using random forest with grey model. PLoSONE 6(9):e24756, Sep. 2011.
Seera, M., and Lim, C. P., A hybrid intelligent system for medical data clas-sication. Expert Syst. Appl. 41(5):2239 2249, Apr. 2014.
Breiman, L., Random forests. Mach. Learn. 45(1):5–32, 2001.
Biau, G., Analysis of a random forests model, J. Mach. Learn. Res., vol. 13, pp. 1063 1095, Apr. 2012.
Breiman, L., Friedman, J. H., Olshen, R., and Stone, C. J., ClassicationandRegression Trees. Belmont, CA, USA: Wadsworth, 1984.
Genuer, R., Poggi, J.-M., and Tuleau, C., Random forests: Some method-ological insights, INRIA, Saclay, France, Res. Rep. RR-6729, Nov. 2008.
Liaw, A., and Wiener, M., Classication and regression by random forest. R Newslett 2(3):18 22, 2002.
Suresh, A., Udendhran, R., Balamurgan, M. et al., J Med Syst 43(165), 2019. https://doi.org/10.1007/s10916-019-1302-9.
Suresh, A., Udendhran, R., and Balamurgan, M., Soft Comput, 2019. https://doi.org/10.1007/s00500-019-04066-4.
Kotu, V., and Deshpande, B., Predictive Analytics and Data Mining. San Mateo, CA, USA: Morgan Kaufmann, 2015, 63 163.
Kavitha, R. and Kannan, E., An efcient framework for heart disease clas-sication using feature extraction and feature selection technique in data mining, in Proc. Int. Conf. Emerg. Trends Eng., Technol. Sci. (ICETETS), Pudukkottai, India, pp. 1 5 2016.
Zhang, C., Li, Y., Yu, Z., and Tian, F., Feature selection of power system transient stability assessment based on random forest and recursive fea-ture elimination, in Proc. IEEE PES Asia Paci c Power Energy Eng.Conf. (APPEEC), Xi’an, China, pp. 1264 1268, 2016.
Guyon, I., Weston, J., Barnhill, S., and Vapnik, V., Gene selection for cancer classication using support vector machines, Mach. Learn., vol. 46, nos. 1 3, pp. 389 422, 2002.
Díaz-Uriarte, R., and de AndrØs, S. A., Gene selection and classication of microarray data using random forest. BMC Bioinf. 7(1):3, Jan. 2006.
Chawla, N. V., Bowyer, K. W., Hall, L. O., and Kegelmeyer, W. P., SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 16(1):321 357, 2002.
Cieslak, D. A., Chawla, N. V., and Striegel, A., Combating imbalance in network intrusion datasets, in Proc. IEEE Int. Conf. Granular Comput., pp. 732 737, 2006.
Fallahi, A., and Jafari, S., An expert system for detection of breast cancer using data preprocessing and Bayesian network. Int. J. Adv. Sci. Technol. 34(9):65 70, 2011.
Liu, Y., Chawla, N. V., Harper, M. P., Shriberg, E., and Stolcke, A., A study in machine learning from imbalanced data for sentence boundary detection in speech. Comput. Speech Lang. 20:468 494, Oct. 2006.
Chase, D. M., Kalouyan, M., and DiSaia, P. J., Colposcopy to evaluate abnormal cervical cytology in 2008. Am. J. Obstet. Gynecol. 200(5):472–480, May 2009. https://doi.org/10.1016/j.ajog.2008.12.025.PMID19375565.
Schiller's test at Who Named It?
Vimal, S., Kalaivani, L., Kaliappan, M., Suresh, A., Gao, X.-Z., and Varatharajan, R., Development of secured data transmission using machine learning based discrete time partial observed markov model and energy optimization in Cognitive radio networks. Neural Comput&Applic, 2018. https://doi.org/10.1007/s00521-018-3788-3.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Patient Facing Systems
Rights and permissions
About this article
Cite this article
Geetha, R., Sivasubramanian, S., Kaliappan, M. et al. Cervical Cancer Identification with Synthetic Minority Oversampling Technique and PCA Analysis using Random Forest Classifier. J Med Syst 43, 286 (2019). https://doi.org/10.1007/s10916-019-1402-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10916-019-1402-6