Abstract
Worldwide, cervical cancer is the leading cause of death among women from cancer. The symptoms of this gynecological disease are difficult to recognize at early stage, especially in those countries that don’t have facility of screening programs. In diagnosis of cervical cancer, machine learning methods can be used to detect the malignous cancer cells at initial stage. The foremost apprehension in disease diagnosis involves data imbalance issue and non-uniform scaling in dataset. In this article, a prevalent oversampling approach Synthetic Minority Oversampling Technique along with fivefold cross-validation is being used on unscaled and scaled data to handle these issues. A promising comparison is been made among the performance of most prevalent machine learning (ML) classifiers such as Naive Bayes, Logistic Regression, K-Nearest Neighbor, Support Vector Machine (SVM), Linear Discriminant analysis, Multi-Layer Perceptron, Decision Tree (DT) and Random Forest (RF) on unscaled data and scaled data obtained by Min–Max scaling, Standard scaling and Normalization. RF, SVM and DT are the top three ML algorithms obtained in cervical cancer diagnosis for which optimization possibilities are explored with feature selection methods as Univariate feature selection and Recursive feature elimination (RFE). Overall performance of Random Forest predictor with RFE (RF-RFE) is superior to all others being implemented.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Among the severe medical emergencies, cancer is the most lethal disease induced by tumor cells. Obsolete development of cells as tumors, is still being a major challenge ahead of technological world today. Cancer treatments of tumor cells involve chemotherapy and endoscopy also has high risks of destroying the healthy tissue cells. Cervical cancer is found as fourth most prominent category of death from cancer in between women. By World Health Organization (WHO), there were 604,127 new cases of cervical cancer found in 2020 with mortality of 341,831 patients, 6.5% of all cancer infected females. In 2020, more than 83% of deaths of cervical cancer infected patients occurred in the low and middle income nations [1]. Here alone in India, in 2020, 18.3% cases of cervix cancer observed in all women cancer patients that are 9.4% of all cancer infected patients. It has found third place in all cancer cases in India, total 123,907 new registered patients along with 77,348 deaths in 2020 [2]. Each type of cancer is categorized into malignant and benign cancer. Due to the lack of, early diagnosis, effective screening and treatment programs, causes it to be as one of the most malignant type of cancer. Cervical cancerous cells are developed in the cervix of female’s uterus and early stage symptoms are abnormal bleeding in vagina, increase in vagina discharge, menopause bleeding after going through menopause, pain during sex, pelvic pain, etc.
Cervical cancer is found in females infected with the Human Papilloma virus (HPV) that causes cervical tissue to be change abnormally. Sexual relations with different partners, early age sexual relationship, long term usage of oral contraceptives; smoking, etc. lead to increase risk of cervical cancer [1, 3]. Most popularly, Pap test and HPV DNA test are recommended for screening of cervical cancer. Pap test (a.k.a. Pap smear test) is a cytology-based screening test in which a sample of cells is taken from cervix of a female and then tests for abnormality in cervix cells with cancer cells and also the cells that causes increase in chances of cervical cancer. HPV DNA test detects any type of HPV within the cells taken from the cervix that are responsible to lead cervical cancer. Pap smear and HPV tests both can be examine at similar time using the similar swab or by second swab. On suspicion of cervical cancer, patients have to go through the detailed diagnosis tests such as biopsy [4]. Presently, along with the conventional medical approaches, computer vision algorithms i.e. machine learning in cyber-physical system, interestingly playing a vital role in various medical applications such as diagnosis of diseases. Here in this paper, we applied some of the most popular machine learning (ML) approaches like NB, LR, KNN, SVM, LDA, MLP, DT and RF on cervical cancer data with some preprocessing methods. Analyzing with all the risk factors in disease diagnosis degrades the efficiency of classification model and also increases its computational complexity. So selection of relevant features also plays a vital role while analyzing the performance of a classification model. This article also realizes some of the popular feature selection methods for getting optimized performance in classification of disease.
2 Background of ML Algorithms Used
2.1 Naive Bayes (NB)
NB is another supervised classification model based on a conditional probabilistic approach utilizing the Bayes theorem to detect all the substances within the information set. This classification method is often suited for high dimensionality datasets [5,6,7]. This approach classifies the problem based on joint posterior probability distribution:
Here, \(p(C/X)\), p(C) and \(p\left( X \right)\) gives the posterior probability, prior probability of class and probability of attributes respectively and X is the vector space of n attributes. Due to statistical independence among features, these classifiers are highly scalable and can utilize limited training data with high dimensional features. In [6], Weighted Principle Component Analysis (WPCA) is used along with NB classifier to achieve improved performance in pap smear cervix cell image classification for Herlev dataset. References [8,9,10,11] compare NB classifier prognosis performance for cervical data with other ML classification models.
2.2 Logistic Regression (LR)
It is a statistical binary classification supervised learning method that fits linear regression algorithm to classify data in terms of discrete binary outputs based on logistic function. It intuits maximum-likelihood estimation by a search procedure to minimize the probability error in predicted model and optimize best coefficients value for the data so that the threshold value for classification can be easily adjusted [12]. The essence of the algorithm involves the minimization of cost function:
Here, \(h_{\theta } \left( {\text{x}} \right)\) denotes the hypothesis for logistic function, \(\log h_{\theta } \left( {{\text{x}}^{{\left( {\text{i}} \right)}} } \right)\) and \({\text{log}}(1 - { }h_{\theta } \left( {{\text{x}}^{{\left( {\text{i}} \right)}} } \right)\) gives the cost function when class y is ‘1’ and ‘0’ respectively for m training examples. Reference [13] proposed a LR classifier with fuzzy inference model utilizing combined grayscale-texture based features for Cervical Intraepithelial Neoplasia (CIN) image classification. Many researches [8, 10, 11, 14] utilized LR as one of the classifier to perform comparative analysis in cervical cancer classification. Reference [15] utilized logistic regression for probability estimation of knowledge, attitude, and perception (KAP) of Human Papillomavirus (HPV) infection and cervical cancer.
2.3 Linear Discriminant Analysis (LDA)
LDA is popularly known as dimensionality reduction approach however it proved an effective algorithm to classify objects in two or more groups or clusters based upon the features measured describing that objects. LDA is alternative of LR when there are more than two classes to be classified as LR has limited to binary classification problems. It comprises statistical characteristics of data determined for each of the class that is used to predict decision based on Bayes theorem [16]. Its objective lies in predicting the class, for an input x with the largest:
Here, \(\pi_{k} = p\left( {y = k} \right)\) exactly, \(\mu_{k}\) and \(\hat{\varepsilon }\) denotes mean and the covariance matrix for class ‘k’. Reference [17] implemented fuzzy-entropy based prime feature discrimination from segmented cell nuclei for Herlev dataset. These segmented features are used with LDA as one of the classification model for abnormal cell detection.
2.4 K-Nearest Neighbor (KNN)
KNN is a non-parametric classification technique that uses feature similarity approach by searching the very similar data points among the available data to categorize them into a class. The KNN finds nearest K data points by determining the Euclidean distance (Other distance measures includes Manhattan, Minkowski and Hamming distance) to the given query point and identifies its class by determining the mainly repeated class label. The value of K is chosen by parameter tuning, providing best suited prediction for the given data [18]. An input x is considered to belong from the class that evident largest probability among all:
Here, I(x) denotes the indicating function that is ‘1’ for argument x is true and 0 otherwise, A is the set of K nearest points of input x. References [9,10,11, 17] utilized KNN classifier for comparative analysis with other classification models for cervical cancer classification.
2.5 Multilayer Perceptron (MLP)
MLP is a neural network based proficient and robust method that is used for finding solution of nonlinear and complex classification problems. It comprises multiple neurons arranged in form of input layer, hidden layers and output layer. Here some the nodes i.e. neurons uses non-linear activation functions so that it can also find solution for the problem that are not linearly separable. Here the most complex task is to determine the hidden layer size [19]. The optimization objective of MLP model is based on minimization:
Here, F denotes transfer function, X is input to the model, W indicates weight matrix, d is desired response, Y denotes computed output vector, N is total count of hidden layers and nN+1 indicates total output layer neurons. As a consequence incorrect estimation of same may results in approximation error, generalization error and overfitting. In [14], MLP classifier is used for performance comparison with other classifiers for two-class classification on risk-factor cancer data using RFE and RF based ensemble method for feature selection.
2.6 Decision Tree (DT)
DT is a supervised learning tree-like structure in which every single node in DT signifies an attribute value within an instance that is to detect for a class and each branch provides a value that assumed by a node [7, 20]. Selection of best split among training samples based on the measures in terms of class distribution:
Here, c is total number of targets and \(p{(}i{|}t)\) is the predicted sample belonging to class i at a specific node t. Conventional DT [8, 10, 21] algorithms as well as types of decision tress like ID3, C4.5, C5.0, CHAID, and CART [9], and J48 [10, 11] standalone and with ensemble approach performed efficiently in cervical cancer detection.
2.7 Support Vector Machine (SVM)
Vapnik introduced the SVM approach to deal with classification as well as regression models. SVM is supervised discriminative linear approach to accomplish binary classification through an explicit hyper plane [7]. Optimization in SVM is based on the minimization of equation:
Here, C is the penalty factor for error, \(z^{\prime}\left( {\theta^{T} x^{\left( i \right)} } \right)\) and \(z^{\prime\prime}\left( {\theta^{T} x^{\left( i \right)} } \right)\) denotes the cost function when class y is equals to ‘1’ and ‘0’ respectively and m indicates number of samples. In [22], SVM, support vector machine-recursive feature elimination (SVM-RFE) and support vector machine-principal component analysis (SVM-PCA) methods were used for cervical cancer detection with 90–94% accuracy for the risk-factor cervical cancer data. At early stage, SVM application was constrained to two-class classification, but afterward, kernel capacities for SVM presented that are valuable in multiclass classification [17, 23, 24]. Reference [17] implemented SVM with linear kernel (SVM-linear) and radial basis function kernel (SVM-RBF) using fuzzy entropy based feature extraction mechanism for abnormal cells detection in pap-smear images. References [8,9,10,11, 14, 25] performed comparative analysis of SVM with other prediction models for cervical cancer prognosis.
2.8 Random Forest (RF)
Random forest by Breiman (2001) is based on ensemble method that is used for both classification and regression problems. Ensemble methods used to group weak learners to shape strong learner and use multiple learning approach to produce enhanced predictive result. It trains multiple numbers of DTs that returns with the class find in majority within the ensemble of overall DTs [26]. In RF, each DT predicts a class for the classification model and among all the predictions most voted class becomes our RF model prediction. Bagging approach in RF involves prediction for sample x′ by taking average of all predicted values obtained from individual DT’s:
Here, N is a parameter gives samples/tree. Generally RF algorithms perform slightly better than SVMs in many classification problems [27]. RF classifier [8, 10, 11, 14, 21, 25] performs efficiently for risk-factor cancer data as well as for pap smear cervix images in cervical cancer detection.
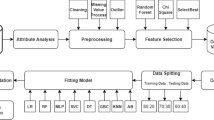
3 Methodology
3.1 Data Description
The cancer patients’ data used here for diagnosis is available at UCI, collected at 'Hospital Universitario de Caracas' in Caracas, Venezuela [28]. The dataset include 858 instances with 36 risk factors including 32 attributes and 4 target categories—Hinselmann, Schiller, Cytology and Biopsy. The description of attributes in the given Cervical Cancer data is shown in Table 1. Hinselmann test uses colposcopy using acetic acid, while colposcopy using Lugol iodine includes Schiller test, Cytology and Biopsy. Malignant infected target described as ‘1’ and benign as ‘0’. In whole of the dataset, around 90–96% data belongs to benign class in each of four target variables. All the attributes values given are either boolean, integer or float type. To build an efficient learning model the data to be fed to it, should be proper and complete. As some of the samples within dataset have missing values, also each attribute have different scaling ranges thus it required to preprocess the data before feeding to a learning algorithm.
3.2 Preprocessing of Data
In preprocessing, we eliminate the Instances and attributes with missing values. After elimination, features 27 and 28 get removed along with samples having at least single missing value and only 668 samples with 30 features left. Variation in scaling range of features may cause a particular feature might take over rest of the features while analyzing the performance in a dataset. If magnitude of attribute’s variance has much high order than the other ones, it may take over the objective function and build an estimator that may incapable of learning from rest of the attributes appropriately as being expected. In the given cervical dataset, attribute ‘age’ has high value of mean, variance and standard deviation as 27.265, 76.168 and 8.727 respectively, while some of the attributes as ‘STDs: cervical condylomatosis’ and ‘STDs: AIDS’ even have zero value of mean, variance and standard deviation. To restrain the weighing effect of the attributes with higher statistical mean and to obtain more numerically stable and improved optimization, all attributes should bring at the same scaling level by using different feature scaling approaches. Scaling methods are also quite helpful in speeding up the rate of computations within an algorithm. Here in this article, ML algorithms are analyzed on unscaled as well as with scaled data. For scaling, we use Min–Max Scaler, Standard Scaler and Normalization on available cancer data [29, 30]. Standard Scaler or Z-score Normalization scales the dataset in standard normalize distribution having unit variance with zero mean. If µand σ indicates mean and standard deviation respectively then standardization aims ~ N (µ, σ2) → ~ N (0, 1) i.e. Z ~ N (0, 1), N stands for normal distribution. Standard score or Z-score for an instance referred by:
Standard scaling is preferred where contribution of distance measures equally requires for all the attributes. It proves much valuable if distribution of attributes is nearly normalized or Gaussian. Min–Max Scaler and Normalization are an alternative to Standard Scaler if the attribute’s distribution is not of Gaussian nature and the attributes may lie within a restricted space. Both Min–Max Scaler and Normalization scales the data between 0 and 1, along with the difference that distribution characteristic is bounded and unit norm respectively for the two. Min–Max scaling conserves the outline of original distribution and finishes up with smaller value of σ that can suppress the consequence of outliers. Normalization scales each instance (row) instead of attributes (column), using Euclidian distance (l2 normalization) or Manhattan distance (l1 normalization).
Here \(x\) is the instance in a feature set (column) and in a sample (row) for Min–Max scaling and Normalization respectively, x’ is normalized value, xmin, \(x_{max}\) and \(x_{mean}\) indicate minimum, maximum and mean value in a given set.
Among all the samples, only around 4–10% found of malignant category in all four target variables in given dataset. Its causes a worst consequence in computation of performance metrics in data analysis and may cause the prediction to bias towards the majority class, when the number of one’s class is much more than the other class, i.e. in case of class imbalanced data [31]. Generally, the data for patients with positive results in the disease diagnosis is quite less than the negative group. The imbalance ratio for each of the target variable in the given dataset is shown in the Table 2. This imbalancing may consequence in prediction of high accuracy for the model even if the minority class being wrongly predicted, due to weightage of majority class in the dataset. To achieve balance distribution among the classes, oversampling and undersampling are the two ways but as the dataset is not being so large; oversampling approach is better than the other one. However, traditional oversampling approach is based on randomly replicating the instances that may cause overfitting hence a hybrid approach, synthetic minority oversampling technique (SMOTE) is used as a preprocessing method [32]. SMOTE is aimed on creating the new minority ‘synthetic’ instances by linear interpolation rather than duplicating them. A new minority instance is generated by SMOTE involves:
Here, \(x\) is one of the minority instances in set of minority class A, for each \(x \in A\), \(rand\left( {0, 1} \right)\) implies any random number between 0 and 1 and \(\left| {x - x_{k} } \right|\) gives the Euclidean distance among the instance x and its kth nearest neighbor (for k = 1,2,…N, where N is sampling rate set in proportion of imbalance).
3.3 Implementation of ML Algorithms
On preprocessed data, we apply the aforementioned ML algorithms with fivefold cross-validation to find performance metrics. Cross-validation is a performance analysis method by reserving a set of samples for testing purpose and train the model by remaining data. Repeatedly perform this for every set of samples in the dataset and compute the performance metrics for each trained model. Finally resultant performance metric is computed by taking mean of metrics for each trained model. K-fold cross-validation splits whole of the dataset in K-subsets, where one fold is used as testing set and others as training set in each of the iteration, and repeated until each of the set in K-fold is used as testing set. Stratified K-folds cross-validation is used in this article available on Scikit-learn library for Python programming [33], for splitting the data into 5-folds (Fig. 1).
Apart from fivefold cross-validation, parameter selection method is being used to determine best parameters for each of the ML algorithm with given data. For each of the algorithm we used a range of values for each parameter and then identify the suitable combination of parameters. GridSearchCV function is available in Scikit-learn that compute finest parameters for each of the ML algorithm. Parameter grid is used while using GridSearchCV that contain all possible parameters from which best are selected. The parameter grids used for simulation of ML algorithms are given in Table 3.
When we come to performance, accuracy is not only the criterion to determine best model. As most of the data belongs to benign category, overall accuracy has more weight for the benign cases. It specifies that overall accuracy can be very good even if the accuracy of malignant category is low. However, for correct diagnosis of disease, the prediction of malignant samples should also be accurate. Hence Precision, Recall, F-score, ROC-curve and AUC are used here for performance analysis to diagnose correctly [34, 35].
Here TP, TN, FP and FN refer to True Positive, True negative, False Positive and False Negative values. True Positive value indicates whose malignant type of cervical cancer detected correctly while True Negative value gives uninfected patient predicted correctly. False Positive value provides uninfected samples find with positive results while False Negative value gives cervical cancer infected patient whose result is found negative. Accuracy provides the proportion of correctly diagnose samples among all. Precision a.k.a. Positive Predictive Value (PPV) gives the ratio of actually infected persons to all positive detected samples value. Recall a.k.a. Sensitivity provides the fraction of correctly detected cancer infected patients to all the samples that are actually infected with cervical cancer. Recall is also called as True Positive Rate (TPR). F-score is the harmonic mean of precision and recall that has best prediction with value 1. Other constraints ROC-curve and AUC are related to each other in the context that ROC-curve is plotted among True Positive Rate (TPR) and False Positive Rate (FPR). AUC score gives area under ROC-curve. FPR is the ratio of positive detected cancer uninfected samples to the total uninfected samples. AUC is as close to 1 indicates a model with much better performance and lie between 0 (worst) and 1(best).
3.4 Feature Selection Methodology
Computation cost of a learning model is directly proportional to dimensionality of attributes. Further performance improvement can be done by selecting the relevant risk factors that has more contribution in evaluation of classification model. Analyzing a learning methodology with irrelevant attributes may cause overfitting and increases computational complexity of model. Thus to create an effective classification model, redundant features should be eliminated from the dataset. Selection of certainly important features can be able to train an accurate model with enhanced performance. Filter method, Wrapper method and Embedded method are the three categories of methods for attribute selection [36, 37]. Among many of the available methods, two popular feature selection metholodologies i.e. Univariate feature selection and Recursive feature elimination has been used here.
3.4.1 Univariate Feature Selection
Univariate feature selection is a type of Filter method based on examining the strongest correlation between attributes and target variables for each risk factor independently. This method works on various statistical tests that select the attributes having more importance and distinct information. Scikit-Learn provide SelectKBest(), SelectPercentile() and GenericUnivariateSelect() as the transforming object for univariate feature selection method. SelectKBest() retain K-maximum scoring attributes while eliminates all others, SelectPercentile() eliminates others except uppermost user-specified proportion scoring attributes and GenericUnivariateSelect() uses configurable tactic to carry out selection of attributes. For classification the univariate feature selection has a popular chi-square test as a statistical tool used with SelectKBest() univariate approach that is being used in this paper for selection of features. Chi-square test is used for discrete set of values and tests the independency among two samples. Chi-square is based on the intuition that a risk-factor is uninformative for classification if it is independent to the class variable. SelectKBest() with Chi-square involves selection of attributes with having K-highest chi-square scores that is computed between attributes and target categories.
Here \(O_{i}\) and \(E_{i}\) are observed and expected values for class i among total n set of features. This approach aims selection of attributes that are highly dependent on the categorical data.
3.4.2 Recursive Feature Elimination (RFE)
RFE is a Wrapper method that involves recursive elimination approach for ruling out the important risk-factors in disease diagnosis. RFE is a kind of backward selection algorithm with the difference that it selects features based on ranking of attributes while backward selection eliminates them on the grounds of p-value score [38]. Dealing with classification problem RFE fits a learning model and retain the specified number of attributes that has highest importance or eliminate the weakest ones. An estimator is made fit on initial set of attributes for recursive selection of appropriate features by removing a few of the features in every loop based on ranking using the attributes ‘coef_’ or ‘feature_importances_’. RFE has the option to select specific number of features or select strongest features by default. Scikit-learn have RFE for recursive feature elimination and RFECV for finding optimized number of attributes using cross-validation approach. RFECV is useful in searching out finest set of attributes ranked based on validation score using K-fold cross-validation.
4 Experimental Analysis
The cancer patient data has four target variables i.e. Hinselmann, Schiller, Cytology and Biopsy with 30, 63, 39 and 45 malignant infected samples respectively out of overall 668 samples. Here, the performance metrics for the ML algorithms NB, LR, KNN, SVM, LDA, MLP, DT and RF are computed with fivefold cross-validation and parameter selection for unscaled and scaled data. Min–Max scaling, Standard scaling and Normalization are the three methods used with oversampled data for getting scaled data of three kinds. A tabular comparison is made between evaluation parameters in terms of accuracy, precision, recall, F-score and AUC listed in Tables 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18 and 19 along with the comparison of ROC curves shown in Figs. 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16 and 17 for all target variables i.e. Hinselmann, Schiller, Cytology and Biopsy. The abbreviations used are as follows:
C, Classifiers; M, Performance metrics; A, Accuracy (%); P, Precision (%); R, Recall (%); F, F score; Ac–AUC.
Evaluation involves that the top three performing ML algorithms for all four targets are RF, SVM and DT, of which RF performance is superior in terms of the aforementioned evaluation metrics. The analysis reveals that RF gives maximum accuracy with Standard scaled data as 97.81%, 93.97%, 95.6% and 96.18% for target variables Hinselmann, Schiller, Cytology and Biopsy respectively. For SVM in standard scaled data, accuracy for all four targets is 93.63%, 90.64%, 91.52% and 92.69% respectively. DT gives accuracy of 93.54%, 88.6%, 90.67% and 90.97% that is highest in Standard scaled observation for the four targets respectively. Other evaluation parameters are also quite good for RF, SVM and DT observed from the tabular data. ROC curve gives visible comparison for all the ML algorithms that gives RF with standard scaled data has maximum AUC scores of 0.99, 0.98, 0.98 and 0.99 for four target variables respectively. Performance of NB classifier is worst among all eight predictors.
Observation shows that ML algorithms performance is finest when data is standard scaled in most of the cases, however unscaled data also provide high-quality result with a little bit difference in performance metrics. Min–Max Scaler also performs nearly Standard Scaler with most of the algorithms. Performance of normalization is worst among all. Concerning computation time,Footnote 1 evaluation for unscaled data has poor computational efficiency rather than scaled data as shown in Fig. 18. In terms of computational cost and performance RF, SVM and DT with standard scaled data are the finest algorithms for cancer diagnosis data when all the risk-factors are involved in computation. Further computation efficiency can be enhanced by eliminating less important features using Univariate feature selection and RFE algorithm.
4.1 Feature Selection Using SelectKBoost
SelectKBoost is a univariate feature selection approach that selects K-risk factors having highest correlation with the target variables. Chi-square statistical test is being used here with SelectKBoost algorithm to determine feature importance as shown in Fig. 19. Top ten attributes obtained by SelectKBoost algorithm for four target categories of disease dataset are shown in Table 20. It is being observed that attributes 6, 13, 14, 29 and 31 are common in all target variables. Table 1 show that Attribute 6 corresponds to smokes in year, Attribute 13 is number of STD diseases, Attribute 14 is STDs related to Condylomatosis, Attribute 29 is radiography test for Cancer disease and attribute 31 is radiography test for HPV disease. Except these attributes 26 and 32 are common in three target variables. To obtain optimized performance top 16 relevant risk-factors are selected using SelectKBoost. The performance analysis is done with these 16 risk factors for previously obtained top three ML algorithms i.e. RF, SVM and DT with standard scaled data using fivefold cross-validation and parameter grid for classifiers as listed in Table 3. Removal of almost half risk factors from the dataset doesn’t affect much on the evaluation metrics. Table 21 shows the implementation results that conclude RF, SVM and DT performance with 16-risk factors is approximately same as that obtained with complete set of attributes.
4.2 Feature Selection Using RFE
RFE is being implemented here with fivefold cross validation for the three ML algorithms i.e. DT, SVM and RF with selection of 16 risk factors. SelectKBoost retains important features based on scores of Chi-square test and then performance is analyzed for ML methods, while RFE is a recursive sequence selective approach for optimal risk factors selection using ML classifier. Top 10 attributes among all 30 risk factors for DT-RFE, SVM-RFE and RF-RFE is shown in Tables 22, 23 and 24.
Table 22 shows DT-RFE gives risk factors 9, 13, 31 and 32 are common for all target variables and attributes 7, 17 and 29 are found in at least three target variables among top 10 attributes. Attributes 3, 7, 9 and 13 are involves in each column of Table 23 among most important 10 risk factors for SVM-RFE. Table 24 for RF-RFE has attribute 6, 7, 9, 13 and 31as found similar in all columns. The implementation results of DT-RFE, SVM-RFE and RF-RFE are shown as Table 25 in terms of performance metrics. An optimized performance achieved with recursive feature elimination (RFE) with reduced 16-risk factors compared to analysis with complete set of 30 attributes. Random Forest (RF) again proves the best classifier ML algorithm in diagnosis of given cervix data. The predictor accuracy is 93.72%, 95.05% and 99.21% for DT-RFE, SVM-RFE and RF-RFE respectively in Hinselmann test. For Schiller test, accuracy of 89.33%, 92.17% and 96.13% is achieved for the three classifiers respectively. For Cytology test, DT-RFE, SVM-RFE and RF-RFE provide accuracy as 91.7%, 92.89% and 97.01% respectively. Accuracy is 91.11%, 93.81% and 98.53% respectively for these ML predictors in Biopsy test. Tabular data shows highest precision of 98.5 for RF-RFE in Hinselmann test. Observation shows that RF-RFE gives highest recall score of 100% for Hinselmann and Biopsy test. Maximum F-score achieved is 0.99 again for RF-RFE in Hinselmann test prediction. RF-RFE also gives highest AUC score of 0.99 in three tests i.e. Hinselmann, Cytology and Biopsy. RF-RFE gives best results in terms of all performance metrics compared to other DT-RFE and SVM-RFE for four target variables.
5 Comparison Analysis
Analysis with complete cervix data shows best results are obtained in RF, SVM and DT classifiers when data is standard scaled. Experimental results show that optimized performance is achieved with elimination of risk factors that are more irrelevant. SelectKBoost and RFE significantly approached with the attributes that are more relevance in prediction. Most significant risk factors in both SelectKBoost and RFE are Attribute 6, 7, 9, 13, 29 and 31 that appear in most of the columns. These risk factors are shown in Table 26 which contributes more in prediction. To make a logical comparison 16 most relevant risk factors are chosen in both SelectKBoost and RFE feature selective approach.
A detailed comprehensive performance comparison is made between the three best ML predictors i.e. RF, SVM and DT in Table 27 with all 30 risk factors, 16 risk factors obtained with SelectKBoost and 16 risk factors obtained for RF-RFE, SVM-RFE and DT-RFE. All these results are obtained on standard scaled cervix data with SMOTE for oversampling, GridSearchCV for parameter selection and fivefold cross-validation for performance scores computation. Tabular data makes a clear comparison among the implemented RF, SVM and DT classifiers with different approaches used for selecting risk factors.
RF-RFE is given great results with accuracy as 99.21%, 96.13, 97.01% and 98.53% for four targets Hinselmann, Schiller, Cytology and Biopsy respectively. Other parameter for RF-RFE gives that precision is 98.5%, 95.59%, 96.27% and 98.07%, recall score is 100%, 98.4%, 98.41% and 100%, F-score is 0.99, 0.95, 0.97 and 0.97, and AUC score is 0.99, 0.98, 0.99 and 0.99 for the four target variables respectively. Figure 20 shows the comparison of accuracy for implemented ML classifiers. Performance metrics with 16 risk factors obtained from SelectKBoost approach are almost same to that obtained with complete set of features. However RFE approach significantly enhances the optimization in parameter metrics for RF, SVM and DT classifiers specifically in accuracy, precision and recall.
6 Conclusion
This paper analyzes the performance of some of the most prominent ML algorithms for cervical cancer data and observes the effect of scaling on performance metrics to efficiently predict the samples of malignant type. NB, LR, KNN, SVM, LDA, MLP, DT & RF are the ML classifiers which made prediction for all 30 risk factors. RF, DT and SVM classifiers are ranked as top three that makes best prediction for all four target category with Standard scaled data, however the performance is not so much get affected with unscaled data and Min–Max scaled data except in case of normalization. Furthermore, classification is made with these predictors using relevant features having more importance in data by feature selection algorithms. RF, SVM and DT classifiers using Univariate feature selection (SelectKBoost) and RFE make the predictors more efficient compared to the classifiers using with complete set of risk factors. There is significant reduction in computational cost and time when low information risk-factors are removed from the disease diagnosis data. RFE is proven better approach than SelectKBoost and the performance of RF-RFE algorithm with 16 risk attributes is superior to other algorithms.
Notes
System Configuration.
Window 10 OS.
Core i3, Integrated Graphics.
4 GB RAM, 1 TB Hard disk.
References
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., & Bray, F. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians, 71(3), 209–249. https://doi.org/10.3322/caac.21660
Globocan 2020: India Factsheet. (2021). The global cancer observatory. Updated March 2021. https://gco.iarc.fr/today/data/factsheets/populations/356-india-fact-sheets.pdf. Retrieved December 3, 2021.
Gadducci, A., Barsotti, C., Cosio, S., Domenici, L., & Riccardo, A. G. (2011). Smoking habit, immune suppression, oral contraceptive use, and hormone replacement therapy use and cervical carcinogenesis: A review of the literature. Gynecological Endocrinology, 27(8), 597–604. https://doi.org/10.3109/09513590.2011.558953
Cervical Cancer Prevention. (2021). PDQ screening and prevention editorial board. MD: National Cancer Institute (US). Updated 14 Oct 2021. https://www.ncbi.nlm.nih.gov/books/NBK65997. Retrieved December 3, 2021.
Kuncheva, L. I. (2006). On the optimality of Naïve Bayes with dependent binary features. Pattern Recognition Letters, 27(7), 830–837. https://doi.org/10.1016/j.patrec.2005.12.001
Dewi, Y. N., Riana, D., & Mantoro, T. (2017). Improving Naïve Bayes performance in single image pap smear using weighted principal component analysis (WPCA). In 2017 International conference on computing, engineering, and design (ICCED) (pp. 1–5). IEEE. https://doi.org/10.1109/CED.2017.8308130
Chauhan, N. K., & Singh, K. (2018). A review on conventional machine learning vs deep learning. In 2018 International conference on computing, power and communication technologies (GUCON) (pp. 347–352). IEEE. https://doi.org/10.1109/GUCON.2018.8675097
Ashraf, F. B., & Momo, N. S. (2019). Comparative analysis on prediction models with various data preprocessings in the prognosis of cervical cancer. In 2019 10th International conference on computing, communication and networking technologies (ICCCNT) (pp. 1–6). IEEE. https://doi.org/10.1109/ICCCNT45670.2019.8944850
Ahishakiye, E., Wario, R., Mwangi, W., & Taremwa, D. (2020). Prediction of cervical cancer basing on risk factors using ensemble learning. In 2020 IST-Africa conference (IST-Africa) (pp. 1–12). IEEE. ISSN 2576-8581.
Ilyas, Q. M., & Ahmad, M. (2021). An enhanced ensemble diagnosis of cervical cancer: A pursuit of machine intelligence towards sustainable health. IEEE Access, 9, 12374–12388. https://doi.org/10.1109/ACCESS.2021.3049165
Alpan, K. (2021). Performance evaluation of classification algorithms for early detection of behavior determinant based cervical cancer. In 2021 5th international symposium on multidisciplinary studies and innovative technologies (ISMSIT) (pp. 706–710). IEEE. https://doi.org/10.1109/ISMSIT52890.2021.9604718
Peng, C. Y., Lee, K. L., & Ingersoll, G. M. (2002). An introduction to logistic regression analysis and reporting. The Journal of Educational Research, 96(1), 3–14. https://doi.org/10.1080/00220670209598786
Liu, J., Peng, Y., & Zhang, Y. (2019). A fuzzy reasoning model for cervical intraepithelial neoplasia classification using temporal grayscale change and textures of cervical images during acetic acid tests. IEEE Access, 7, 13536–13545. https://doi.org/10.1109/ACCESS.2019.2893357
Ahmed, M., Kabir, M. M. J., Kabir, M., & Hasan, M. M. (2019). Identification of the risk factors of cervical cancer applying feature selection approaches. In 2019 3rd international conference on electrical, computer & telecommunication engineering (ICECTE) (pp. 201–204). IEEE. https://doi.org/10.1109/ICECTE48615.2019.9303554
Omone, O. M., Gbenimachor, A. U., Kovács, L., & Kozlovszky, M. (2021). Knowledge estimation with HPV and cervical cancer risk factors using logistic regression. In 2021 IEEE 15th international symposium on applied computational intelligence and informatics (SACI) (pp. 000381–000386). IEEE. https://doi.org/10.1109/SACI51354.2021.9465585
McLachlan, G. J. (1992). Discriminant analysis and statistical pattern recognition. New York: Wiley. https://doi.org/10.1002/0471725293
Saha, R., Bajger, M., & Lee, G. (2019). Prior guided segmentation and nuclei feature based abnormality detection in cervical cells. In 2019 IEEE 19th international conference on bioinformatics and bioengineering (BIBE) (pp. 742–746). IEEE. https://doi.org/10.1109/BIBE.2019.00139
Islam, M. J., Wu, Q. J., Ahmadi, M., & Sid-Ahmed, M. A. (2007). Investigating the performance of naive-Bayes classifiers and k-nearest neighbor classifiers. In 2007 International conference on convergence information technology (ICCIT 2007) (pp. 1541–1546). IEEE. https://doi.org/10.1109/ICCIT.2007.252
Kotsiantis, S. B. (2007). Supervised machine learning: A review of classification techniques. Informatica, 31(3), 249–268.
Rokach, L., & Maimon, O. (2005). Top-down induction of decision trees classifiers: A survey. IEEE Transactions on Systems, Man, and Cybernetics, Part C (Applications and Reviews), 35(4), 476–487. https://doi.org/10.1109/TSMCC.2004.843247
Akter, L., Islam, M. M., Al-Rakhami, M. S., & Haque, M. R. (2021). Prediction of cervical cancer from behavior risk using machine learning techniques. SN Computer Science, 2(3), 1–10. https://doi.org/10.1007/s42979-021-00551-6
Wu, W., & Zhou, H. (2017). Data-driven diagnosis of cervical cancer with support vector machine-based approaches. IEEE Access, 5, 25189–25195. https://doi.org/10.1109/ACCESS.2017.2763984
Shen, Y., Wu, C., Liu, C., Wu, Y., & Xiong, N. (2018). Oriented feature selection SVM applied to cancer prediction in precision medicine. IEEE Access, 6, 48510–48521. https://doi.org/10.1109/ACCESS.2018.2868098
Diez-Olivan, A., Pagan, J. A., Khoa, N. L. D., Sanz, R., & Sierra, B. (2018). Kernel-based support vector machines for automated health status assessment in monitoring sensor data. The International Journal of Advanced Manufacturing Technology, 95, 327–340. https://doi.org/10.1007/s00170-017-1204-2
Deng, X., Luo, Y., & Wang, C. (2018). Analysis of risk factors for cervical cancer based on machine learning methods. In 2018 5th IEEE international conference on cloud computing and intelligence systems (CCIS) (pp. 631–635). IEEE. https://doi.org/10.1109/CCIS.2018.8691126
Breiman, L. (2001). Random Forests. Machine Learning, 45, 5–32. https://doi.org/10.1023/A:1010933404324
Biau, G. (2012). Analysis of a random forests model. The Journal of Machine Learning Research, 13, 1063–1095.
Fernandes, K., Cardoso, J. S., & Fernandes, J. (2017). Transfer learning with partial observability applied to cervical cancer screening. In Iberian conference on pattern recognition and image analysis (pp. 243–250). Springer. https://doi.org/10.1007/978-3-319-58838-4_27
Patro, S. G., & Sahu, K. K. (2015). Normalization: A preprocessing stage. IARJSET, 2(3), 20–22. https://doi.org/10.17148/IARJSET.2015.2305
Han, J., Pei, J., & Kamber, M. (2012). Data mining: Concepts and techniques. Amsterdam: Elsevier. https://doi.org/10.1016/C2009-0-61819-5
Japkowicz, N., & Stephen, S. (2002). The class imbalance problem: A systematic study. Intelligent Data Analysis, 6(5), 429–449. https://doi.org/10.3233/IDA-2002-6504
Chawla, N. V., Bowyer, K. W., Hall, L. O., & Kegelmeyer, W. P. (2002). SMOTE: Synthetic minority over-sampling technique. Journal of Artificial Intelligence Research, 16(1), 321–357. https://doi.org/10.1613/jair.953
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V., Thirion, B., Grisel, O., et al. (2018). Scikit-learn: Machine learning in Python. Journal of Machine Learning Research, 12, 2825–2830.
Hajian-Tilaki, K. (2013). Receiver operating characteristic (ROC) curve analysis for medical diagnostic test evaluation. Caspian Journal of Internal Medicine, 4(2), 627–635.
Powers, D. M. (2020). Evaluation: from precision, recall and F-measure to ROC, informedness, markedness and correlation. arXiv preprint arXiv:2010.16061
Jović, A., Brkić, K., & Bogunović, N. (2015). A review of feature selection methods with applications. In 2015 38th international convention on information and communication technology, electronics and microelectronics (MIPRO) (pp. 1200–1205). IEEE. https://doi.org/10.1109/MIPRO.2015.7160458
Bagherzadeh-Khiabani, F., Ramezankhani, A., Azizi, F., Hadaegh, F., Steyerberg, E. W., & Khalili, D. (2016). A tutorial on variable selection for clinical prediction models: Feature selection methods in data mining could improve the results. Journal of Clinical Epidemiology, 71, 76–85. https://doi.org/10.1016/j.jclinepi.2015.10.002
Darst, B. F., Malecki, K. C., & Engelman, C. D. (2018). Using recursive feature elimination in random forest to account for correlated variables in high dimensional data. BMC Genetics, 19(1), 1–6. https://doi.org/10.1186/s12863-018-0633-8
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
This research is not supported under any funding. The authors declare that they have no conflict of interest.
Human and Animal Rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chauhan, N.K., Singh, K. Performance Assessment of Machine Learning Classifiers Using Selective Feature Approaches for Cervical Cancer Detection. Wireless Pers Commun 124, 2335–2366 (2022). https://doi.org/10.1007/s11277-022-09467-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11277-022-09467-7