Abstract
The classification and analysis of data is an important issue in today’s research. Selecting a suitable set of features makes it possible to classify an enormous quantity of data quickly and efficiently. Feature selection is generally viewed as a problem of feature subset selection, such as combination optimization problems. Evolutionary algorithms using random search methods have proven highly effective in obtaining solutions to problems of optimization in a diversity of applications. In this study, we developed a hybrid evolutionary algorithm based on endocrine-based particle swarm optimization (EPSO) and artificial bee colony (ABC) algorithms in conjunction with a support vector machine (SVM) for the selection of optimal feature subsets for the classification of datasets. The results of experiments using specific UCI medical datasets demonstrate that the accuracy of the proposed hybrid evolutionary algorithm is superior to that of basic PSO, EPSO and ABC algorithms, with regard to classification accuracy using subsets with a reduced number of features.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The production of data throughout the world every second of every day puts us in the era of big data. In the past, medical data, such as X-ray images, the medical history of patients, disease characteristics, and gene sequences had to be collected and analyzed manually [1]. However, recently data mining techniques have been developed to identify the relevance of features in medical datasets in a rapid and efficient manner. Data mining is particularly effective in obtaining valuable information from huge quantities of data [2]. Machine learning is widely applied in data mining for tasks involving search and analysis [3]. These developments have largely eliminated the need to perform these actions manually.
Raw data may contain non-essential features, which can increase computational complexity and decrease classification. This explains why feature selection [4] is so important in machine learning in fields such as image recognition, medical information [5], botnet characteristics [6] and the classification of files [7]. Feature selection [8] makes it possible to focus on data with high correlation characteristics, thereby improving the efficiency of classifiers, increasing classification accuracy, and reducing computational costs. The purpose of feature selection is to identify the characteristics of data within multiple dimensions. We used continuous selection and screening to collect features with a high degree of correlation to produce feature subsets with the aim of improving efficiency in the classification of data.
An increase in the number of data dimensions leads to an exponential increase in the complexity and subsequently the difficulty of assembling an effective feature subset. Thus, we employ evolutionary computation to assemble the feature subset, in which an adequate feasible solution can be found using a wrapper under certain conditions. The evolutionary approach is intended to overcome problems of optimization. Evolutionary algorithms are based on natural phenomena, in which a random search process is guided by the goal of improving search results from one generation to the next, ultimately leading to an optimal solution. The most famous example is the genetic algorithm (GA) [9], based on Darwin’s theory of natural selection. Another technique involves simulated annealing (SA) [10], similar to a process found in the atomic lattice structure of metallurgy.
In this study, we developed a hybrid evolutionary algorithm based on endocrine-based particle swarm optimization (EPSO) and the artificial bee colony (ABC) algorithm using a support vector machine (SVM) for the selection of an optimal feature subset to facilitate the classification of data.
In Section 2, we introduce the concept of SVM and evolutionary algorithms. In Section 3, we outline our hybrid algorithm based on EPSO and the ABC algorithm with SVM. Section 4 presents our experiment methods, results, and the dataset used in the experiment. Conclusions are drawn in Section 5.
Related work
Support vector machine
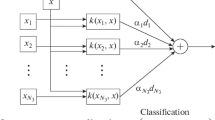
Support vector machine (SVM) is a supervised learning method that was proposed by Vapnik in 1995 [11]. It is widely used in regression analysis and statistical classification. SVM involves the construction of a multi-dimensional hyper-plane for the classification of data. As shown in Fig. 1, the distance between the hyper-plane and the data is referred to as the margin. This space may also exist on multiple hyper-planes; however, a single plane provides the greatest margin to the nearest data.
Difficulties in the classification of labels are generally characterized according to the dimensions and complexity of the data; i.e., not all of the data are linearly separable. In the case of linearly inseparable problems, the kernel of the support vector machine maps the input space into a Vapnik-Chervonenkis dimension, thereby rendering problems that were linearly non-separable in the original input space as linearly separable problems in a high-dimensional space. Before building a classification model using the support vector machine, we must select the type of core functions and their parameters as well as penalty factor C. Three common core functions are radial-based functions (RBF), polynomial functions, and sigmoid functions, as shown in Eqs. (1), (2), and (3):
RBF kernel:
Polynomial kernel:
Sigmoid kernel:
Kernel parameters are selected based on the core functions. For example, the RBF kernel used in this experiment includes parameter γ, which must be adjusted to find the optimized hyper-plane. Penalty factor C affects the complexity and ultimately the accuracy of the model. When the value of C is relatively small, the accuracy of the model will tend to be poor; however, generalizability will be improved. Conversely, a higher value for C will increase the accuracy of the model, the tolerated error will become smaller, and the generalizability will decline. Thus, we also searched for C and γ during the search for feature subsets in order to maximize the effectiveness of the classification model.
Evolutionary intelligence
Evolutionary intelligence is a behavior of natural or artificial organization among individuals. This notion was inspired by observation of natural phenomena, wherein individual entities share information for the sake of survival. One classic example of this behavior is the division of labor among bees in a hive. Similarly, in ant colonies information related to the location of food is shared through the secretion of chemicals as a form of signal transmission. Flocks of birds and schools of fish are other examples of group behavior among animals. The algorithm proposed in this study is intended to simulate the foraging behavior of animals involving the search for biological formation.
Particle swarm optimization
The particle swarm optimization algorithm [12] was proposed by Kennedy and Eberhart in 1995. PSO has been successfully applied in a variety of fields to obtain optimal solutions more rapidly than can be achieved using other algorithms, such as GA or SA. PSO involves a group of potential solutions called particles, which move around within a search space at a specific velocity in search of better solutions. Each particle has a memory of the best location it has previously visited, which is regarded as its individual best (pbest). In cases where this solution is the best within the entire population, then it is denoted as the global best (gbest).
Endocrine-based particle swarm optimization
Endocrine-based PSO [13] combines particle swarm intelligence with the mechanism involved in the regulation of human hormones. Particles with poor adaptability are provided hormones aimed at increasing the amount of variation in the displacement of the particle, thereby increasing its search range. In contrast, particles displaying better adaptability receive only a small amount of hormone and thereby undergo only subtle changes. The particles in an endocrine system are simultaneously affected by the global best particle as well as neighboring particles, the effects of which can be calculated as follows:
EM (S) represents the hormone concentration of particle, f max is the best global fitness, f i is the fitness of the i-th particle, f avg is the average fitness of all particles. Where fun 1() = atan(x), when the fitness f i becomes worse, the value of fun 1 increases the hormone concentration of particle i. In cases where f i is less than f avg , the amount of administered hormone is reduced. fun 2() = atan(−x), wherein the value of the i-th particle is affected by the nearest particle. The equation used for hormone updating is as follows:
The hormone concentration of next generation E i (k + 1) equals the sum of the hormones concentration of this generation E i (k), and the variation of EM(S); C3 is the hormone updating constant, C4 is the inertia constant, and rand ranges between 0 and 1.
Equation (6) used for updating speed in PSO is also used for this purpose for EPSO and the equation for updating position in EPSO is as follows:
Artificial bee colony
The artificial bee colony (ABC) algorithm [14] is an intelligence algorithm proposed by Dervis Karaboga in 2005. It was inspired by the concept of bees looking for nectar and their hierarchical behavior. The ABC algorithm regards the solution space as foraging space, in which each solution (food source) symbolizes a quantity of nectar and bees exchange messages in order to guide one another to a better source of food.
A number of parameters must initially be set, such as the number of food sources, trial limit, and termination conditions. In the initialization phase, n food sources are generated at random and expressed as solution x i , where i ∈ {1, 2, …, N}; x i is a vector with D dimensions, with D determined by the problem. At the same time, x i denotes the ith food source. Each food source is produced as follows:
Here x max and x min are the upper and lower boundaries of x ij . The trial limit here is initially a constant during the initialization phase for all of the bees (employed bees as well as onlookers). When the number of times that food source has not been updated reached this number, the food source is abandoned.
In the second stage, only employed bees search for a food source in order to generate new food v ij using the following equation:
where j ∈ {1,2, …, D} and k ∈ {1,2, …, N}, j and k are not identical. A greedy choice is made between x ij and the new food source v ij . If the fitness of v ij is better than that of x ij , v ij will replace as x ij the food source. Conversely, when the fitness of v ij does not exceed that of x ij , the number of the failed attempts of the current food source is increased by 1.
In the third stage, employed bees provide onlooker bees with information related to the location of food sources. The onlooker bees follow the food source according to probability function P i (10) based on the fitness of the food and decide whether to search randomly for food sources.
When x i is selected by onlooker bees, the bees search around x i in accordance with Eq. (10) in order to generate new v i and perform the same greedy choice phase and update the number of failed attempts as the employer bees do.
In the following phase, scout bees determine whether food sources need to be abandoned. In cases where the number of failures is equal to or greater than the pre-set number of allowable attempts, scout bees abandon the associated food source and randomly generate a new solution in accordance with the new food Eq. (8).
Hybrid methods
Multiple technologies can be merged in order to solve problems. For example, when figuring out the best answer to a problem and proving that it is the best solution, using a single approach can significantly increase time costs. Data transmission in networks, for instance, may be achieved via a number of protocols: wired, wireless, packages, routing, and form management, which present a hybrid solution for data transmission applications.
The various calculation methods in evolutionary algorithms possess distinct advantages and disadvantages. Simulated annealing and the Tabu search method deal with local search properties, whereas genetic algorithms take an evolutionary approach to the problem of performing global searches [15]. This study proceeded under the assumption that these methods could complement one another or make up for the shortcomings inherent in simpler approaches. Hybrid methods can help to improve the convergence efficiency, robustness and reliability [16] of the algorithm.
The PSO and ABC algorithms have proven effective in a number of optimization applications; however, a number of problems have yet to be eliminated. In a large dimensional space, the ability of the PSO of find the optimal solution begins to drop into local optimums, with a negative effect on the accuracy of the final results [17]. During searching processes with the ABC algorithm, the direction in which the solution moves is generated based on random neighbors, which enables the search to converge in the search area more easily [18]. Literature has also indicated that the ABC algorithm converges more slowly in some unimodels and easily falls into local optimums in some complex multi-models [19].
Many hybrid search methods have been proposed to overcome these shortcomings by compensating for the deficiencies of a single algorithm. In [20], the population is divided into two subgroups in each iteration. One of the subgroups evolves via ABC while the other evolves via PSO. A search is then performed to find the optimum solution for each of the respective subgroups, whereupon the solution with the best fitness is identified through a comparison of these candidate solutions. ABC is effective in local searches but weak in global searches. The PSOABC chain [21] uses multiple algorithms in the search for a global optimum. We employed the differential evolution method (DE) in the employed bee phase and included the PSO global optimum reference method in the onlooker bee phase so that the ABC algorithm maintains a local search capacity in the employed bee phase. Global development effects are also then enhanced in the onlooker bee phase [22].
Proposed hybrid evolutionary algorithm based on EPSO and ABC
This study developed a hybrid approach combining the EPSO and ABC algorithms. EPSO is used to search through the entire solution space and then output individual best positions as well as a global best position. The initial locations of the food sources in the employed bee phase of the ABC method originate from the individual bests in the EPSO phase. In subsequent steps, onlooker bees and scout bees continue their search to improve the quality of the overall solutions. Figure 2 presents the pseudo code of the EPSO_ABC algorithm.
Initialize population
Generate the initial velocity of each population as well as the initial position (solution) and the initial hormone content. Then calculate the fitness of the initial population by recording the individual as well as global fitness values.
Calculate and update the velocity and position of each population
The changes in the hormones of the particles under the influence of surrounding particles are as shown in Eq. (4). The changes in their speed under the influence of the global and local optimums are as shown in Eq. (6), and the formula for the final position update is as shown in Eq. (7).
Evaluate the fitness of each population
Using the evaluation function to evaluate the fitness.
Update the best solution obtained from each individual population
If the fitness level of the current individual population is superior to the previous individual fitness value, then the current location of the individual population is identified as the optimal individual population x pbest .
Update the global optimum
The global optimum of this generation is calculated with the fittest value among the global optimum of previous generation and the local optimum of each individual particle for this generation. In subsequent steps, x pbest is set as the food source for further search procedures.
Employed bees find new food sources
Each employed bee corresponds to only one food source, and it searches the vicinity of its food source based on Eq. (9). The new food sources replace the current food sources if they are better. Otherwise, the failure count is updated.
Onlooker bees find new food sources
Onlooker bees search for food based on the roulette method using Eq. (9). They search for new food sources based on Eq. (8) and perform greedy comparisons. If the new solution is better than the existing solution, then it is adopted as a replacement. Otherwise, the failure count is updated.
Scout bees abandon food source
Determine whether any of the food sources have reached the maximum number of allowable failures. If the limit has been reached, then that particular food source is discarded. Equation (10) is used to randomly generate a new food solution.
Update the best food source
The best food source is updated after performing a comparison with the entire population. The above process is repeated until the termination conditions are reached, at which point the calculation results are output.
SVM classifier with feature selection based on EPSO_ABC
In this study, we adopted SVM as a classifier for the classification of known types of data for the prediction of unknown data. However, these data may contain noise or redundant features, which could disturb the classification process. This is also why we need another method to perform the feature selection and leave only the features with higher degrees of correlation. We used the EPSO_ABC algorithm to increase or reduce the number of features in order to produce a variety of feature subsets for the training of SVM. The best feature subset is then output when the termination conditions are fulfilled.
Initialization phase
In the initialization phase, the particles were randomly encoded as shown in Fig. 3. F1 to Fn refer to features 1 to n; c and γ are the parameters used in SVM. The range of the variables in each feature subset falls between 0 and 1. The parameters vary with the feature subset and influence the classification model. The velocity and location of the particles as well as c and γ are continuous values.
EPSO_ABC algorithm phase
The particles search for the feature subset according to the EPSO_ABC procedure. All features, c and γ are searched step by step in EPSO_ABC. c and γ are a part of the population; these parameters will be searched in the same way as features in this phase. The evaluation of fitness is conducted after the search process has been completed.
Fitness evaluation
In the fitness evaluation phase, the evaluation is performed based on the selected feature subset and parameters. The population is transformed to binary type with the boundary set to 0.5. As shown in Figs. 4 and 5, a feature that is less than 0.5 is not selected, whereas a feature that is equal to or exceeding 0.5 is selected.
In this part, only the selected data is kept, and the remainder is discarded. Next, the data is divided into two parts: a training set and a testing set. The SVM reads the training set and establishes a classification model. The testing set is then applied to the completed classification set.
Output optimized subset of features
The algorithm continues searching until the termination conditions are fulfilled. The best solutions are iteratively updated with new populations presenting superior fitness values. In cases where the fitness of the new population equals the current population but with fewer features, it also replaces the current population. When the termination conditions are reached, the outputs are yielded, including the best feature subset, parameters c and γ, and the accuracy of classification. Figure 6 presents a detailed flowchart of the EPSO_ABC with SVM.
Experiments
In this section, we describe the experiments used to verify the effectiveness of the EPSO_ABC, including the computer equipment, the development environment, and the datasets. We also compare the proposed approach with the PSO and EPSO algorithms.
The experiments were conducted on a computer with an Intel Core i7 CPU running Windows 7 at 3.0GHz, with memory 4GB. The development environment is Dev C++ combined with LibSVM [23] libraries and RBF as the SVM kernel function. The datasets in Table 1 have been used in variety of different fields for many years and were obtained from the University of California, Irvine (UCI) [24].
Experiment parameters
We employed k-fold cross-validation [25] to test the search ability of the algorithms. In k-fold cross-validation, the original sample is randomly partitioned into k equally sized subsamples. Here k was set to five, which means 80 % of the original data are randomly selected as training data and the remaining are used as testing data. The parameter settings are presented in Table 2. The termination condition is set to prevent updating within 1 h. In the experiment, the population size was set to 20, constants c1 and c2 were set to 1.5 (usually between 1 and 2), and the parameter weight was 0.7 (usually between 0.4 and 0.9) [26]. Constants c3 and c4 were set to 0.05 [13] and the limit in ABC phase was set to 100. The range of c and γ are preset parameters of SVM.
Experiment results
Table 3 illustrates the difference between performance of the proposed hybrid EPSO_ABC algorithm obtained with and without feature selection. The experiment results clearly illustrate the superiority of the SVM classifier with feature selection mechanism with regard to classification accuracy using subsets with fewer features (about 3.4 ~ 6.2). In fact, the classification accuracy with the Parkinsons dataset was improved to 100 % using only 5.2 features. The classification accuracy of Parkinsons, CTGs, and SPECT increased, and the number of features decreased from 22 to 4.0, 6.2, and 5.6, respectively.
Table 4 presents a comparison of the proposed EPSO_ABC-SVM and other algorithms pertaining to classification accuracy and the number of selected features. Overall, the EPSO_ABC-SVM provides better classification accuracy and fewer selected features. Using the Ecoli dataset, the accuracy of the proposed method was the same as that of EPSO; however, fewer features were selected. Using the Breast and Heart datasets, the EPSO_ABC provided higher classification accuracy with fewer selected features. Using the Parkinsons dataset, EPSO_ABC found 4.0 features and the accuracy was superior to that of the other algorithm (100.00 % vs. 99.49 %). Even when the classification accuracy was equal, EPSO_ABC was able to assemble feature subsets using fewer features. We also used the Friedman test to verify our experiment result which shows at the bottom of Table 4. The p-value is 0.00657 lower than 0.05 which means there are significant differences between the methods.
Table 5 presents the relationship between the datasets and the number of times that features are selected, ranging between 0 and 5. In the Ecoli dataset, the most important features are as follows: chg and aac (selected 5 times) and sequence name (selected 4 times). In the dataset Breast, uniformity of cell shape was selected 4 times and clump thickness was selected 3 times. These two important feature can help to determine whether cancer is benign or malignant. In Heart, the number of major vessels was selected 5 times, and feature 13 was selected 3 times. Among the 22 features in the Parkinsons dataset, only feature 1 was identified 3 times and all others were selected fewer than 3 times. CTGs include measurements of fetal heart rate (selected 5 times) and uterine contraction features on cardiotocograms in which mean value of long-term variability was selected 3 times and tendency was selected 3 times. In the SPECT dataset, all features are partial diagnosis or binary. Features 16 and 22 were selected 3 times and all others were selected fewer than 3 times.
Conclusion
This study presents a hybrid evolutionary algorithm based on EPSO and ABC for feature selection and the optimization of parameters for SVM. Experiment results demonstrate the superiority of the SVM classifier with feature selection mechanism with regard to classification accuracy using subsets with fewer features (3.4 ~ 6.2), compared to the SVM classifier without feature selection. Experiment results also demonstrate the superiority of the proposed hybrid evolutionary algorithm with regard to classification accuracy using subsets with fewer features than in the basic PSO, EPSO and ABC algorithms. Moreover, as the experiment results shown in Table 5, the selected feature subset can help the clinical diagnosis.
References
Raghupathi, W., Data mining in health care. Health. Informat. Improv. Efficienc. Productiv. 211–223, 2010.
Piateski, G., and Frawley, W., Knowledge discovery in databases. MIT press: Cambridge, MA, USA, 1991.
Mannila, H., Data mining: machine learning, statistics, and databases. In: Eighth International Conference on Scientific and Statistical Database Systems (pp. 2–9). IEEE Computer Society: Stockholm, 1996.
Dash, M., and Liu, H., Feature selection for classification. Intell. Data Anal. 1(3):131–156, 1997.
Furey, T. S., Cristianini, N., Duffy, N., Bednarski, D. W., Schummer, M., and Haussler, D., Support vector machine classification and validation of cancer tissue samples using microarray expression data. Bioinformatics 16(10):906–914, 2000.
Livadas, C., Walsh, R., Lapsley, D., and Strayer, W. T., Using machine learning techniques to identify botnet traffic. In: The 31st IEEE Conference on Local Computer Networks (pp. 967–974). IEEE: Tampa, FL, 2006.
Shin, C., Doermann, D., and Rosenfeld, A., Classification of document pages using structure-based features. Int. J. Doc. Anal. Recog. 3(4):232–247, 2001.
Liu, H., and Motoda, H., Feature selection for knowledge discovery and data mining. Kluwer Academic Publishers: Norwell, MA, USA, 1998.
Holland, J. H., Adaptation in natural and artificial systems: an introductory analysis with applications to biology, control, and artificial intelligence. MIT Press: Cambridge, MA, USA, 1992.
Kirkpatrick, S., and Vecchi, M. P., Optimization by simmulated annealing. Science 220(4598):671–680, 1983.
Cortes, C., and Vapnik, V., Support-vector networks. Mach. Learn. 20(3):273–297, 1995.
Kennedy, J., Particle swarm optimization. Encyclopedia of Machine Learning (pp. 760–766), Springer, US, 1995.
Chen, D. B., and Zhao, C. X., Particle swarm optimization based on endocrine regulation mechanism. Contr. Theor. Appl. 24(6):126–134, 2007.
Karaboga, D., An idea based on honey bee swarm for numerical optimization (Vol. 200). Technical report-tr06, Erciyes University, Engineering Faculty, Computer Engineering Department, 2005.
Youssef, H., Sait, S. M., and Adiche, H., Evolutionary algorithms, simulated annealing and tabu search: a comparative study. Eng. Appl. Artif. Intel. 14(2):167–181, 2001.
Wang, X., Hybrid nature-inspired computation methods for optimization. TKK Dissertations, Doctoral Dissertation, Helsinki University of Technology, 2009.
Zhi-gang, W., Hybrid optimization algorithm based on particle swarm optimization and artificial bee colony algorithm. Sci. Technol. Eng. 12(20):4921–4925, 2012.
Guo, Z., A hybrid optimization algorithm based on artificial bee colony and gravitational search algorithm. Int. J. Dig. Cont. Tech. Appl. 6(17):620–626, 2012.
Karaboga, D., and Akay, B., A comparative study of artificial bee colony algorithm. Appl. Math. Comput. 214(1):108–132, 2009.
Liu, J., Zhang, X., and Ning, A., Hybrid optimization algorithm of PSO and ABC. Comput. Eng. Appl. 47(35):32–34, 2011.
Altun, O., and Korkmaz, T., Particle Swarm Optimization–Artificial Bee Colony Chain (PSOABCC): A hybrid meteahuristic algorithm. Scientific Cooperations International Workshops on Electrical and Computer Engineering Subfields (pp. 22–23). Istanbul, Turkey: Koc University, 2014.
Kong, X., Liu, S., and Wang, Z., A new hybrid artificial bee colony algorithm for global optimization. Int. J. Comp. Sci. 10(1), 2013.
Hsu, C. W., Chang, C. C., and Lin, C. J., A practical guide to support vector classification. Technical report, Department of Computer Science, National Taiwan University, 2003.
Lichman, M., UCI repository of machine learning databases. Irvine, CA: University of California, School of Information and Computer Science. [http://www.ics.uci.edu/∼mlearn/MLRepository.html], 2013.
Salzberg, S. L., On comparing classifiers: Pitfalls to avoid and a recommended approach. Data Min. Knowledg. Discov. 1(3):317–328, 1997.
Shi, Y., and Eberhart, R. C., Empirical study of particle swarm optimization. In Proceedings of the Congress on Evolutionary Computation (CEC '99) (pp. 1945–1950). IEEE Service Center, Piscataway, NJ, USA, 1999.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of the Topical Collection on Systems-level Quality Improvement
Rights and permissions
About this article
Cite this article
Lin, KC., Hsieh, YH. Classification of Medical Datasets Using SVMs with Hybrid Evolutionary Algorithms Based on Endocrine-Based Particle Swarm Optimization and Artificial Bee Colony Algorithms. J Med Syst 39, 119 (2015). https://doi.org/10.1007/s10916-015-0306-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10916-015-0306-3