Abstract
Machine learning is used as an effective support system in health diagnosis which contains large volume of data. More commonly, analyzing such a large volume of data consumes more resources and execution time. In addition, all the features present in the dataset do not support in achieving the solution of the given problem. Hence, there is a need to use an effective feature selection algorithm for finding the more important features that contribute more in diagnosing the diseases. The Particle Swarm Optimization (PSO) is one of the metaheuristic algorithms to find the best solution with less time. Nowadays, PSO algorithm is not only used to select the more significant features but also removes the irrelevant and redundant features present in the dataset. However, the traditional PSO algorithm has an issue in selecting the optimal weight to update the velocity and position of the particles. To overcome this issue, this paper presents a novel function for identifying optimal weights on the basis of population diversity function and tuning function. We have also proposed a novel fitness function for PSO with the help of Support Vector Machine (SVM). The objective of the fitness function is to minimize the number of attributes and increase the accuracy. The performance of the proposed PSO-SVM is compared with the various existing feature selection algorithms such as Info gain, Chi-squared, One attribute based, Consistency subset, Relief, CFS, Filtered subset, Filtered attribute, Gain ratio and PSO algorithm. The SVM classifier is also compared with several classifiers such as Naive Bayes, Random forest and MLP.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 INTRODUCTION
In general, the heart is found to be a most important organ of human body. Thus, heart diseases are considered as a significant health issue in day-to-day life. Many reports state that the cardiovascular diseases are the root cause of sudden death of individuals in industrialized countries [1]. The increased death in industrialized countries affects the individuals' health and finances and budget of the countries [2]. The following diseases are found to be most important risk factors for the cardiovascular disease it includes diabetes, high saturated fat, family history, fatness, smoking and high cholesterol. Nowadays, newborn babies are also affected by cardiovascular diseases. Hence, checking of cardiovascular diseases is very common in day-to-day life.
Moreover, chest pain and fatigue are considered as the most familiar symptoms of getting the heart disease [3, 4].
In order to overcome this issue, a number of feature selection methods are identified by the modern computational researchers [7, 10, 14, 16]. In this paper, recent advancements in feature selection and frontiers in heart disease predictions are discussed in detail. More commonly, feature selection algorithms are classified into two types namely consistency filter-based feature selection and correlation-based feature selection. Correlation-based feature selection is developed on the basis of filter based feature selection while consistency filter-based feature selection methods select the more important features based on their consistency values of each feature. In correlation-based feature selection, a simple heuristic evaluation function is used to rank the features subsets [17, 18].
Hence, this heuristic evaluation function significantly identifies the more significant features on the basis of their high correlation values. Thus, low correlation between the features is removed not only from the training dataset but also testing dataset [19, 20]. Moreover, the correlation-based feature selection methods are also used to remove the redundant features from the training dataset and testing dataset. The consistency filter-based feature selection methods use the consistency value of each feature to select the more important features. In addition, a consistency filter-based feature selection method identifies a random subset S from the number of features [21]. The current best position is updated on the basis of the comparison between the number of features of S and the current best features. Afterwards, the selected features are randed based on the various feature randing methods such as Information Gan, ReliefF and etc. [22, 23].
Relief algorithm and ReliefF algorithm are the two familiar methods used to rank the multiclass features with incomplete and noisy data. The ReliefF algorithm is extended based on the original Relief algorithm [25]. In many cases, the Relief algorithm is used when the communication among features is small and taking of features with local dependencies. In addition to Relief algorithm and ReliefF algorithm, Information Gain is used more commonly in various applications [38–41]. A user-defined threshold value is used in the Information Gain method for selecting the more significant features. The threshold value is selected on the basis of entropy method.
In this paper, we have presented a novel feature selection method on the basis of PSO algorithm with SVM. The traditional PSO algorithm cannot find the ideal weight to update the velocity and position of the particles. To overcome this issue, we have proposed a novel function to find the optimal weights on the basis of population diversity function and tuning function. In order to minimize the number of attributes and increase the accuracy, an SVM classifier with constant value λ is used as a fitness function of the proposed PSO algorithm. In the proposed PSO algorithm, the large inertia weight is used for a global search while a small inertia weight is used for a local search. A constant value λ is altered on the basis of a total number of features used in the PSO. In the proposed PSO algorithm, if the number of features present in the subset is high then the fitness function reduce the attributes. Otherwise, if the number of features present in the subset id low then the fitness function improves the accuracy.
2 RELATED WORK
An effective rule reduction algorithm is proposed by Carlos Ordonez [3]. This algorithm is applied to the real-time medical records for identifying the early stage of heart attacks. An objective of this algorithm is to reduce the number of association rules in the mining process. Hence, the resultant rules generated from this algorithm is used for classifying the heart disease effectively. In addition, the performance of the resultant rules is evaluated based on the following performance evaluation metrics it includes sensitivity, accuracy, precision, specificity, true positive rate and false positive rates [26]. These parameters are calculated for a given set of independent test cases. In recent years, this rule generation algorithms are effectively used for predicting the heart attacks in an early stage.
Similarly, a novel feature selection algorithm is proposed by K.C. Tan et al. [8] with the help of wrapper method based Genetic Algorithms (GAs). The objective of this method is to find the more significant attributes of this method is to find the more significant attributes that independently classify the heart disease in an efficient manner. The fitness function is used in order to indentify the more appropriate features from a set of given features. More similarly, Jesmin Nahar et al. [9] have developed a novel intelligent feature extraction method on the basis of various meta-heuristic algorithms it includes Predictive Apriori and Tertius. The extracted features can effectively classify the heart disease with an improved accuracy rate of 87%. The performance evaluation parameters are used to prove the good performance of the proposed meta-heuristic based feature selection algorithm [27].
A novel Kemel F-score Feature Selection (KFFS) methods proposed by Kemal Polat [11] is used to remove the irrelevant and redundant features and select the more significant features. The proposed algorithm is tested with a large size electronic health record and the performance of this algorithm is evaluated with the help of sensitivity, accuracy, precision, specificity, true positive rate and false positive rates [28]. The function of the proposed DFFS algorithm is divided into two types, namely, F-score method and Radial Basis Function (RBF) kernel functions. The vital role of the RNF kernel method is to convert the features of medical datasets into a kernel space [29–31]. In order to calculate the F-score values with high dimensional feature space, the F-score technique is initially used in the proposed framework [37, 38].
Moreover, the differential evolution classifier with Principal Component Analysis (PCA) is used by Pasi Luukka [15] to develop more efficient classification rules for early detection of heart disease. The vital role of this PCA method is to develop the classification model and removes the irrelevant and redundant features from not only training dataset but also testing dataset. They have used the classical Electronic Medical Record (EMR) to evaluate the performance of the proposed method, where a range of patient demographical attributes are considered for predicting the diabetes it includes symptoms and behavior of the patients, heartbeat rate, history of coronary infarction, angina, Electro-CardioGraphy (ECG), blood glucose level, blood pressure and blood insulin level and so on [32, 33].
A novel genetic algorithm based feature selection method has proposed by Hongmei Yan et al. for identifying the more important attributes to get heart disease. This algorithm tested with a variety of heart disease dataset it includes coronary heart disease, hypertension, congenital heart disease, rheumatic valvular and chronic pulmonale heart disease. In addition to the above methods, a number of artificial intelligence methods and machine learning procedures are used for predicting and classifying the heart diseases effectively [34]. These methods are also employed for identifying the more important features [35]. The following research works have used various methods for predicting the heart diseases it includes machine learning method [44], artificial intelligence technique [12], Genetic Algorithm (GA) with Binary Particle Swarm Optimization (BPSO) [13] and Case-based reasoning (CBR) method [24].
In addition, a variety of regression techniques and classification algorithms have employed in [21] for developing a predictive model that used to discover the hidden values from the heart disease dataset. Iftikhar has used SVM and PSO to develop a healthcare analytic model [42]. The proposed model is used to identify the risk factors of heart disease. They have used Cleveland heart disease database to evaluate the performance of the proposed healthcare analytic model. Shah et al. have developed a novel feature selection approach based on Probabilistic Electronic Medical Record Principal Component Analysis (PPCA) [43]. The vital role of PPCA is used to extract the more important features for predicting the heart disease. In addition, hybrid neural network-Genetic algorithm is proposed to classify the heart disease in effective manner [44].
This method also improves the performance of neural network by about 10% through augmenting its the heart disease. The experimental results are compared initial weights. This weight changing task is done with the help of genetic algorithm which proposes better weights for neural network. The proposed method achieved accuracy, sensitivity and specificity rates of 93.85, 97, and 92% respectively, on Z-Alizadeh Sani dataset [44]. Li et al. proposed a kernel extreme learning machine (KELM) and improved grey wolf optimization (IGWO) method for heart disease diagnosis. The objective of IGWO method is to find the optimal features from the heart disease dataset [45]. Vivekanandan et al. have proposed a adapted differential evolution (DE) algorithm to select the more significant attributes to get heart disease. The proposed algorithm tested with a diversity of heart disease dataset it comprises hypertension, congenital heart disease, coronary heart disease, chronic pulmonale and rheumatic valvular heart disease [46].
Jabbar et al. have proposed a classification method which uses chi square and random forest for predicting the heart disease. The performance of the proposed algorithm is evaluated on UCI machine learning heart disease data sets. The experimental results demonstrate that the proposed method produced high classification accuracy compared to other classification methods [47]. Paul et al. have presented a genetic algorithm based fuzzy decision support system to predict the heart disease. The proposed method consists of following steps: preprocessing, feature selection, fuzzy rule generation based on genetic algorithm and predicting the heart disease [48].
Inbarani et al. have proposed a novel supervised feature selection method on the basis of hybridization of Particle Swarm Optimization (PSO). The proposed method is verified with a large size of electronic health record and the performance of this algorithm is evaluated with the help of various performance evaluation parameters such as ROC analysis, accuracy, sensitivity, precision, specificity, true positive rate and false positive rates [49]. Tomar et al. have proposed a feature selection based Least Square Twin Support Vector Machine (LSTSVM) for early diagnosis of heart diseases. This method uses F-score statistics for identifying the weight of each feature. The proposed LSTSVM model is used to identify the risk factors of heart disease [50].
Reddy et al. have used rule-based fuzzy logic (RBFL) and oppositional firefly with BAT method for predicting the heart disease. The hybrid OFBAT-RBFL heart disease diagnosis method is also proposed on the basis of RBFL and oppositional firefly with BAT method. The performance evaluation parameters are used to prove the good performance of the hybrid OFBAT-RBFL algorithm [51].
Pimentel has used Naive Bayes (NB) to classify with the variety of classifiers such as Alternating Decision Tree (ADT), Support Vector Machine (SVM), Random Forest (RF), Random Tree (RT) and K-Nearest Neighbour (KNN). The extracted features can efficiently classify the heart disease with an improved accuracy rate of 82%. The performance evaluation parameters such as ROC and accuracy are used to prove the effectiveness of the Naive Bayes based feature selection algorithm.
3 ENHANCED PARTICLE SWARM OPTIMIZATION WITH SVM FOR FEATURE SELECTION
The objective of the metaheuristic algorithms is to classify the best solution with less time. The performance metaheuristic algorithm is good when compared with traditional deterministic algorithms. The PSO is one of the familiar metaheuristic algorithms used to identify the optimal solution in an efficient manner. In this method, the particles are selected on the basis of random approach. Moreover, the particles present in PSO moves randomly from source to destination. In each movement and iteration, a fitness function is calculated to reach the best solution. The fitness function is found to be a most important method in PSO. The performance and efficiency of the PSO algorithm are calculated based on the effectiveness of the fitness function. In this algorithm, the best particles are selected based on their fitness value.
The traditional PSO algorithm has an issue in selecting the optimal weight to update the velocity and position of the particles. Hence, the large inertia weight is used for a global search while a small inertia weight is used for a local search [51]. More commonly, the inertia weight of the PSO algorithm is change between 0 and 1. In order to find the optimal weights for a global search and local search, a population diversity function and tuning function are proposed in this paper for the PSO.
3.1 Population Diversity Function
In general, all the particles present in the PSO will meet at a certain time. If the particles meet at a specific time then the population diversity will fall down. Afterwards, the consistency of the algorithm will reduce. In order to overcome this issue, the proposed PSO algorithm uses population diversity function. The objective of the population diversity function is to help algorithm out of local optimum. The population diversity function is defined by, population diversity function
where
#N = total size of all swarm,
#\(f(x_{k}^{t})\) = particle fitness at kth position,
#favg = current average of the fitness of the swarm.
The notation “E” represents the “aggregation” level of all particles it is also called a range of the group particles. If the group fitness E is small, then the group particles (diversity) will also be small. Otherwise, the group particles (diversity) will be high.
3.2 Tuning Function
In all most all the swarm intelligence algorithms group the individuals in a specific place, then the population diversity function αt should decrease with the number of iteration is increased. When the swarm fitness E approaches zero, the particles are meet in a specific place. Hence, the number of iteration is increasing the population diversity function should reduce. In order to maintain this level, a tuning function is proposed for the PSO algorithm. The tuning function is defined by,
where
T = Total number of iteration,
t = Current iteration.
More commonly, the inertia weight is found to be a more significant option to maintain the balance between the local and global search solution. In PSO algorithm, the objective of the inertia weight is used to update the previous velocity on the current velocity. In the end of the PSO algorithm, all the particles are meeting in a specific place and also the velocities of the particles will decrease slowly to zero. To solve this issue, the proposed PSO algorithm use the diversity function and tuning function. The proposed combined function for calculating the inertia weight is defined as follows:
where
w = represents the inertia weight,
wmin = represents the minimum inertia weight,
wmax = represents the maximum inertia weight,
αt = denotes the range function,
βt = denotes the tuning function.
3.3 Proposed Fitness Function Using SVM Classifier
Algorithm: Proposed Particle Swarm Optimization with SVM Classifier
1. Function PSO()
2. Begin
a. Generating the initial population
b. For k = 1 to n (where, n represents the population size)
i. Particle[k].best = C_P (where, C_P represents the current position)
ii. Particle[k].bestFitness = C_F (where, C_F represents the current fitness)
1. Train the SVM classifier by training set
2. Calculate the Fitness function
3. Classification accuracy of SVM and the number of selected features are used to construct a fitness function.
4. classification accuracy of SVM = \(\frac{{TP + TN}}{{TP + TN + FP + FN}}\).
5. Fitness Function Fx(I) = (100 – accuracy) + \(\mu \frac{{Feature\;in\;Subset}}{{Total\;No.\;of\;Features}}\)
where μ = 10log(total No. of Features) + 1.
c. End for
d. gBest = particle.best with lowest fitness
e. For t = 1 to m (Where m represents the maximum iteration)
i. For k = 1 to n (Where n represents the population size)
1. \({v}_{k}^{{t + 1}} = w{v}_{k}^{{t + 1}} + {{c}_{1}}{{r}_{1}} + (p_{k}^{t} - x_{k}^{t}) + {{c}_{2}}{{r}_{2}} + (p_{g}^{t} - x_{k}^{t})\)
2. \(x_{k}^{{t + 1}} = x_{k}^{t} + {v}_{k}^{{t + 1}}\)
#\(x_{k}^{t}\) = The kth particles in a D-dimensional vector
#\({v}_{k}^{t}\) = the space of flight speed
#r1, r2 = random numbers between 0 and 1
#c1, c2 = learning factors
#w = intertia weight
3. \({{w}_{t}} = {{w}_{{\min }}} + {{\alpha }_{t}}{{\beta }_{t}}({{w}_{{\max }}} - {{w}_{{\min }}})\)
#wmin = inertia weight at minimum level,
#wmax = inertia weight at maximum level,
#αt = population diversity function,
#βt = tuning function.
4. population diversity function \({{\alpha }_{t}} = 1 - \left( {\frac{2}{\pi }} \right)\arctan (E)\)
where \(E = 1 - \frac{1}{N}\sum\limits_{1 = 1}^N {{{{(f(x_{k}^{t}) - {{f}_{{{\text{avg}}}}})}}^{2}}.} \)
#N = swarm size
#\(f(x_{k}^{t})\) = fitness of the particle at k
#favg = current average of the fitness of the swarm
5. \({{f}_{{{\text{avg}}}}} = \frac{1}{N}\sum\limits_{1 = 1}^N {f(x_{k}^{t})} \)
6. tuning function \({{\beta }_{t}} = {{e}^{{( - {{t}^{2}}/2\sigma )}}}\)
#\(\sigma = T{\text{/}}3,\)
#T = Total number of iteration
#t = Current iteration
7. If current_fitness < particle[k] best_Fitness
a. Particle[k]_best = current_position
b. Particle[k]_best_Fitness = current_fitness
8. End if
ii. End for
iii. gBest = particle_best with lowest_fitness
f. End for
g. Return gBest
End
The PSO algorithm uses a set of possible solutions to obtain a suitable solution for a given problem. As PSO is one of the optimization methods, the goal of this method is to find the global optimum of a real-valued (fitness) function defined in a given space. The proposed PSO algorithm uses SVM classifier and its accuracy as a fitness function to calculate the fitness value of the particles.
SVM is originally proposed by Vapnik [6] for regression, classification and density estimation problems (Fig. 1). The linear SVM score function is
where
x = represents the observation,
β = represents the vector comprises the coefficients that describe an orthogonal vector to the hyperplane,
b = represents the bias term,
2/||β|| = represents the optimal margin length.
Equation (4) is used as a classifier in the fitness function of proposed PSO. It is a classifier defined by a separating hyperplane. In other words, given labeled training data (supervised learning), the algorithm outputs an optimal hyperplane which categorizes new examples. On the basis of classification results the fitness function is calculated in the proposed PSO.
The maximum margin length is defined by the SVM, whereas keeping observations in the positive (y = 1) and negative (y = –1) classes.
classification accuracy of
where TP = True Positive, TN = True Negative, FP = False Positive, FN = False Negative.
where μ = 10log(total No. of Features) + 1.
The objective of the fitness function is to minimize the number of attributes and increase the accuracy. A value μ is changed on the basis of a total number of features used in the PSO. In the proposed PSO algorithm, if the number of features present in the subset is high then the fitness function reduce the attributes, otherwise, it improves the accuracy.
4 RESULTS AND DISCUSSION
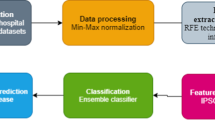
The performance of the proposed PSO-SVM is compared with the various existing feature selection algorithms such as Info gain, Chi-squared, One attribute based, Consistency subset, Relief, CFS, Filtered subset, Filtered attribute, Gain ratio and PSO algorithm (Fig. 4). As shown in Fig. 2, the proposed PSO-SVM based features selection method uses the heart disease dataset collected from Cleveland heart disease database [36]. The good performance of the SVM classifier is proved on the basis of ROC analysis. The ROC curve for the classifiers SVM, Naive Bayes, Random Forest and MLP are presented in Fig. 3. The experimental results are performed in the MATLAB environment. As shown in Fig. 5, the SVM classifier is compared with the various familiar classifier methods such as Naive Bayes, Random Forest and MLP. In order to perform the ROC analysis, the confusion matrix is calculated based on Table 1. Tables 2 and 3 depict the Accuracy of classifiers for the original features and selected attributes respectively. The accuracy of different classifiers such as Naïve Bayes, Random Forest, MLP, SVM are compared on the basis of different feature selection algorithms such as Info gain, Chi-squared, One attribute based, Consistency subset, Relief, CFS, Filtered subset, Filtered attribute, Gain ratio and PSO algorithm and the results are depicted in Table 4.
As shown in Table 4, SVM classifiers produces 88.22% of accuracy when using the significant features originally identified by the proposed PSO with SVM based feature selection method. However, the traditional PSO feature selection method with SVM classifier classifies the heart disease with the accuracy of 85.13%. The proposed algorithm effectively increases the accuracy of 3.09% when using the proposed weight selection method and SVM based fitness function. The features (3, 7, 8, 9, 10, 13) identified by using PSO with SVM based feature selection method are used by the various classifiers such as Naive Bayes, Random Forest and MLP to prove the significance of the selected features. The experimental results proved that the SVM based classifier outperformed well when compared with other classifiers.
5 CONCLUSIONS
The proposed PSO with SVM algorithm based feature selection method identifies 6 significant features it includes sex, fasting blood sugar level, resting electrocardiographic result, maximum heart rate, exercise-induced angina real and a number of major vessels (3, 7, 8, 9, 10, 13) to classify the heart disease. The resultant features are supplied to SVM for finding the accuracy. The SVM produces 79.35% of accuracy when classifying the heart disease with the whole features. However, the SVM classifier produces 84.36% of accuracy when classifying the heart disease with the selected features. We have evaluated the performance of the proposed PSO-SVM method on the basis of comparison between the various existing feature selection algorithms such as Info gain, Chi-squared, One attribute based, Consistency subset, Relief, CFS, Filtered subset, Filtered attribute, Gain ratio and PSO algorithm. The good performance of the SVM classifier is proved on the basis of Receiver Operating Characteristic (ROC) analysis. We have used the Cleveland heart disease database for the demonstration of the proposed PSO-SVM based features selection method to predict the heart disease. The MATLAB environment is used to perform the experimental evaluation. The SVM classifier is also compared with the various familiar classifier methods such as Naïve Bayes, Random Forest and MLP to prove the significance of the selected features.
REFERENCES
Imran Kurt, Mevlut Ture, et al., Comparing performances of logistic regression, classification and regression tree, and neural networks for predicting coronary artery disease, J. Expert Syst. Appl., 2008, vol. 34, pp. 366–374.
Hongmei Yan, Jun Zheng, et al., Selecting critical clinical features for heart diseases diagnosis with a real-coded genetic algorithm, J. Appl. Soft Comput., 2008, vol. 8, pp. 1105–1111.
Carlos Ordonez, Association rule discover with the train and test approach for the heart disease prediction, IEEE Trans. Inf. Technol. Biomed., 2006, vol. 10, no. 2, pp. 334–343.
Kusiak, A., Caldarone, Ch.A., et al., Hypo plastic left heart syndrome knowledge discovery with a data mining approach, J. Comput. Biol. Med., 2006, vol. 36, no. 1, pp. 21–40.
Babaoglu, I., Kaan Baykan, O., et al., Assessment of exercise stress testing with artificial neural network in determining coronary artery disease and predicting lesion localization, J. Expert Syst. Appl., 2009, vol. 36, pp. 2562–2566.
Rajeswari, K., Vaithiyanathan, V., et al., Feature selection in ischemic heart disease identification using feed forward neural networks, Int. Symposium on Robotics and Intelligent Sensors, 2012, vol. 41, pp. 1818–1823.
Mu-Jung Huang, Mu-Yen Chen, et al., Integrating data mining with case-based reasoning for chronic diseases prognosis and diagnosis, J. Expert Syst. Appl., 2007, vol. 32, pp. 856–867.
Tan, K.C., Teoh, E.J., et al., A hybrid evolutionary algorithm for attribute selection in data mining, J. Expert Syst. Appl., 2009, vol. 36, pp. 8616–8630.
Jesmin Nahar, Tasadduq Imam, et al., Association rule mining to detect factors which contribute to heart disease in males and females, J. Expert Syst. Appl., 2013, vol. 40, pp. 1086–1093.
Austin, P.C., Tu, J.V., et al., Using methods from the data-mining and machine-learning literature for disease classification and prediction: A case study examining classification of heart failure subtypes, J. Clin. Epidemiol., 2013, vol. 66, pp. 398–407.
Kemal Polat and Salih Gunes, A new feature selection method on classification of medical datasets: Kernel F-score feature selection, J. Expert Syst. Appl., 2009, vol. 36, pp. 10367–10373.
Babaoglu, I., Findik, O., et al., A comparison of feature selection models utilizing binary Particle Swarm Optimization and genetic algorithm in determining coronary artery disease using Support Vector Machine, J. Expert Syst. Appl., 2010, vol. 37, pp. 3177–3183.
Jesmin Nahar, Tasadduq Imam, et al., Computational intelligence for heart disease diagnosis: A medical knowledge driven approach, J. Expert Syst. Appl., 2013, vol. 40, pp. 96–104.
Setiawan, N.A. et al., A comparative study of imputation methods to predict missing attribute values in coronary heart disease data set, J. Dep. Electr. Electron. Eng., 2009, vol. 21, pp. 266–269.
Luukka, P. and Lampinen, J., A classification method based on Principal Component Analysis and differential evolution algorithm applied for prediction diagnosis from clinical EMR heart data sets, J. Comput. Intell. Optimization Adaption, Learn. Optim., 2010, vol. 7, pp. 263–283.
Das, R., Turkoglu, I., et al., Effective diagnosis of heart disease through neural networks ensembles, J. Expert Syst. Appl., 2009, vol. 36, pp. 7675–7680.
Das, R., Turkoglu, I., et al., Diagnosis of valvular heart disease through neural networks ensembles, J. Comput. Methods Programs Biomed., 2009, vol. 93, pp. 185–191.
Chang-Sik Son, Yoon-Nyun Kim, et al., Decision-making model for early diagnosis of congestive heart failure using rough set and decision tree approaches, J. Biomed. Inf., 2012, vol. 45, pp. 999–1008.
Laercio Brito Gonçalves, Marley Maria Bernardes Rebuzzi Vellasco, et al., Inverted hierarchical neuro-fuzzy BSP system: A novel neuro-fuzzy model for pattern classification and rule extraction in databases, J. IEEE Trans. Syst., Man, Cybernetics, 2006, vol. 36, no. 2.
Kemal Polat and Salih Gunes, A hybrid approach to medical decision support systems: Combining feature selection, fuzzy weighted pre-processing and AIRS, J. Comput. Methods Progr. Biomed., 2007, vol. 88, pp. 164–174.
Kemal Polat, Seral Sahan, et al., Automatic detection of heart disease using an Artificial Immune Recognition System (AIRS) with fuzzy resource allocation mechanism and k-nn (nearest neighbor) based weighting preprocessing, J. Expert Syst. Appl., 2007, vol. 32, pp. 625–631.
Akin Ozcift and Arif Gulten, Classifier ensemble construction with rotation forest to improve medical diagnosis performance of machine learning algorithms, J. Comput. Methods Progr. Biomed., 2011, vol. 104, pp. 443–451.
Chih-Lin Chi, Nick Street, W., et al., A decision support system for cost-effective diagnosis, J. Artif. Intell. Med., 2010, vol. 50, pp. 149–161.
Yoon-Joo Park, Se-Hak Chun, et al., Cost-sensitive case-based reasoning using a genetic algorithm: Application to medical diagnosis, J. Artif. Intell. Med., 2011, vol. 51, pp. 133–145.
Debabrata Pal, Mandana, K.M., et al., Fuzzy expert system approach for coronary artery disease screening using clinical parameters, J. Knowl. Based Syst., 2012, vol. 36, pp. 162–174.
Kahramanli, H. and Allahverdi, N., Design of a hybrid system for the diabetes and heart diseases, J. Expert Syst. Appl., 2008, vol. 35, pp. 82–89.
Vahid Khatibi and Gholam Ali Montazer, A fuzzy-evidential hybrid inference engine for coronary heart disease risk assessment, J. Expert Syst. Appl., 2010, vol. 37, pp. 8536–8542.
Goekmen Turan, R., Bozdag, I., et al., Improved functional activity of bone marrow derived circulating progenitor cells after intra coronary freshly isolated bone marrow cells transplantation in patients with ischemic heart disease, J. Stem Cell Rev. Rep., 2011, vol. 7, pp.646–656.
Karsdorp, P.A., Kindt, M., et al., False heart rate feedback and the perception of heart symptoms in patients with congenital heart disease and anxiety, Int. J. Behav. Med., 2009, vol. 16, pp. 81–88.
Carlosnasillo/Hybrid-Genetic-Algorithm, 2017. GitHub. https://github.com/carlosnasillo/Hybrid-Genetic-Algorithm. Retrieved October 22, 2017.
Muthukaruppan, S. and Er, M.J., A hybrid Particle Swarm Optimization based fuzzy expert system for the diagnosis of coronary artery disease, J. Expert Syst. Appl., 2012, vol. 39, pp. 11657–11665.
Anooj, P.K., Clinical decision support system: Risk level prediction of heart disease using weighted fuzzy rules, J. Comput. Inf. Sci., 2012, vol. 24, pp. 27–40.
Tsipouras, M.G., Exarchos, T.P., et al., Automated diagnosis of coronary artery disease based on data mining and fuzzy modeling, J. IEEE Trans. Inf. Technol. Biomed., 2008, vol. 12, no. 4.
Paredesa, S. et al., Long term cardiovascular risk models’ combination, J. Comput. Methods Progr. Biomed., 2011, vol. 101, pp. 231–242.
Swati Shilaskar et al., Feature selection for medical diagnosis: Evaluation for cardiovascular diseases, J. Expert Syst. Appl., 2013, vol. 40, pp. 4146–4153.
UCI Machine Learning Repository: Heart Disease Data Set. Archive.ics.uci.edu. http://archive.ics.uci. edu/ml/datasets/Heart+Disease. Retrieved October 22, 2017.
Zhao, M., Fu, C., Ji, L., Tang, K., and Zhou, M., Feature selection and parameter optimization for Support Vector Machines: A new approach based on genetic algorithm with feature chromosomes, Expert Syst. Appl., 2011, vol. 38, no. 5, pp. 5197–5204.
Li-Na Pu, Ze Zhao, et al., Investigation on cardiovascular risk prediction using genetic information, J. IEEE Trans. Inf. Technol., Biomed., 2012, vol. 16, no. 5.
Pfister, R., Barnes, D., et al., Individual and cumulative effect of type 2 diabetes genetic susceptibility variants on risk of coronary heart disease, J. Diabetologia, 2011, vol. 54, pp. 2283–2287.
Nazri Mohd Nawi, Rozaida Ghazali, et al., The development of improved back-propagation neural networks algorithm for predicting patients with heart disease, in Proceedings of the First International Conference ICICA, 2010, vol. 6377, pp. 317–324.
Jae-Hong Eom, Sung-Chun Kim, et al., AptaCDSS-E: A classifier ensemble-based clinical decision support system for cardiovascular disease level prediction, J. Expert Syst. Appl., 2008, vol. 34 2465, p. 2479.
Iftikhar, S., Fatima, K., Rehman, A., Almazyad, A.S., and Saba, T., An evolution based hybrid approach for heart diseases classification and associated risk factors identification, Biomed. Res., 2017, vol. 28, no. 8.
Shah, S.M.S., Batool, S., Khan, I., Ashraf, M.U., Abbas, S.H., and Hussain, S.A., Feature extraction through parallel probabilistic Principal Component Analysis for heart disease diagnosis, Phys. A: Statistical Mechanics and Its Applications, 2017, vol. 482, pp. 796–807.
Arabasadi, Z., Alizadehsani, R., Roshanzamir, M., Moosaei, H., and Yarifard, A.A., Computer aided decision making for heart disease detection using hybrid neural network-Genetic algorithm, Comput. Methods Progr. Biomed., 2017, vol. 141, pp. 19–26.
Li, Q., Chen, H., Huang, H., Zhao, X., Cai, Z., Tong, C., and Tian, X., An enhanced grey wolf optimization based feature selection wrapped kernel extreme learning machine for medical diagnosis, Comput. Math. Methods Med., 2017.
Vivekanandan, T. and Iyengar, N.C.S.N., Optimal feature selection using a modified differential evolution algorithm and its effectiveness for prediction of heart disease, Comput. Biol. Med., 2017, vol. 90, pp. 125–136.
Jabbar, M.A., Deekshatulu, B.L., and Chandra, P., Prediction of heart disease using random forest and feature subset selection, in Innovations in Bio-Inspired Computing and Applications, Cham.; Springer, 2016, pp. 187–196.
Paul, A.K., Shill, P.C., Rabin, M.R.I., and Akhand, M.A.H., Genetic algorithm based fuzzy decision support system for the diagnosis of heart disease, in Informatics, Electronics and Vision (ICIEV), 2016 5th International Conference, IEEE, 2016, pp. 145–150.
Inbarani, H.H., Azar, A.T., and Jothi, G., Supervised hybrid feature selection based on PSO and rough sets for medical diagnosis, Comput. Methods Progr. Biomed., 2014, vol. 113, no. 1, pp. 175–185.
Tomar, D. and Agarwal, S., Feature selection based Least Square Twin Support Vector Machine for diagnosis of heart disease, Int. J. Bio-Sci. Bio-Technol., 2014, vol. 6, no. 2, pp. 69–82.
Reddy, G.T. and Khare, N., An efficient system for heart disease prediction using hybrid OFBAT with rule-based Fuzzy Logic Model, J. Circuits, Syst. Comput., 2017, vol. 26, no. 04, p. 1750061.
Pimentel, A., Coronary heart disease prognosis using machine-learning techniques on patients with type 2 Diabetes Mellitus, in Ubiquitous Machine Learning and Its Applications, IGI Global, 2017, pp. 89–112.
Author information
Authors and Affiliations
Corresponding author
Additional information
The article is published in the original.
Rights and permissions
About this article
Cite this article
Vijayashree, J., Sultana, H.P. A Machine Learning Framework for Feature Selection in Heart Disease Classification Using Improved Particle Swarm Optimization with Support Vector Machine Classifier. Program Comput Soft 44, 388–397 (2018). https://doi.org/10.1134/S0361768818060129
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0361768818060129