Abstract
Epichloid endophytes are well known symbionts of many cool-season grasses that may alleviate environmental stresses for their hosts. For example, endophytes produce alkaloid compounds that may be toxic to invertebrate or vertebrate herbivores. Achnatherum robustum, commonly called sleepygrass, was aptly named due to the presence of an endophyte that causes toxic effects to livestock and wildlife. Variation in alkaloid production observed in two A. robustum populations located near Weed and Cloudcroft in the Lincoln National Forest, New Mexico, suggests two different endophyte species are present in these populations. Genetic analyses of endophyte-infected samples revealed major differences in the endophyte alkaloid genetic profiles from the two populations, which were supported with chemical analyses. The endophyte present in the Weed population was shown to produce chanoclavine I, paspaline, and terpendoles, so thus resembles the previously described Epichloë funkii. The endophyte present in the Cloudcroft population produces chanoclavineI, ergonovine, lysergic acid amide, and paspaline, and is an undescribed endophyte species. We observed very low survival rates for aphids feeding on plants infected with the Cloudcroft endophyte, while aphid survival was better on endophyte infected plants in the Weed population. This observation led to the hypothesis that the alkaloid ergonovine is responsible for aphid mortality. Direct testing of aphid survival on oat leaves supplemented with ergonovine provided supporting evidence for this hypothesis. The results of this study suggest that alkaloids produced by the Cloudcroft endophyte, specifically ergonovine, have insecticidal properties.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wild grasses have evolved symbiotic relationships with endophytic fungi that cope with multiple abiotic and biotic stresses (Cheplick and Faeth 2009; Kannadan and Rudgers 2008). The most well studied of these fungal endophytes are Epichloë species that systemically infect many cool-season pooid grasses. The most pronounced and well known effect of these endophytes is the production of bioactive alkaloids that can protect their host from vertebrate and invertebrate herbivores and pathogens (Clay 1996; Crawford et al. 2010). Epichloid alkaloids are grouped into four classes: ergot alkaloids (e.g., chanoclavine, ergonovine, and ergovaline), lolines (e.g., N-acetylnorloline and N-formylloline), indole-diterpenes (e.g., terpendole C and lolitrem B), and peramine. Each has varying biological activity against vertebrate or invertebrate herbivores (Panaccione et al. 2014). A given endophyte may produce alkaloids from one or more classes, and multiple alkaloids from within each class (Schardl et al. 2013a, c). Genome sequencing of multiple Epichloë species has indicated that the source of variation for the type of alkaloid produced stems mainly from the remarkable variation in presence of alkaloid genes among Epichloë species and strains (Schardl et al. 2013c). Hybrid Epichloë species, by the nature of arising from multiple progenitors, have the potential to increase genetic variation for alkaloid production (Schardl and Craven 2003; Schardl et al. 2012, 2013a, c). However, environmental factors, such as soil nutrients or herbivore grazing, may modulate alkaloid levels (Bultman et al. 2004; Hunt et al. 2005). Accumulating evidence also indicates that Epichloë species and strains vary greatly not only among grass species but also within a single grass species. For example, Hordelymus europaeus (Oberhofer and Leuchtmann 2012), Festuca arizonica (Sullivan and Faeth 2008), and Bromus laevipes (Charlton et al. 2014) can harbor both hybrid and nonhybrid Epichloë species.
Achnatherum robustum (formerly Stipa robusta) is native to mountainous areas of the southwestern USA, and is commonly known as sleepygrass because of its long-recognized toxic and narcotic effects on livestock (Jones et al. 2000). Indeed, sleepygrass is one of the relatively few epichloid-infected native grasses known to be highly toxic to vertebrates (Faeth 2002b). The toxic effects are due presumably to ergot alkaloids (ergonovine and lysergic acid amide) (Petroski et al. 1992) produced by an asexual, seed-borne epichloid endophyte. Infected grasses with high levels of ergot alkaloids occur in a restricted range of A. robustum near Cloudcroft, NM in the Lincoln National Forest (Faeth et al. 2006). Endophyte-infected A. robustum from this location show very high levels of the ergot alkaloids ergonovine (EN), lower levels of lysergic acid amide (LAA), isolysergic amide, and much lower levels of ergonovinine. The presence of these alkaloids could explain the toxic effects of sleepygrass on livestock, such as narcotized sleep, elevated body temperature, weakness, frequent urination, dizziness, hyper salivation, diarrhea, and potential death (Miles et al. 1996; Petroski et al. 1992). Cytotoxic effects to animal muscle tissue also have been described for ergonovine and ergonovinine (Zhang et al. 2014). Although less well studied, ergot alkaloids also may have deterrent and toxic effects on invertebrate herbivores (Panaccione et al. 2014; Schardl et al. 2013a). In contrast to the Cloudcroft population, endophyte-infected A. robustum from other nearby and distant populations do not produce ergot alkaloids such as those found in the Cloudcroft population. One of these populations is located within 22 km from Cloudcroft in the Lincoln National Forest near Weed, NM, USA (Faeth et al. 2006).
It is likely that A. robustum is a host for more than one endophyte species, based upon dramatic differences in alkaloids produced among different endophyte-infected plants (Faeth et al. 2006). Presently, only one endophyte species, Epichloë funkii (formerly Neotyphodium funkii) has been described from A. robustum (Leuchtmann et al. 2014; Moon et al. 2007). Epichloë funkii, a hybrid endophyte with E. elymi and E. festucae ancestral progenitors, was described based on a single plant collection in Colorado, USA (Moon et al. 2007). Recent draft genome sequence of E. funkii indicates the presence of EAS biosynthesis genes required for production of chanoclavine I, an early ergot alkaloid pathway intermediate, IDT/LTM biosynthesis genes required for production of terpendoles from the indole-diterpene pathway, and the perA gene required for peramine production (Schardl et al. 2013c). To date, however, only chanoclavine I has been detected from E. funkii-infected plant tissues, while peramine and indole-diterpenes have not been analyzed (Schardl et al. 2013a, c). The alkaloid genetic profile of E. funkii does not support the production of ergonovine, yet ergonovine is found at high levels in endophyte-infected sleepygrass plants from the Cloudcroft population. Therefore, this evidence suggests that a different Epichloë species with the capability to produce ergonovine also infects A. robustum.
Our goal was to examine the variation in epichloid endophytes, their alkaloid genes and products, and the ecological consequences for herbivores, in two disjunct, but nearby A. robustum populations (Cloudcroft and Weed). We tested whether the endophytes and their associated alkaloids differentially affected herbivores via a standard insect bioassay with aphids. To test for a mechanism underlying the observed variation in aphid resistance, we tested the anti-herbivore properties of a specific ergot alkaloid, ergonovine, in controlled experiments.
Methods and Materials
Field Plants
To study endophyte infection status (E), variation in production of the alkaloids ergonovine and lysergic acid amide (A) and alkaloid levels, we sampled A. robustum plants established in 2005 in an experimental plot at the Arboretum of Flagstaff, Flagstaff, Arizona (Faeth et al. 2010). The experimental plot included three groups: uninfected plants (E-A-), endophyte-infected not producing ergonovine (E+A-), and endophyte-infected producing ergonovine (E+A+). Seed used for this plot (Table 1), originated from Cloudcroft and Weed natural populations, New Mexico, USA collected during 2001–2004 (Faeth et al. 2006). Each group was organized from multiple seedlings grown from 1 to 2 maternal plants. In September 2011, one tiller per plant was collected, checked for endophyte infection, and stored at −20 °C for alkaloid analyses. In May 2013, we recollected several plant samples from each group and tested for endophyte infection, ergot alkaloid production, and endophyte alkaloid genotype. Achnatherum robustum is an obligate outcrossing species, and plants were allowed to naturally pollinate each other within the experimental plot to produce seed.
Maintenance of Greenhouse Plants
To establish plants for herbivory experiments, chemotyping, and genotyping, second generation seeds originating from the Cloudcroft and Weed populations (2010 collection from the experimental plot) were planted on January 5, 2011 in potting mix soil (Timberline, USA) in 300 ml pots. Seedlings were grown in the greenhouse at 25 °C/ 22 °C day/night temperatures and natural light conditions and fertilized with 20:20:20 soluble fertilizer with minor elements (Southern Agricultural Insecticides, Inc., Hendersonville, NC, USA) twice a month. One tiller was sampled prior to the herbivory experiment (April 2011) to determine endophyte infection status and alkaloid production. This sampling was repeated after the herbivory experiment (January 2012) to confirm endophyte infection. Samples for genetic studies were taken in December 2012.
Detection of Endophyte Infection Status
The Phytoscreen Immunoblot Kit “Neotyphodium Field Tiller” (Agrinostics, Ltd. Co, GA, USA) was used to determine the infection status of all plant samples. One tiller per plant was tested by imprinting the base of the tiller onto nitrocellulose paper to detect endophyte presence by immunoblot analysis, while the remainder of the tiller was retained for chemical analysis. Fresh samples were used from greenhouse plants and frozen samples from field plants.
Alkaloid Extraction
Leaf samples were freeze-dried and extracted with 95 % methanol (40 mg in 1 ml) at 5 °C for 48 h. The extract was filtered through a 0.22 μm spin filter (Corning Inc.), air dried, and re-dissolved in 17 % aqueous methanol. The resulting extracts were stored at 5 °C until time of analysis.
Lysergic Acid Amide and Ergonovine Analysis
To detect and quantify ergot alkaloids, HPLC-HESI-MS analyses were performed on a triple quadruple mass spectrometer (TSQ Quantum, Thermo, San Jose, CA, USA) interfaced to an HPLC system with photodiode array detector (monitored at 300 nm) and quaternary pump (Agilent HP1100 series). A binary solvent composition of aqueous 0.1 % formic acid (solvent A) and 0.1 % formic acid in methanol (solvent B) was employed with a flow rate of 0.20 ml/min on a C18 column (50 × 2.1 mm, 3 μm particle size, Prevail packing, Grace, Deerfield, IL, USA). Separation was achieved using a linear gradient that initiated at 95%A:5%B (v/v) and remained isocratic from 0 to 4 min; decreased linearly from 95%A:5%B to 90%A:10%B from 4 to 5 min; from 90 %A:10 %B to 70 %A:30 %B from 5 to 11.5 min; from 70 %A:30 %B to 10 %A:90 %B from 11.5 to 11.6 min; remained isocratic at 10 %A:90 %B from 11.6 to 16 min; increasing from 10 %A:90 %B to 95 %A:5 %B from 16 to 16.1 min; and remained isocratic at 95 %A:5 %B from 16.1 to 24 min.
Aqueous solutions of the ergot alkaloids lysergic acid amide tartrate (98 % pure by LC-MS) and ergonovine maleate (Sigma-Aldrich, 100 % pure by TLC) were employed as standards for quantitation. The mass spectrometer was operated in the positive ion mode with a 0.1 s scan time and a scan width of 0.5 m/z. Quantification was performed using selected reaction monitoring (SRM) with a 268 to 208 transition for lysergic acid amide and a 326 to 223 transition for ergonovine. Alkaloid quantities were calculated by linear regression of the relevant calibration curves.
Analysis of N-acetylnorloline, Chanoclavine I, and Peramine
Loline alkaloids, chanoclavine I, and peramine were analyzed by using ultra performance liquid chromatography – high resolution mass spectrometry (UPLC-HRMS) on an Orbitrap mass spectrometer with electrospray ionization (ESI) source (LTQ Orbitrap XL, Thermo, San Jose, CA, USA) coupled to Acquity UPLC (Waters Corp., Milford, MA, USA). A hydrophilic interaction chromatography (HILIC) column (150 × 2.1 mm, 5 μm particle size, 120 Å pore size, Alltima packing, Grace, Deerfield, IL, USA) was utilized for the analysis of all extracts, with a 0.3 ml/min flow rate and a 3 μl injection volume. The samples were analyzed using the following gradient composition, where A = 0.1 % formic acid in (acetonitrile) and B = 0.1 % formic acid in (water), 95.1 % A from 0 to 8 min. Mass spectrometric detection was conducted in the positive ion mode with a scan range of 75–300 m/z. Capillary temperature was 275 °C, sheath gas pressure was 20 (arbitrary units), and spray, capillary, and tube lens voltages were 4.5 kV, 20 V, and 100 V, respectively. For comparison, this method was applied to the analysis of endophyte-infected Elymus canadensis (strain NFe746), and the alkaloids N-acetylnorloline, peramine, and chanoclavine I were all detected, consistent with previous literature (Charlton et al. 2012; Clay and Schardl 2002; Schardl et al. 2013c). A synthetic standard of N-acetylnorloline also was analyzed as a positive control.
Indole-Diterpenes Chemical Analysis
Indole-diterpene analyses were performed by AgResearch in New Zealand using LC-MS/MS according to Rasmussen et al. (2012).
DNA Extraction and Chemoprofiling
Tillers from greenhouse and field plants were evaluated for the presence of associated Epichloë species, and the endophyte was characterized using PCR. DNA was isolated from plant material with MagAttract 96 DNA Plant Core Kit (QIAGEN Inc.) according to manufacturer’s instructions. PCR with six multiplex primers sets were used to determine endophyte infection status, mating type, and genes present at each alkaloid loci as described in Charlton et al. (2014). In addition, the multiplex three primer set included primers, dmaW818(311 + 21)d (5′-AACCCATCAACGGAGCAACTG) and dmaW818(1068 + 21)u (5′-GCCAAACACTGTGAAATACACCTG), designed to the E. gansuensis var. inebrians e818 dmaW EN gene required for ergonovine production (L. Chen, C. L. Schardl unpublished).
Aphid Biological Assay
An aphid bioassay was employed to test the effects of endophytic alkaloids from different endophyte-infected A. robustum on herbivore resistance (e.g., Cheplick and Faeth 2009). In total, 101 greenhouse grown plants originating from the Cloudcroft population and 54 plants from the Weed population were evaluated. Twenty seven plants from the Cloudcroft population with total ergonovine plus lysergic acid amide (EN+LAA) ergot alkaloid levels greater than 26.7 μg/kg (at the age of 3 months) were selected for one group, and 26 infected plants from the Weed population with no detectable ergonovine and lysergic acid amide alkaloids were selected for the other group. Two Rhopalosiphum padi L. aphid populations were used for this experiment: wild NC (North Carolina) origin (collected in Greensboro, NC) and NY (New York) origin (obtained from the UNC-Chapel Hill collection). The NY population has been observed to be more tolerant to endophytic alkaloids (M. Dekker, pers. communication). Rhopalosiphum padi has been used commonly to bioassay the effects of endophytic alkaloids on herbivores (Leuchtmann et al. 2000; Saari et al. 2014). Aphids were reared on oat (Avena sativa) plants, so they were naïve to fungal alkaloids (oats do not produce alkaloids). This experiment continued for 30 day in October-November 2011 when plants were 10 month old. Initially, three aphids were placed on A. robustum plants enclosed with clear plastic cups and thin fabric secured on top for air exchange. Every 3 days, wingless and winged aphid numbers were recorded, and an additional three aphids were added to each plant to maintain populations. Both wingless and winged forms were recorded because aphids may produce winged forms when host plant quality deteriorates (Braendle et al. 2006; De Barro 1992).
Bioassay to Test Anti-Herbivore Activity of Ergonovine
To test the direct effects of the ergot alkaloid ergonovine on aphid herbivores, 20 one-wk-old oat (Avena sativa) seedlings (seed material from Nasco, Fort Atkinson, WI, USA) were cut at soil level and placed into an aqueous ergonovine solution (1.5 ppm, 1 ml) in a microcentrifuge tube covered with aluminum foil. Each leaf was secured in the tube with a small piece of sponge. Ergonovine was adsorbed naturally due to transpiration. A 15 ml clear plastic centrifuge tube with the end cut off was inverted to cover the leaf in the microfuge tube, and the hole was closed with a small roll of KimWipes to allow some gas exchange. Five R. padi (NY) aphids of 3rd and 4th instar were added to each leaf. For the control group, deionized water was used in place of the ergonovine solution. Plants were placed in a growth chamber at 25 °C with 16 h of L/D for 4 days. All aphids were counted, and leaves were freeze-dried to determine the ergonovine concentration. Extraction and LC-MS analysis of ergonovine levels in three control and 20 ergonovine treated leaves was performed as described above. We did not have sufficient lysergic acid amide, the second candidate for insecticidal properties, to test the direct effects on aphids.
Statistical Analysis
RGui 32-bit software with R Commander Package was used for statistical analyses. For ergot alkaloid concentration measurements, averages and population standard deviations were determined. For the ergonovine testing bioassay, we used aphid means with SE counts; one-way ANOVA test was performed to determine the difference between the treatment groups, and a simple linear regression model was used to test the effect of ergonovine concentration on aphid numbers. Data from the aphid biological assay was non-normally distributed, so we used rank transformation and Wilcoxon nonparametric tests for comparing the differences at each of ten measurements between two plant and two aphid populations. Because of repeated measures, overall aphid numbers between populations also were compared with Hotelling’s T 2 test for ranked data. To test differences in the collective number of wingless and winged forms over all time periods, we used the Pearson’s Chi-square test.
Results
Infection Status and Ergot Alkaloid Levels in Seedlings from Cloudcroft and Weed Populations
Differences were observed in alkaloid content and endophyte infection status between 3 month old seedlings originating from Cloudcroft and Weed populations. When the endophyte infection status was determined by immunoblot analysis for 155 greenhouse three-month old seedlings, only the Weed population tested positive for endophyte infection, while all Cloudcroft seedlings appeared to be endophyte free. However, chemical analysis revealed the presence of ergot alkaloids (ergonovine and lysergic acid amide) at varying levels in 74 out of 101 Cloudcroft population seedlings (Table 2) despite negative immunoblot results. All 54 plants from the Weed population seedlings tested negative for the presence of ergot alkaloids, ergonovine, and lysergic acid amide.
Infection Status and Ergot Alkaloid Production in Field Plot Plants
Endophyte infection status and ergot alkaloid analysis of 105 adult plants originating from all four mother plants from the Cloudcroft and Weed populations were determined (Table 3). Endophyte infection was detected by the immunoblot method from the adult plants for both populations. We detected seven endophyte-free plants out of 59 Cloudcroft plants and six endophyte-free plants out of 46 plants from the Weed population. Surprisingly, the purported E-A- group (Faeth et al. 2006) from Weed mother plant 5–91 had only four uninfected plants from the total of 21 plants (Table 3), suggesting that the original mother plant was mistakenly identified as uninfected. The original infection status of the majority (23 of 25) of the E+A- group plants was confirmed by immunoblot. As expected, the ergot alkaloids ergonovine and lysergic acid amide were detected only from plants that originated from Cloudcroft, E+A+ group. Ergonovine levels in dry plant tissues ranged from 0 to 2.67 μg/g, and lysergic acid amid levels ranged from 0 to 1.18 μg/g (Table 3).
Genetic and Chemical Variation of Endophytes from Two Populations
Infection status, mating type, and alkaloid gene profiles were determined for 26 Cloudcroft and nine Weed samples that originated from all mother plants used in our aphid experiments. Within each population, endophytes from all mother plants had the same genetic profiles represented in Fig. 1. However, the endophytes from the Weed and Cloudcroft populations are genetically distinct from each other (Fig. 1). The endophyte from the Weed population resembles E. funkii (Schardl et al. 2013c), whereas the endophyte from the Cloudcroft population is distinct in mating type and alkaloid gene profiles.
The endophytes from each of the locations have different mating types (Fig. 1, Table 4). The Cloudcroft endophyte contains both mating type idiomorphs, MTA and MTB, which indicates a hybrid origin. The endophyte from the Weed population has one mating type MTB, but probably is also a hybrid where both ancestral progenitors were MTB. In addition, the endophytes from each location contained different alkaloid gene profiles that suggest they are capable of producing different alkaloids (Fig. 1). The presence or absence of key pathway genes allowed us to predict the likelihood of an alkaloid being produced based on our knowledge of the associated biosynthetic pathways (Schardl et al. 2013b).
Two dmaW markers were used to identify variation at the EAS locus. The dmaW ERV and dmaW EN markers were designed to different dmaW alleles identified within Epichloë species. The dmaW ERV marker was designed for species that are able to produce chanoclavine (e.g., E. elymi E56) or ergovaline (e.g., E. festucae Fl1); while dmaW EN is specific for ergonovine producers such as E. gansuensis var. inebrians from Achnatherum inebrians (Schardl et al. 2013b). The presence of only dmaW ERV and easC in the Weed population endophyte is suggestive of a chanoclavine producer, as markers to the later EAS pathway genes were not detected. The endophyte present in the Cloudcroft population has the dmaW EN and easC markers (Fig. 1, Table 4), and this profile has been associated with ergonovine and lysergic acid amide producers (L. Chen, C. L. Schardl unpublished). It is likely that other ergonovine pathway specific genes exist in the Cloudcroft endophyte but these were not tested for in our study.
Markers for the perA gene encoding peramine synthetase were detected only in endophyte-infected plants from the Weed population. Three of the four perA markers produced PCR bands, while the expected band for the perA reductase domain was faint, so it is unclear if this gene is likely to encode a functional protein, and if peramine would be produced (Fig. 1). The complete absence of all PER markers in the Cloudcroft population endophyte indicates this endophyte likely lacks the perA gene and would be unable to make peramine (Table 4).
Variation also was identified within the IDT/LTM locus of the endophytes from each population (Fig. 1). The endophyte from the Weed population contained the markers for idtG, idtF, and idtQ. Based on this genetic profile, we would predict this endophyte could produce early indole-diterpene products, such as paspaline, paxiline, and some terpendoles. The endophyte present in the Cloudcroft population contained the markers idtG and ltmF. The absence of a product for idtQ suggests that the indole-diterpene pathway for this endophyte may be blocked early, which would result in the biosynthesis of paspaline (Fig. 1, Table 4).
Neither the Cloudcroft nor the Weed population endophytes have the potential for loline alkaloid production. PCR products for the LOL markers were not detected from samples of endophyte-infected material from the Weed population. Although the endophyte in the Cloudcroft population contained lolA, a gene associated with the LOL gene cluster, the presence of this gene alone will not support synthesis of any loline compounds (Fig. 1, Table 4) (Schardl et al. 2013b).
Genetic analysis to detect the presence of key genes from each alkaloid locus provides knowledge of alkaloids that could be produced within the different endophyte-infected populations (Table 5). To date, ergot alkaloids were expected only from endophyte-infected plants from the Cloudcroft population, as ergonovine and lysergic acid amide have previously been detected (Faeth et al. 2006). However, the genetic profile of the endophyte from the Weed population indicated the capacity of this endophyte to produce chanoclavine, which was confirmed by chemical analysis. Peramine production was not detected in the Weed population, thus supporting the likelihood that the perA gene is not functional. Chemical analysis for indole-diterpenes confirmed their presence in plant tissues from both populations. Paspaline was detected in infected plants from both populations. Furthermore, as predicted by genetic analysis, terpendoles E, I, J, and C were detected only from Weed population plants. As expected based upon genetic profiles, no lolines, which are well known insecticides (e.g. Panaccione et al. 2013; Siegel et al. 1990), were detected in infected plants from either population.
Response of Aphids to Endophyte-Infected Plants
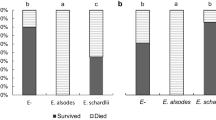
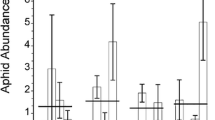
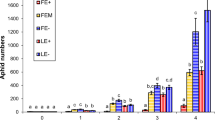
Aphid numbers were significantly lower on the Cloudcroft population than Weed population plants at each of the 10 sampling periods (P < 0.001) and across all dates (Hotelling’sT 2 P < 0.001). Aphids on most of the Cloudcroft plants did not survive at all during sample periods, so it was necessary to add more aphids to all plants after each aphid count. In contrast, aphid numbers increased over time on the Weed population plants (Fig. 2). The two aphid strains did not differ (Hotelling’s T 2 P = 0.39) in overall numbers on plants from each of the Weed and Cloudcroft populations, although the NY aphid strain had higher mean population sizes, especially on Weed plants. Aphids reared on the endophyte-infected Weed population plants produced more winged forms (20 and 11 % for NY and NC aphids, respectively), than on Cloudcroft plants (13 and 4 % for NY and NC aphids, respectively). However, increased proportion of winged forms likely stems from denser aphids populations on Weed plants due to decreased host plant quality (Braendle et al. 2006; De Barro 1992) rather than host toxicity, since very few aphids survived on any Cloudcroft plants.
Aphid Performance From the Ergonovine Insecticidal Bioassay
Aphids reared on oat leaves supplemented with ergonovine had reduced mean numbers (8.4 ± 1.0 SE) when compared to aphids reared on control plants (12.2 ± 1.3 SE) (ANOVA F 1,38 = 5.348, P = 0.026). The mean concentration of ergonovine in the oat leaves after treatment was 0.123 ± 0.05 μg/kg, approximately 8.3 times lower than the ergonovine concentration measured from field plants (Table 3). However, in a linear regression model of actual concentrations vs. aphid numbers, ergonovine concentration measurements at the end of the treatment had marginally negative relationship on aphid numbers (P = 0.087). Because experimental levels of ergonovine were low relative to naturally-infected plants, our assay suggests that even very low levels of ergonovine may reduce aphid numbers, presumably via reduced aphid performance and survival.
Discussion
It is well established that systemic Epichloë species infecting native grasses are genetically diverse among grass species (Leuchtmann et al. 2014; Schardl et al. 1997; Schardl and Phillips 1997). More recent evidence suggests that endophytes also can be highly variable within wild grass species, whereby endophyte diversity identified within a single host species can be due to different endophyte species or different strains of the same endophyte species representing different alkaloid potential (Charlton et al. 2012, 2014; Iannone et al. 2012; Kang et al. 2011; Moon et al. 2004; Oberhofer and Leuchtmann 2012; Takach et al. 2012; Takach and Young 2014; Wali et al. 2007). Genetic variation among endophytes can lead to phenotypic changes in host grasses that may be greater than that caused by endophyte infection per se (Morse et al. 2007; Oberhofer et al. 2014). These phenotypic changes caused by endophyte genetic variation, especially in alkaloid production, can then profoundly affect competing plant species, herbivores, and natural enemies of herbivores (Cheplick and Faeth 2009).
Our results indicate two genetically distinct endophytes inhabit two A. robustum populations in close proximity (22 km apart). Based on genotype and chemotype, the endophyte from the Weed population resembles E. funkii, described by Moon et al. (2007) from an A. robustum Colorado population and analyzed for alkaloid gene diversity by Schardl et al. (2013a, c). Phylogenetic data are needed to confirm that the Weed endophyte is indeed E. funkii. Interestingly, the Cloudcroft endophyte that likely has been responsible for the name “sleepygrass” due to its well-renowned toxicity to livestock is an undescribed new Epichloë species. A forthcoming paper (M. Oberhofer, T. Shymanovich, C. Young, and S. Faeth, unpublished data) will include detailed genetic and morphological data that will describe this endophyte species. Notably, this endophyte appears to be restricted to the Cloudcroft region in the distribution range of A. robustum, whereas the endophyte identified from the Weed population is more widespread based upon absence of ergonovine production (Faeth et al. 2006; Jones et al. 2000) and endophyte phylogeny (Moon et al. 2004, 2007). Similarity can be seen between the A. robustum Cloudcroft population endophyte with E. gansusensis var. inebrians from A. inebrians hosts in China. Each is known to produce ergonovine and lysergic acid amide, although E. gansusensis var. inebrians also can produce lysergic acid α-hydroxyethylamide (Schardl et al. 2013b). Similarly, A. inebrians also is known to be a host for two different endophytes, E. gansusensis var. inebrians, the likely causal agent of “drunken horse grass”, and E. gansuensis that is unable to produce ergot alkaloids (Moon et al. 2007; Schardl et al. 2013b).
As we predicted, A. robustum hosts two different endophytes, which have different effects on insect herbivore performance and survival. Aphid survival and abundances were reduced on infected plants from the Cloudcroft population in comparison to endophyte-infected plants from the Weed population. The main difference observed in the endophyte-infected Cloudcroft population as compared to the endophyte-infected Weed population was the presence of the ergot alkaloids ergonovine and lysergic acid amide. Ergot alkaloids are thought to be effective mainly against vertebrate herbivores (Jackson et al. 1987; Zavos et al. 1987; Zhang et al. 2014). Nonetheless, it seems that ergonovine and possibly lysergic acid amide, produced in the Cloudcroft population, are likely candidates for anti-herbivore effects against aphids. Moreover, there is growing evidence that some ergot alkaloids from different groups, ergopeptines, clavines, and simple amides of lysergic acid possess insecticidal and nematicidal activities (Panaccione et al. 2014; Potter et al. 2008). Ergonovine causes feeding inhibition of Japanese beetle, Popillia japonica, grubs (Patterson et al. 1991). The adult black lawn beetle, Heteronychus arator, showed a moderate reduction of artificial feed consumption in the presence of ergonovine but not lysergic acid amide (Ball et al. 1997). Likewise, ergonovine caused weight reduction in fall armyworm, Spodoptera frugiperda, larvae but not reduction in leaf area consumed (Clay and Cheplick 1989). Consistent with these findings, our bioassays with ergonovine-treated A. sativa leaves confirmed that the ergot alkaloid ergonovine reduced aphid survival and reproduction. Our study is the first to indicate that ergonovine has insecticidal activity against sucking insects such as aphids. Moreover, reduction in aphid number occurs even when ergonovine is at very low levels. Because endophyte-infected A. robustum plants had ergonovine levels more than eight times higher than our experimental assay, we would expect even much stronger effects of ergonovine in plants in natural populations. Although ergonovine seems like a probable candidate for the reduced aphid numbers on Cloudcroft plants, other alkaloidal and non-alkaloidal differences (e.g., nutritional or water content) cannot be ruled out.
There have been two previous studies (Faeth et al. 2010, Jani et al. 2010) on the effects of infection in sleepygrass on herbivory or herbivore abundances and species richness. Most relevant to the current study, Faeth et al. (2010) showed that infected plants from the Weed population reduced seed dry biomass and reproductive effort under ambient herbivory treatments compared to conditions of greatly reduced herbivory. In contrast, seed production and reproductive effort of infected plants from the Cloudcroft population were equivalent under ambient and reduced herbivory, suggesting a protective effect of infection and alkaloids in this population. However, Jani et al. (2010) found that natural enemies of herbivores also may be affected by alkaloids in infected plants in the Cloudcroft population. They found that abundances and species richness of herbivores and natural enemies was greater and lower, respectively, on sleepygrass plants with high ergot alkaloids compared to plants with low or no ergot alkaloids. They concluded that high alkaloid plants may provide “enemy-reduced” space for specialist herbivores and, thus, herbivory could be greater on infected grasses with high alkaloid levels. Therefore, whether endophytes that produce ergonovine or other alkaloids in sleepygrass reduce, increase, or have no effect on herbivory in nature likely depends on the herbivore species and the presence of natural enemies.
Alkaloid synthesis is energetically and nutritionally costly because alkaloids contain nitrogen that is often limiting in southwestern USA soils (Faeth 2002a; Faeth and Sullivan 2003). From this perspective, production of alkaloids that are diverse yet part of the same biosynthetic pathway and effective against both vertebrate and invertebrate herbivores may be more efficient at protecting the host than alkaloids produced from multiple pathways. The ergot alkaloids ergonovine and lysergic acid amide are known to have toxic effects on vertebrates (Oliver et al. 1993; Schiff 2006), and ergonovine, according to our study and other sources (Ball et al. 1997; Clay and Cheplick 1989; Patterson et al. 1991), has insecticidal or insect deterring properties. The Cloudcroft endophyte is devoid of LOL and PER genes required for loline and peramine production, and although it is capable of producing indole-diterpenes, the IDT pathway is greatly reduced. Thus, the endophyte from the Cloudcroft population may provide host protection against both vertebrate and invertebrate herbivores through the production of alkaloids from a single biosynthetic pathway.
Although the endophyte present in the Weed population also is capable of producing alkaloids from the EAS and IDT pathways, the compounds produced were different from the Cloudcroft endophyte and were not efficient at providing protection against aphids. Moreover, the perA gene encoding peramine synthetase for the insect feeding deterrent peramine (Tanaka et al. 2005) is present in the Weed endophyte, but it appears to be non-functional, and peramine was not detected in endophyte-infected samples from the Weed population. We predict that the endophyte in the Weed population affords less protection against both invertebrate and vertebrate herbivores based upon its alkaloid potential (chanoclavine and terpendole production). The environment could influence host fitness benefits provided by the endophyte. If herbivory is reduced or resources are more limiting within an environment, reduction of alkaloid pathways may lower the metabolic cost of maintaining alkaloid defenses. Interestingly, the Weed and Cloudcroft populations are only a short distance apart, yet they vary in rainfall and soil nutrients. The Weed location has lower rainfall and fewer nutrients than Cloudcroft (Tong Jia et al. unpublished). Thus, it is possible that the Weed environment selected for persistence of an endophyte that produced fewer alkaloids due to higher costs and lower benefits, or that this endophyte provides another yet to be discovered benefit.
Epichloid endophytes that associate with A. robustum are challenging to work with compared to other infected grass species. Traditional detection methods such as microscopic examination of leaf tissue and seeds or culturing the endophyte for identification have been unreliable. For example, the endophyte within the Cloudcroft population is slow growing and has only been isolated successfully from the seeds after a 5 month growth period (M. Oberhofer et al. unpublished). Similarly, endophyte infection in 3-month-old Cloudcroft plants could not be detected by the more reliable and commonly used tissue-print immunoblot method. Additionally, vertical transmission rates appear much lower (T. Shymanovich, personal obs.) than other asexual endophytes (Afkhami and Rudgers 2008). However, unlike detection of the endophyte itself, we could reliably detect ergot alkaloids in endophyte-infected Cloudcroft plant material in 3-month-old seedlings. In contrast, the Weed endophyte was easily detected by the immunoblot method. By using multiple detection methods, genotypic profiling, and alkaloid analyses, we are confident of the endophyte infection status and genotype of endophyte-infected material used in this study.
Our study shows that natural populations of cool-season grasses can harbor genetically different endophyte species or genotypic variants. In turn, these endophytes can have different effects on host plant phenotypes through the production of bioactive alkaloids. In our study, two genetically distinct endophytes from A. robustum produced different alkaloid compounds, resulting in varying resistance to aphid herbivores. Although ergot alkaloids are traditionally viewed as active against vertebrates, at least one ergot alkaloid, ergonovine, from the Cloudcroft population has insecticidal properties against aphids, even at low levels.
References
Afkhami ME, Rudgers JA (2008) Symbiosis lost: imperfect vertical transmission of fungal endophytes in grasses. Am Nat 172:405–416. doi:10.1086/589893
Ball OJP, Miles CO, Prestidge RA (1997) Ergopeptine alkaloids and Neotyphodium lolii-mediated resistance in perennial ryegrass against adult Heteronychus arator (Coleoptera: Scarabaeidae). J Econ Entomol 90:1382–1391
Braendle C, Davis GK, Brisson JA, Stern DL (2006) Wing dimorphism in aphids. Heredity 97:192–199. doi:10.1038/sj.hdy.6800863
Bultman TL, Bell G, Martin WD (2004) A fungal endophyte mediates reversal of wound-induced resistance and constrains tolerance in a grass. Ecology 85:679–685. doi:10.1890/03-0073
Charlton ND, Craven KD, Mittal S, Hopkins AA, Young CA (2012) Epichloë canadensis, a new interspecific epichloid hybrid symbiotic with Canada wildrye (Elymus canadensis). Mycologia 104:1187–1199. doi:10.3852/11-403
Charlton ND, Craven KD, Afkhami ME, Hall BA, Ghimire SR, Young CA (2014) Interspecific hybridization and bioactive alkaloid variation increases diversity in endophytic Epichloë species of Bromus laevipes. FEMS Microbiol Ecol. doi:10.1111/1574-6941.12393
Cheplick GP, Faeth SH (2009) Ecology and evolution of the grass-endophyte symbiosis. Oxford University Press, Oxford
Clay K (1996) Interactions among fungal endophytes, grasses and herbivores. Res Popul Ecol 38:191–201. doi:10.1007/Bf02515727
Clay K, Cheplick GP (1989) Effect of ergot alkaloids from fungal endophyte-infected grasses on fall armyworm (Spodoptera-Frugiperda). J Chem Ecol 15:169–182. doi:10.1007/Bf02027781
Clay K, Schardl C (2002) Evolutionary origins and ecological consequences of endophyte symbiosis with grasses. Am Nat 160:S99–S127. doi:10.1086/342161
Crawford KM, Land JM, Rudgers JA (2010) Fungal endophytes of native grasses decrease insect herbivore preference and performance. Oecologia 164:431–444. doi:10.1007/s00442-010-1685-2
De Barro PJ (1992) The role of temperature, photoperiod, crowding and plant quality on the production of alate viviparous females of the bird cherry-oat aphid, Rhopalosiphum padi. Entomol Exp Appl 65:205–214. doi:10.1111/j.1570-7458.1992.tb00673.x
Faeth SH (2002) Are endophytic fungi defensive plant mutualists? Oikos 99:200–200. doi:10.1034/j.1600-0706.2002.990122.x
Faeth SH, Sullivan TJ (2003) Mutualistic asexual endophytes in a native grass are usually parasitic. Am Nat 161:310–325. doi:10.1086/345937
Faeth SH, Gardner DR, Hayes CJ, Jani A, Wittlinger SK, Jones TA (2006) Temporal and spatial variation in alkaloid levels in Achnatherum robustum, a native grass infected with the endophyte Neotyphodium. J Chem Ecol 32:307–324. doi:10.1007/s10886-005-9003-x
Faeth SH, Hayes CJ, Gardner DR (2010) Asexual endophytes in a native grass: tradeoffs in mortality, growth, reproduction, and alkaloid production. Microb Ecol 60:496–504. doi:10.1007/s00248-010-9643-4
Hunt MG, Rasmussen S, Newton PCD, Parsons AJ, Newman JA (2005) Near-term impacts of elevated CO2, nitrogen and fungal endophyte-infection on Lolium perenne L. growth, chemical composition and alkaloid production. Plant Cell Environ 28:1345–1354. doi:10.1111/j.1365-3040.2005.01367.x
Iannone LJ, Novas MV, Young CA, De Battista JP, Schardl CL (2012) Endophytes of native grasses from South America: biodiversity and ecology. Fungal Ecol 5:357–363. doi:10.1016/j.funeco.2011.05.007
Jackson JA, Hemken RW, Bush LP, Boling JA, Siegel MR, Zavos PM, Yates SG (1987) Physiological-responses in rats fed extracts of endophyte infected tall fescue seed drug. Chem Toxicol 10:369–379. doi:10.3109/01480548709042993
Jani AJ, Faeth SH, Gardner D (2010) A sexual endophytes and associated alkaloids alter arthropod community structure and increase herbivore abundances on a native grass. Ecology Letters 13:106-117. doi:10.1111/j.1461-0248.2009.01401.x
Jones TA, Ralphs MH, Gardner DR, Chatterton NJ (2000) Cattle prefer endophyte-free robust needlegrass. J Range Manag 53:427–431. doi:10.2307/4003755
Kang Y, Ji YL, Zhu KR, Wang H, Miao HM, Wang ZW (2011) A new Epichloë species with interspecific hybrid origins from Poa pratensis ssp pratensis in Liyang, China. Mycologia 103:1341–1350. doi:10.3852/10-352
Kannadan S, Rudgers JA (2008) Endophyte symbiosis benefits a rare grass under low water availability. Funct Ecol 22:706–713. doi:10.1111/j.1365-2435.2008.01395.x
Leuchtmann A, Schmidt D, Bush LP (2000) Different levels of protective alkaloids in grasses with stroma-forming and seed-transmitted Epichloë/Neotyphodium endophytes. J Chem Ecol 26:1025–1036. doi:10.1023/A:1005489032025
Leuchtmann A, Bacon CW, Schardl CL, White JF, Tadych M (2014) Nomenclatural realignment of Neotyphodium species with genus Epichloë. Mycologia 106:202–215. doi:10.3852/106.2.202
Miles CO et al (1996) High levels of ergonovine and lysergic acid amide in toxic Achnatherum inebrians accompany infection by an Acremonium-like endophytic fungus. J Agric Food Chem 44:1285–1290. doi:10.1021/Jf950410k
Moon CD, Craven KD, Leuchtmann A, Clement SL, Schardl CL (2004) Prevalence of interspecific hybrids amongst asexual fungal endophytes of grasses. Mol Ecol 13:1455–1467. doi:10.1111/j.1365-294X.2004.02138.x
Moon CD, Guillaumin JJ, Ravel C, Li C, Craven KD, Schardl CL (2007) New Neotyphodium endophyte species from the grass tribes Stipeae and Meliceae. Mycologia 99:895–905
Morse LJ, Faeth SH, Day TA (2007) Neotyphodium interactions with a wild grass are driven mainly by endophyte haplotype. Funct Ecol 21:813–822. doi:10.1111/j.1365-2435.2007.01285.x
Oberhofer M, Leuchtmann A (2012) Genetic diversity in epichloid endophytes of Hordelymus europaeus suggests repeated host jumps and interspecific hybridizations. Mol Ecol 21:2713–2726. doi:10.1111/j.1365-294X.2012.05459.x
Oberhofer M, Gusewell S, Leuchtmann A (2014) Effects of natural hybrid and non-hybrid Epichloë endophytes on the response of Hordelymus europaeus to drought stress. New Phytol 201:242–253. doi:10.1111/Nph.12496
Oliver JW, Abney LK, Strickland JR, Linnabary RD (1993) Vasoconstriction in bovine vasculature induced by the tall fescue alkaloid lysergamide. J Anim Sci 71:2708–2713
Panaccione DG, Beaulieu WT, Cook D (2013) Bioactive alkaloids in vertically transmitted fungal endophytes. Funct Ecol 28:299–314
Patterson CG, Potter DA, Fannin FF (1991) Feeding deterrency of alkaloids from endophyte-infected grasses to Japanese-beetle grubs. Entomol Exp Appl 61:285–289
Petroski RJ, Powell RG, Clay K (1992) Alkaloids of Stipa robusta (sleepygrass) infected with an Acremonium endophyte. Nat Toxins 1:84–88
Potter DA, Stokes JT, Redmond CT, Schardl CL, Panaccione DG (2008) Contribution of ergot alkaloids to suppression of a grass-feeding caterpillar assessed with gene knockout endophytes in perennial ryegrass. Entomol Exp Appl 126:138–147. doi:10.1111/j.1570-7458.2007.00650.x
Rasmussen S, Lane GA, Mace W, Parsons AJ, Fraser K, Xue H (2012) The use of genomics and metabolomics methods to quantify fungal endosymbionts and alkaloids in grasses. Methods Mol Biol 860:213–226. doi:10.1007/978-1-61779-594-7_14
Saari S, Richter S, Robbins M, Faeth SH (2014) Bottom-up regulates top-down: the effects of hybridization of grass endophytes on an aphid herbivore and its generalist predator. Oikos 123:545–552. doi:10.1111/j.1600-0706.2013.00690.x
Schardl CL, Craven KD (2003) Interspecific hybridization in plant-associated fungi and oomycetes: a review. Mol Ecol 12:2861–2873
Schardl CL, Phillips TD (1997) Protective grass endophytes: where are they from and where are they going? Plant Dis 81:956–956
Schardl CL, Leuchtmann A, Chung KR, Penny D, Siegel MR (1997) Coevolution by common descent of fungal symbionts (Epichloe spp) and grass hosts. Mol Biol Evol 14:133–143
Schardl CL, Young CA, Faulkner JR, Florea S, Pan J (2012) Chemotypic diversity of epichloae, fungal symbionts of grasses. Fungal Ecol 5:331–344. doi:10.1016/j.funeco.2011.04.005
Schardl CL, Florea S, Pan J, Nagabhyru P, Bec S, Calie PJ (2013a) The epichloae: alkaloid diversity and roles in symbiosis with grasses. Curr Opin Plant Biol 16:480–488. doi:10.1016/j.pbi.2013.06.012
Schardl CL et al (2013b) Plant-symbiotic fungi as chemical engineers: multi-genome analysis of the clavicipitaceae reveals dynamics of alkaloid loci. PLoS Genet 9:e1003323. doi:10.1371/journal.pgen.1003323
Schardl CL et al (2013c) Currencies of mutualisms: sources of alkaloid genes in vertically transmitted epichloae. Toxins 5:1064–1088. doi:10.3390/toxins5061064
Schiff PL Jr (2006) Teachers’ topics: ergot and its alkaloids. Am J Pharm Educ 70:98
Siegel MR et al (1990) Fungal endophyte-infected grasses - alkaloid accumulation and aphid response. J Chem Ecol 16:3301–3315. doi:10.1007/Bf00982100
Sullivan TJ, Faeth SH (2008) Local adaptation in Festuca arizonica infected by hybrid and nonhybrid Neotyphodium endophytes. Microb Ecol 55:697–704. doi:10.1007/s00248-007-9312-4
Takach JE, Young CA (2014) Alkaloid genotype diversity of tall fescue endophytes. Crop Sci 54:667–678. doi:10.2135/cropsci2013.06.0423
Takach JE, Mittal S, Swoboda GA, Bright SK, Trammell MA, Hopkins AA, Young CA (2012) Genotypic and chemotypic diversity of Neotyphodium endophytes in tall fescue from Greece. Appl Environ Microbiol 78:5501–5510
Tanaka A, Tapper BA, Popay A, Parker EJ, Scott B (2005) A symbiosis expressed non-ribosomal peptide synthetase from a mutualistic fungal endophyte of perennial ryegrass confers protection to the symbiotum from insect herbivory. Mol Microbiol 57:1036–1050. doi:10.1111/j.1365-2958.2005.04747.x
Wali PR, Ahlholm JU, Helander M, Saikkonen K (2007) Occurrence and genetic structure of the systemic grass endophyte Epichloë festucae in fine fescue populations. Microb Ecol 53:20–29. doi:10.1007/s00248-006-9076-2
Zavos PM, Varney DR, Jackson JA, Siegel MR, Bush LP, Hemken RW (1987) Effect of feeding fungal endophyte (Acremonium-coenophialum)-infected tall fescue seed on reproductive-performance in Cd-1 mice through continuous breeding. Theriogenology 27:549–559. doi:10.1016/0093-691x(87)90242-1
Zhang X, Nan Z, Li C (2014) Cytotoxic effect of ergot alkaloids in Achnatherum inebrians infected by the Neotyphodium gansuense endophyte. J Agric Food Chem. doi:10.1021/jf502264j
Acknowledgments
We thank Melissa Robbins for aphid counts; Miranda Weavil and David Zich for technical assistance with alkaloid analysis; The Samuel Roberts Noble Foundation and Bradley Hall, Ginger Swoboda for help with PCR analysis; Li Chen and Dr. Christopher L. Schardl (University of Kentucky) for designing and providing primers to the dmaW EN; Dr. Wade Mace and Kristy Baker at AgResearch, New Zealand for indole-diterpene analyses; Dr. Jonathan Sheerer of the College of William and Mary for synthesizing N-acetylnorloline, Dr. David E. Nichols of Purdue University for synthesis of lysergic acid amide; University of North Carolina at Chapel Hill (Charles Mitchell lab) for supplying the NY aphid strain; Jonathan Newman for constructive comments. Supported by NSF grant DEB 0917741 to SHF and NBC.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shymanovich, T., Saari, S., Lovin, M.E. et al. Alkaloid Variation Among Epichloid Endophytes of Sleepygrass (Achnatherum robustum) and Consequences for Resistance to Insect Herbivores. J Chem Ecol 41, 93–104 (2015). https://doi.org/10.1007/s10886-014-0534-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10886-014-0534-x