Abstract
Epichloë species are systemic fungal endophytes that usually specialize in a certain group of related grass species. We examined the infection frequency of Epichloë festucae in populations of two fine fescue species (Festuca rubra and F. ovina) in natural and seminatural habitats at 86 study sites (total = 2514 plants) across Finland and northern Norway. Infection incidence varied significantly among grass species and populations. A substantial number of the F. rubra and F. ovina populations (53 out of 77 and 25 out of 30, respectively) were either endophyte-free or had very low (<20%) infection frequencies. The highest infection frequencies were found in subarctic areas. Moreover, infection incidence differed between habitats. In the area with the highest infection frequencies, we used microsatellite markers to study genetic diversity and the rates of gene flow of E. festucae among 12 F. rubra populations. Twenty out of the 25 fungal genotypes detected with four microsatellite markers were carrying multiple alleles in at least one locus, indicating multiple infections or vegetative hybridization of the fungus. One dominant genotype occurred in all 12 populations, representing 63.5% of all isolates. We found a moderate level of average genotypic variation and a low level of genetic differentiation (F st = 0.0814). There was no correlation between infection frequency and genotypic diversity. Although the existence of a dominant genotype and the detected linkage disequilibrium suggest that the fungus is mainly asexual and vertically transmitted, the multiallelic loci and variation of genetic diversity among populations indicate occasional contagious spread and sexual or parasexual recombination of the fungus in some populations. Furthermore, the genotypes carrying multiallelic loci suggest the possibility of multiple infections or hybridization of the endophyte.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Epichloë festucae (Leuchtm., Schardl, & Siegel) is an endophytic fungus of fine fescue grasses (Festuca subg. festuca) that belongs to the Neotyphodium/Epichloë complex (Ascomycota; Clavicipitaceae). Sexual Epichloë and their asexual forms, Neotyphodium spp., are endophytic grass symbionts, which haploid hyphae invades systematically and asymptomatically the host plant foliage. While growing into the seeds of the host plant, the fungus is transmitted vertically to the next grass generation [43, 58]. Because fungi have partly (some Epichloë species) or entirely (Neotyphodium species) lost their horizontal transmission via sexual spores, the fitness of endophytic fungi is closely linked to that of their host plants. These interactions are generally assumed to evolve toward mutualism (e.g., [43, 49]). Empirical evidence appears to support this. First, Neotyphodium/Epichloë endophytes have been shown to benefit the host in many ways [14, 43, 49]. Second, they are commonly detected in grasses worldwide (e.g., [16, 40]). Third, both reach locally very high infection frequencies (e.g., [5, 30, 42, 46, 63]). However, benefits from the endophytes are achieved only with associated costs, because the fungus receives all its nutrition from the host plant (e.g., [2, 29, 45]). The most pronounced cost of Epichloë endophytes is the decreased seed production of the host caused by the sexual reproduction of the fungus (choke disease) (e.g., [49]). Thus, endophyte–plant interactions can be complex and labile ranging from antagonistic to mutualistic depending on whether the benefits to the host plant exceed the associated costs (e.g., [45]).
According to the geographic mosaic theory of coevolution, the natural selection on interaction varies among communities. The combination of gene flow, genetic drift, and extinction/colonization dynamics constantly mix and remix the range of coevolving traits, creating a geographic mosaic of populations, in which host–symbiont interactions range from antagonistic to mutualistic [54]. The net result of interaction between endophytes and grasses may vary spatially and temporally and among different genotypes of the participants [45]. Thus, the varying infection frequency of vertically transmitted endophytes in grass populations should be detected geographically and locally among different environmental conditions. In old mutualistic populations in stable environments, high infection frequencies and dominance of one or a few endophyte genotypes should be detected as a result of the clonal spread of endophytes through effective vegetative spread and the abundant seed production of infected perennial host grasses [45]. In populations where interspecific interaction is neutral or antagonistic, lower infection incidences or endophyte-free populations should be found. Regardless of the outcome of interactions between symbionts, in young and/or disturbed grass populations, infections may be only occasional and the genetic diversity of the endophyte is strongly influenced by the founder effect and genetic drift.
The population genetics of fungal populations have been widely explored in plant pathogens (e.g., [25, 36, 53]. Because clonality plays an important role in many fungal life cycles, one of the main goals has been to estimate the amount of asexual reproduction and frequency of recombination (see, e.g., [53]). Greater genotypic diversity [22, 36] and random association of alleles at different loci [33, 36] should be detected in sexual compared to clonal fungal populations. In strictly vertically transmitted endophytes, the gene flow of the fungus is restricted by the host plant's ability to disperse by seeds, which should lead to marked differentiation among endophyte populations [3, 51]. Studies of the genetic diversity and structure of grass endophyte populations are few, but the results follow the predictions above. In comparisons of sexual and asexual populations of Epichloë species, greater genetic or genotypic diversity has been detected in sexual compared to asexual populations, and linkage disequilibrium (indicating nonrandom association of alleles) has only been observed in asexual populations [3, 7, 9, 34, 51]. However, only scant attention has been given to grass endophytes outside the temperate regions (but see [5]).
In contrast to past studies in the temperate region, our research provides new information on fine fescue endophytes in the edge of their northern distribution range in subarctic latitudes. In this study, (1) we examined the frequencies of endophytic E. festucae in natural and seminatural populations of two fine fescue species Festuca rubra L. sensu lato, and F. ovina L. in Finland and northern Norway in different environments, and (2) in the case of 12 F. rubra populations, determined the genotypic variation and genetic structure of the endophyte, and estimated the prevalence of recombination of the fungus. The fruiting body formation of E. festucae is reported to be occasional, and the completed sexual life cycle (fruiting body with perithecia) is extremely rare in native grass species [3, 5, 31, 40, 46, 56, 63]. Thus, the fungus is thought to be mainly asexual and vertically transmitted via seeds and vegetative propagation of the host plant [46, 47]. Although E. festucae is capable of sexual reproduction, it is thought to be a mutualistic symbiont of fine fescues (e.g., [47], but see [45]). Therefore, high infection frequencies of E. festucae in fine fescue populations are expected, and low to moderate amount of genotypic variation together with marked genetic differentiation among endophyte populations and linkage disequilibrium among microsatellite loci should be detected.
Material and Methods
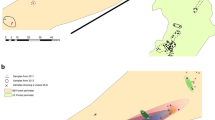
To study the frequency of infected grass individuals, we collected mature inflorescences of fine fescues from 86 study sites. Altogether, 77 F. rubra and 30 F. ovina populations were examined across Finland and in northern Norway during 1999–2000 (Figs. 1 and 2). We sampled 4–68 individuals from each grass population, depending on the number of flowering plants and size of the population. The total sample size was 2514 plant individuals. In dense meadow populations, the plants were verified to be separate individuals by morphological characters (e.g., color of leaves and hairiness of spikelets). The examined populations were actively explored for fruiting bodies of E. festucae during seed collections. The highly infected subarctic populations were checked for fruiting bodies during additional three growing seasons.
The study sites for habitat comparisons and population genetic analysis were located in the Teno river valley in subarctic Finland. To compare infection incidences in different environments, we classified the 29 F. rubra populations into three habitat categories (natural sandy riverbanks, seminatural meadows, and seminatural roadsides) and the 14 F. ovina populations into two (open cliffs or open low-alpine sites and seminatural meadows) habitat categories. The seminatural meadows had been abandoned from intensive agricultural use 15–50 years previously (personal communication from the landowners). The roadsides had been sown in 1979 or 1987 with a seed mixture containing endophyte-free F. rubra ssp. rubra cultivar Echo (Denmark) (Finnish National Road Administration), after which these sandy roadsides had been colonized naturally.
We verified the infection status of F. rubra and F. ovina by staining [41] and microscopic examination of at least five seeds of each plant individual. The fungi were identified as E. festucae by comparing the rDNA sequences with Blast searches of GeneBank http://www.ncbi.nlm.nih.gov). Polymerase chain reaction (PCR) amplification of rDNA regions ITS1, 5.8S rRNA, and ITS2 was performed by using primers ITS1 and ITS4 [60]. One fungal isolate from F. rubra was also identified by Dr. A. Leuchtmann (pers. comm.).
To study the genotypic variation of E. festucae, we chose 12 F. rubra populations from the Teno river valley, all containing more than five endophyte-infected grass individuals (Fig. 1). We germinated three seeds of each infected F. rubra individual and isolated haploid fungi from one randomly selected seedling by plating one surface-sterilized leaf sheath in potato dextrose agar (5% PDA). Pure cultures were established from the edge of the mycelia growing out of the leaf sheath cuttings. We obtained 6–67 isolates from each population (total of 189 isolates) (Tables 1 and 2). The sample sizes of the fungus varied among the populations, depending on the size and infection frequency of the grass populations.
PCR Protocol.
DNA was extracted from pure cultures of E. festucae with the Qiagen Plant Maxi kit. The primers for PCR amplification of microsatellites—B1, B6, B9a, and B9b loci [37]—were obtained from TAG Copenhagen (Copenhagen, Denmark). For automated fragment analysis, one primer of each locus was labeled with fluorescent dye. The primer B1.1 was labeled with 4,7,2′,4′,7′-hexacholoro-6- carboxyfluorescein (HEX), B9.1 with 4,7,2′,7′-tetracholoro-6-carboxyfluorescein (TET), and B6.1 with 6-carboxyfluorescein (6-FAM). PCR reactions (25 μm) were run in an Eppendorf Mastercycler thermocycler. Each reaction contained 10 mM Tris–HCl, 1.5 mM MgCl2, 50 mM KCl, and 50 mM of each deoxynucleotide triphosphate and 0.5 units of the Dynazyme Taq polymerase. Final concentration of each primer was 200 nM.
The thermocycling profiles for loci B1 and B9a were as follows: 5 min denaturation at 95°C, 36 cycles at 94°C for 45 s, 65°C for 1 min, 72°C for 45 s, followed by 72°C for 10 min. The thermocycling profiles for loci B6 and B9b included 6 min 30 s denaturation at 94°C, 35 cycles at 94°C for 1 min, 58°C for 2 min, and 72°C for 1 min followed finally by 72°C for 10 min.
The DNA concentrations of the amplification products were adjusted for automated fragment analysis (50–100 ng DNA/reaction). PCR products were separated in 5% polyacrylamide gels by an ABI Prism 377 DNA sequencer. A portion (1.5 μL) of the PCR product was added to 2.5 μL formamide, 0.5 μL 5% blue dextran, and 0.5 μL GS-500 TAMRA, internal lane standard. We loaded 2 μL of this mixture for each lane. Fragment sizes were estimated with Genescan Analysis 2.1 software. In contrast to the original description of the primers [37], we treated the loci B9a (primers B9.1–B9.2) and B9b (primers B9.1–B9.4) as separate loci, because the results of PCR amplifications with these two pairs of primers (the amount of separate peaks and the sequence lengths) did not correlate in this data set (Table 1).
Data Analysis.
Statistical analyses of fungal infection frequencies were performed with the SAS software package v. 8.02 (SAS Institute, Cary, NC, USA; 1999–2001). Five F. rubra and three F. ovina populations were sampled both in 1999 and 2000. Because no differences were found between the years (proc MIXED: F = 0.18, p = 0.6922), the data were pooled in the final analysis. We examined the differences in infection frequencies between the habitats using probability function distribution. The analysis was conducted with Proc GENMOD (number of infected grasses/sample size of population as dependent variable, binomial distribution and logit link function).
The different allele combinations of the four microsatellite markers were regarded as different genotypes. For the genomewide multilocus analysis of linkage disequilibrium, the data were coded as diploid data, where single-allelic loci were considered to be homozygous. Random association among loci was tested using the index of association (I A) [8, 33] and its modification (r d). The observed values of I A and r d, the simulated sampling distribution and the significances (with 1000 randomizations) were obtained for the total (pooled) data (with and without clone correction [33]) and for all populations separately (without clone correction) using the Multilocus ver. 1.2. software [1]. r d was also calculated for all possible pairs of loci from the total data without clone correction.
Genotypic diversity is defined here as the probability that two randomly chosen individuals have different genotypes [1]. Analysis of molecular variance (AMOVA) was used to examine the genetic differentiation of populations (F st). Genotype distances were estimated by counting the number of allele differences. Significance was tested with 1023 random permutations. AMOVA, pairwise F st of the populations, and gene flow as estimates of migration (N m) were calculated by using Arlequin v. 2.00 [49].
The relationship between pairwise F st and the geographic distance between populations was analyzed by using Mantel's permutation test [32] with Fstat, v. 2.9.3.2 [20]. The geographic distances were log-transformed for the analysis. The relationship between the infection frequencies and genotypic diversities \( \widehat{H} \)of the populations and the relationship between sample sizes and genotypic diversities\( \widehat{H} \) were analyzed with Spearman's correlation coefficient (proc CORR, SAS 1999–2001). The difference in \( \widehat{H} \)genotypic diversity between habitats was tested with t test (proc TTEST, SAS 1999–2001).
Results
We found 62% of F. rubra and 37% of F. ovina populations to have endophyte infections. In these F. rubra populations, the infection frequencies ranged from 4% to 87%, but only 9 out of the 49 infected populations had an infection frequency higher than 50%, and in 25 populations infection frequency was less than 20%. In F. ovina, the highest infection frequency was 36%, and 8 out of the 13 infected populations had infection frequencies lower than 20%. In both grass species, the infections were detected only occasionally in hemiboreal and boreal areas, whereas populations with high infection frequencies were frequently found in subarctic areas (Figs. 1 and 2). However, fruiting body production was never detected in subarctic area. In studied areas, fruiting bodies of E. festucae have been reported only in SW Finland, 1300 km south from the hot spots of infections in subarctic study area (in this study, [28, 56]). In both grass species, infection frequencies were significantly higher in meadows compared to other habitats (F. rubra: χ 2 df=2 = 171.03, p < 0.0001, deviance = 119.7194, df = 26; F. ovina: χ 2 df=1 = 31.24, p < 0.0001, deviance 25.9478, df = 12) (Fig. 3).
Infection frequency (%) (estimates of least squares means ± SD) of Epichloë festucae in (a) F. rubra and (b) F. ovina populations. Numbers above the bars represent the number of replicates (populations) per habitat. The different letters indicate the significant differences between habitats (p < 0.001, χ 2 test).
Overall, we detected 25 different fungal endophyte genotypes in the subarctic F. rubra populations (n = 12) (Table 1). Five of the genotypes contained a single allele at each of the four loci, whereas 20 genotypes had two alleles at least at one locus. Nine genotypes (two single-allelic and seven multiallelic) were found in more than one population, whereas the rest of the genotypes were unique for some population. There were four common genotypes (one single-allelic and three multiallelic), which were found in at least four populations. The single-allelic genotype (H4) was a dominant genotype, as it was found in all of the 12 populations examined (16.7–100% frequency). The population P1 was monomorphic for the dominant genotype. The total number of isolates, the number of genotypes, genotypic diversity, and the frequency of the common and unique genotypes in each population are presented in Table 2.
When all of the 12 populations were combined, moderate genotypic diversity was detected, but genotypic diversity varied greatly in individual populations (Table 2). No habitat difference was found, because the meadow and riverside populations did not differ in terms of genotypic diversity [mean (SD) meadows: \( \widehat{H} \) 0.652 (0.218) and riverbanks: \( \widehat{H} \) 0.4492 (0.4232); t = 0.98, p = 0.3678). Neither genotypic diversity and infection frequency (Spearman correlation: r S = 0.0210, p = 0.9484) nor genotypic diversity and sample size (Spearman correlation: r S = −0.3498, p = 0.2650) correlated significantly. The AMOVA results revealed a low level of population differentiation (Table 3). Geographic and genetic distance (estimated as pairwise F st) correlated positively, but the correlation was only marginally significant (Mantel test: r = 0.27454, p = 0.0754).
Significant linkage disequilibrium indicating highly linked loci was detected by using multilocus association tests when all isolates were pooled together (Table 4). When the isolates were partitioned into populations, a significant association was detected in three of the nine populations (Table 4). Three populations (P1, P7, and P11) did not contain enough variation to allow analysis. When the total (pooled) data were analyzed as clone-corrected, there was no significant linkage disequilibrium (Table 4). When the index of association (r d) was calculated for all pairwise combinations of loci over the whole data, significant linkage disequilibrium (p < 0.05) was detected in four of the six possible pairwise combinations.
Discussion
We found highly variable infection frequencies of E. festucae at a geographical scale and between different habitats locally, suggesting that the selective advantage of the fungus to the host may vary between environments and dispersal of the fungus may be restricted. The presumed predominant clonality of the fungus was supported by (1) the absence of sexual structures, (2) the dominance of one endophyte genotype in all examined populations, and (3) linked microsatellite loci. However, wide variation in genetic diversity and a low degree of structuring were detected among the populations, indicating gene flow among populations and occasional sexual or parasexual recombination of the fungus.
Spatial Differences in Endophyte Prevalence.
Observed infection frequencies were lower than expected based on fine fescue endophytes in Europe [5, 59, 63], and if the interaction between E. festucae and the host grass is strongly mutualistic throughout the study area. Infected F. rubra and F. ovina individuals were detected throughout the survey area, but highly infected populations were rare. The highest infection incidences were detected in the subarctic river valleys and in one river valley; infection frequencies differed between habitats. The meadow populations had higher infection frequencies than cliff and hillside populations (F. ovina) and the sparsely infected riverbank populations (F. rubra). The dispersal of grass seeds is restricted by isolation and long distances between the populations. However, seed dispersal fails to explain the differences in endophyte frequencies between the riverbank and meadow populations. Distance between many meadow and riverbank populations is short and the river occasionally floods the examined populations during the spring, allowing effective long-distance dispersal of the floating seeds of F. rubra with hairy glumes. The striking difference between these habitats is that the riverbank populations are disturbed nearly annually and destroyed regularly by the violent debacle in the spring. In contrast to these sandy riverbanks, meadows are more stable and fertile environments, and their grass populations are older and well established mainly by the clonal spread of the host grasses [23]. The relatively high endophyte frequency of the two grass species, F. rubra and F. ovina, in meadows is in concordance with the idea that endophytes may provide selective benefits to the host in some environments [11, 13, 15], as suggested by the geographic mosaic theory of evolution [54].
Genetic Diversity and Structure of Fungal Populations.
We detected marked variation in genotypic diversity and low genetic differentiation (F st = 0.0814), indicating high gene flow among endophyte populations. Despite the lack of correlation between genotypic diversity and infection frequency and the lack of any significant difference in genotypic diversity among habitats, the lowest genetic diversities were seen in riverbank populations with low infection frequencies. Young and small populations typically have low genetic and genotypic diversity (e.g., [23, 62]). Plausible explanations for the many riverbank populations having only one or two fungal genotypes include genetic drift and founder effect due to heavy disturbance or strong selection by harsh growing conditions. These fungal genotypes in riverbank populations were detected in several grass populations across the study area, indicating abundant gene flow via host seeds by river flow or by reindeer farming. However, the genetic distance of populations tended to correlate positively with geographic distance (see also [17]). Despite the presence of a dominant genotype in our data, we detected wider genotypic variation in some populations than expected for a strictly or predominantly asexual fungus. This finding is consistent with many other population-genetic studies reporting unpredictably high genetic diversities among presumably asexual fungi (e.g., [3, 10, 21, 27, 55]). Conventionally, high genotypic diversity has been explained by sexual recombination, but in the case of fungi, other mechanisms are also possible. For example, mitotic or parasexual recombination (e.g., somatic hybridization) (e.g., [38, 52, 55]), mutation accumulation [52], and hypervariable microsatellite loci [53] are suggested to be possible sources of genetic variation.
Northern E. festucae Populations are Asexual?
During 5 years of intensive fieldwork in the subarctic study area, we never found sexual structures (fruiting bodies) of endophytes on fine fescues. Furthermore, the presence of one common and widespread genotype and linked microsatellite loci (linkage disequilibrium) indicate that, similar to many other organisms in marginal habitats (see, e.g., [6]), E. festucae is primarily clonal at the edge of its distribution range in the subarctic area. Three mechanisms, either alone or in interaction, may explain the lack of fruiting body formation in E. festucae in northern areas: (1) environmental factors and (2) genotypes of endophyte or host may prevent fruiting body formation [7, 9, 35], or (3) the sexual strains of E. festucaë may have limited dispersal capability [56]. In subarctic areas, the growing season is short, about 105 days [18], but day length is extremely long (24 h for most of the growing season). Such a short and intensive growing period favors fast growth of the host grass, possibly constraining horizontal transmission of the fungus by sexual spores, if rapidly growing grasses can produce seeds before the Epichloë endophyte completes its sexual life cycle [14, 26]. Although the dominance of clonal spread of E. festucae was detected, recombination of the fungus cannot be ruled out. Linkage equilibrium was detected in clone-corrected total data, and population-specific linkage equilibrium was found in six out of 12 populations, indicating occasional recombination. In Epichloë endophytes, horizontal transmission occurs via ascospores, but in some species, contagious spread is also suggested to be possible via asexual conidia and/or epiphyllous mycelium through leaf tissue [4, 34, 39, 44] and through cuts on flowering stems [57], which makes multiple infections and parasexual recombination possible even without the sexual stage of the fungus. Alternatively, detected genetic structure may be explained by past or parasexual recombination events [19, 53], and hypervariable microsatellite loci may partly explain the linkage equilibrium detected [53].
Multiallelic Loci Indicate Hybridization?
Hybridization plays an important role in the speciation of Epichloë/Neotyphodium endophytes (e.g., [38, 48]). The mechanisms of hybridization are unclear, but it is suggested to occur via anastomosis (hyphal fusion followed by nuclear fusion), resulting in uninucleate hypha (e.g., [48]). In all studies dealing with the molecular taxonomy of Epichloë/Neotyphodium endophytes, E. festucae has been reported to be a nonhybrid, haploid fungus with a single copy of gene, and E. festucae is suggested to be one partner in the hybridizations leading to speciation of asexual Neotyphodium endophytes [16, 31, 38, 47]. In F. rubra, we found one or more multiallelic loci in 63 out of 189 E. festucae isolates. The detected multiple loci may be a result of (1) multistrain infections of the plants or (2) hybridization of the fungus. Multiple Neotyphodium/Epichloë strain infections have been documented in natural populations [34] and are obtained artificially, but in artificially multiple-infected grasses, individual grass tillers usually only contain a single fungal genotype [12, 61].
The detected marked variability of the four microsatellite loci within one endophyte species and within a relatively small geographic area is important, because it contradicts with the lack of sexual structures. The abundant genotypic variation together with the multiallelic loci call for more experimental and molecular studies to reveal the mechanisms of transmission and recombination of these endophytic fungi. In particular, the presence of both single-allelic and multiallelic genotypes within one species and within single populations suggests a need to reassess the occurrence and frequency of possibilities of intra- and interspecies hybridizations in Epichloë/Neotyphodium endophytes. The rapidly evolving molecular techniques are expanding our understanding of how the genetic diversity of fungi and the phenotypic plasticity of fungal life history traits, plus the fungus and the host plant individually or as phenotypic units respond to changing environmental conditions. Thus, combining empirical fungal ecology with molecular approaches provides feasible visions for biologists who are interested in coevolutionary processes and the evolution of sex and species concepts.
References
Agapow, PM, Burt, A (2000) Multi-locus 1.2. Dept of Biology, Imperial College, Silwood Park, Ascot, Berks, SL57PY, UK
Ahlholm, JU, Helander, M, Lehtimäki, S, Wäli, P, Saikkonen, K (2002b) Vertically transmitted fungal endophytes: different responses of host–parasite systems to environmental conditions. Oikos 99: 173–183
Arroyo García, R, Martínez Zapater, JM, García Criado, B, Zabalgogeazcoa, I (2002) Genetic structure of natural populations of the grass endophyte Epichloë festucae in semiarid grasslands. Mol Ecol 11: 355–364
Bacon, CW, Hinton, DM (1991) Microcyclic conidiation cycles in Epichloë typhina. Mycologia 83: 743–751
Bazely, DR, Vicari, M, Emmerich, S, Filip, L, Lin, D, Inman, A (1997) Interactions between herbivores and endophyte-infected Festuca rubra from Scottish islands of St. Kilda, Benbecula and Rum. J Appl Ecol 34: 847–860
Bierzychudeck, P (1985) Patterns in plant parthenogenesis. Experientia 41: 1255–1264
Brem, D, Leuchtmann, A (2003) Molecular evidence for host-adapted races of the fungal endophyte Epichloë bromicola after presumed host shifts. Evolution 57: 37–51
Brown, AHD, Feldman, MW Nevo E (1980) Multi-locus structure of natural populations of Hordeum spontaneum. Genetics 96: 523–536
Bucheli, E, Leuchtmann, A (1996) Evidence for genetic differentiation between choke-inducing and asymptomatic strains of the Epichloe grass endophyte from Brachypodium sylvaticum. Evolution 50: 1879–1887
Burt, A, Carter, DA, Koenig, GL, White, TJ, Taylor, JW (1996) Molecular markers reveal cryptic sex in the human pathogen Coccidioides immitis. Proc Natl Acad Sci USA 93: 770–773
Cheplick, GP (1989) Interactions between infection by endophytic fungi and nutrient limitation in the grasses Lolium perenne and Festuca arundinacea. New Phytol 111: 89–97
Christensen, MJ, Simpson, WR, Al Samarrai, T (2000) Infection of tall fescue and perennial ryegrass plants by combinations of different Neotyphodium endophytes. Mycol Res 104: 974–978
Clay, K (1987) Effects of fungal endophytes on the seed and seedling biology of Lolium perenne and Festuca arundinacea. Oecologia 73: 358–362
Clay, K (1993) The ecology and evolution of endophytes. Agric Ecosyst Environ 44: 39–64
Clay, K, Holah, J (1999) Fungal endophyte symbiosis and plant diversity in successional fields. Science 285: 1742–1744
Clay, K, Schardl, C (2002) Evolutionary origins and ecological consequences of endophyte symbiosis with grasses. Am Nat 160: S99–S127
Dybdahl, MF, Lively, CM (1996) The geography of coevolution: comparative population structures for a snail and its trematode parasite. Evolution 50: 2264–2275
Elamo, P (2002) Birch rust and endophytic fungi in birch leaves: effects of host plant genetic background and environmental factors. Annales Universitatis Turkuensis. ser. aII. tom. 131. Univ. of Turku, Turku, Finland
Geiser, DM, Pitt, JI, Taylor JW (1998) Cryptic speciation and recombination in the aflatoxin-producing fungus Aspergillus flavus. Proc Natl Acad Sci USA 95: 388–393
Goudet, J (2002) Fstat ver. 2.9.3.2. Institute of Ecology, Biology Building, UNIL, CH-1015, Lausanne, Switzerland
Gräser, Y, Volovsek, M, Arrington, J, Schönian, G, Presber, W, Mitchell, TG, Vilgalys, R (1996) Molecular markers reveal that population structure of the human pathogen Candida albicans exhibits both clonality and recombination. Proc Natl Acad Sci USA 93: 12473–12477
Halkett, F, Simon JC, Balloux F (2005) Tackling the population genetics of clonal and partially clonal organisms. Trends Ecol Evol 20: 194–201
Harberd, DJ (1961) Observations on population structure and longevity of Festuca rubra L. New Phytol 60: 184–192
Hedrick, PW (2005) Genetics of Populations, 3rd edn. Jones and Bartlett Publishers, Sudbury, MA, USA
Keiper, FJ, Hayden, MJ, Park, RF, Wellings, CR (2003) Molecular genetic variability of Australian isolates of five cereal rust pathogens. Mycol Res 107: 545–556
Kirby, EJM (1961) Host–parasite relations in the choke disease of grasses. Trans Br Mycol Soc 44: 493–503
Kohli, Y, Kohn, LM (1998) Random association among alleles in clonal populations of Sclerotinia sclerotiorum. Fungal Genet Biol 23: 139–149
Koponen, H, Mäkelä, K (1976) Phyllachora graminis, P. silvatica, Epichloë typhina and Acrospermum graminum on grasses in Finland. Karstenia 15: 46–55
Lehtonen, P, Helander, M, Saikkonen, K (2005) Are endophyte-mediated effects on herbivores conditional on soil nutrients? Oecologia 142: 38–45
Leuchtmann, A, Clay, K (1997) The population biology of grass endophytes. In: Carroll, GC, Tudzynski, P (Eds.) The Mycota. V. Plant relationships, Part B. Springer-Verlag, Berlin, pp 185–204
Leuchtmann, A, Schardl, CL, Siegel, MR (1994) Sexual compatibility and taxonomy of a new species of Epichloë symbiotic with fine fescue grasses. Mycologia 86: 802–812
Mantel, N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27: 209–220
Maynard Smith, J, Smith, NH, O'Rourke, M, Spratt, BG (1993) How clonal are bacteria? Proc Natl Acad Sci USA 90: 4384–4388
Meijer, G, Leuchtmann, A (1999) Multistrain infections of the grass Brachypodium sylvaticum by its fungal endophyte Epichloë sylvatica. New Phytol 141: 355–368
Meijer, G, Leuchtmann, A (2001) Fungal genotype controls mutualism and sex in Brachypodium sylvaticum infected by Epichloë sylvatica. Acta Biol Hung 52: 249–263
Milgroom, MG (1996) Recombination and the multi-locus structure of fungal populations. Annu Rev Phytopathol 96: 10518–10523
Moon, CD, Tapper, BA, Scott, B (1999) Identification of Epichloë endophytes in plants by a microsatellite-based PCR fingerprinting assay with automated analysis. Appl Environ Microb 65: 1268–1279
Moon, CD, Craven, KD, Leuchtmann, A, Clements, SL, Schardl, CL (2004) Prevalence of interspecific hybrids amongst asexual fungal endophytes of grasses. Mol Ecol 13: 1455–1467
Moy, M, Belanger, F, Duncan, R, Freehoff, A, Leary, C, Meyer, W, Sullivan, R, White, JF Jr (2000) Identification of epiphyllous mycelial nets on leaves of grasses infected by Clavicipitaceous endophytes. Symbiosis 28: 291–302
Saha, DC, Johnson-Cicalese, JM, Halisky, PM, van Heemstra, MI, Funk, CR (1987) Occurrence and significance of endophytic fungi in the fine fescues. Plant Dis 71:1021–1024
Saha, DC, Jackson, MA, Johnson-Cicalese, JM (1988) A rapid staining method for detection of endophytic fungi in turf and forage grasses. Phytopathol 78: 237–239
Saikkonen, K, Ahlholm, J, Helander, M, Lehtimäki, S, Niemeläinen, O (2000) Endophytic fungi in wild and cultivated grasses in Finland. Ecography 23: 360–366
Saikkonen, K, Faeth, SH, Helander, M, Sullivan, TJ (1998) Fungal endophytes: a continuum of interactions with host plants. Annu Rev Ecol Syst 29: 319–343
Saikkonen, K, Helander, M, Faeth, SH (2004) Fungal endophytes: hitchhikers of the green world. In: Gillings, M, Holmes, A (Eds.) Plant Microbiology. BIOS Scientific Publishers Ltd., Oxford, pp 77–95
Saikkonen, K, Wäli, P, Helander, M, Faeth, SH (2004) Evolution of endophyte–plant symbioses. Trends Plant Sci 9: 275–280
Sampson, K (1933) The systemic infection of grasses by Epichloë typhina (Pers.) Tul. Trans Br Mycol Soc 18: 30–47
Schardl, CL (2001) Epichloë festucae and related mutualistic symbionts of grasses. Fungal Genet Biol 33: 69–82
Schardl, CL, Craven, KD (2003) Interspecific hybridization in plant-associated fungi and oomycetes: a review. Mol Ecol 12: 2861–2873
Schardl, CL, Leuchtmann, A, Spiering, MJ (2004) Symbiosis of grasses with seedborne fungal endophytes. Annu Rev Plant Biol 55: 315–340
Schneider, S, Roessli, D, Excoffier, L (2000) ARLEQUIN ver. 2.00: A software for population genetic data analysis. Genetics and Biometry Laboratory, Dept. of Anthropology and Ecology, University of Geneva, Switzerland
Sullivan, TJ, Faeth, SH (2004) Gene flow in the endophyte Neotyphodium and implications for coevolution with Festuca arizonica. Mol Ecol 13: 649–656
Talbot, NJ, Salch, YP, Ma, M, Hamer, JE (1993) Karyotypic variation within clonal lineages of the rice blast fungus, Magnaporthe grisea. Appl Environ Microb 59: 585–593
Taylor, JW, Jacobson, DJ, Fisher, MC (1999) The evolution of asexual fungi: reproduction, speciation and classification. Annu Rev Phytopathol 37: 197–246
Thompson, JN (1999) The raw material for coevolution. Oikos 84: 5–16
Vandenkoornhuyse, P, Leyval, C, Bonnin, I (2001) High genetic diversity in arbuscular mycorrhizal fungi: evidence for recombination events. Heredity 87: 243–253
Väre, H, Itämies, J (1995) Phorbia phrenione (Seguy) (Diptera: Anthomyiidae) in Finland. Sahlbergia 2: 119–124
Western, JH, Cavett, JJ (1959) The choke disease of cocksfoot (Dactylis glomerata) caused by Epichloë typhina (Fr.) Tul. Trans Br Mycol Soc 42: 298–307
White, JF, Morgan-Jones, G, Morrow, AC (1993) Taxonomy, life cycle, reproduction and detection of Acremonium endophytes. Agricult Ecosyst Environ 44: 13–37
White, JF, Lewis, G, Sun, S, Funk, CR Jr (1993) A study of distribution of Agremonium in populations of red fescue in southwest England and in vitro comparisons to isolates from North American collections. Sydowia 45: 388–394
White, TJ, Burns, T, Lee, S, Taylor, J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, MA, Gelfand, DH, Sninsky, JJ, White TJ (Eds.) PCR Protocols: A Guide to Methods and Amplifications. Academic Press, San Diego, pp 315–322
Wille, PA, Aeschbacher, RA, Boller, T (1999) Distribution of fungal endophyte genotypes in doubly infected host grasses. Plant J 18: 349–358
Wrigth, S (1969) Evolution and the Genetics of Populations: A Treatise in Three Volumes. Vol. 2: The Theory of Gene Frequencies. University of Chicago Press
Zabalgogeazcoa, I, Vázquez de Aldana, BR, García Criado, B, García Ciudad, A (1999) The infection of Festuca rubra by the fungal endophyte Epichloë festucae in permanent Mediterranean grassland. Grass For Sci 54: 91–94
Acknowledgments
Dr. Pia Mutikainen and anonymous reviewers provided valuable comments on the manuscript. We thank Mr. Ilkka Blomqvist for preparing the maps. This study was funded by Academy of Finland and by Finnish Cultural Foundation.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table 1
Location, habitat endophyte infection frequency and sample size of Festuca rubra populations (PDF 78 kb)
Rights and permissions
About this article
Cite this article
Wäli, P.R., Ahlholm, J.U., Helander, M. et al. Occurrence and Genetic Structure of the Systemic Grass Endophyte Epichloë festucae in Fine Fescue Populations. Microb Ecol 53, 20–29 (2007). https://doi.org/10.1007/s00248-006-9076-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-006-9076-2