Abstract
Macrocycles represent a potentially vast extension of drug chemical space still largely untapped by synthetic compounds. Sampling of flexible rings is incorporated in the ICM-dock protocol. We tested the ability of ICM-dock to reproduce macrocyclic ligand–protein receptor complexes, first in a large retrospective benchmark (246 complexes), and next, in context of the D3R Grand Challenge 4 (GC4), where we modeled bound complexes and predicted activities for a series of macrocyclic BACE inhibitors. Sub-angstrom accuracy was achieved in ligand pose prediction both in cross-docking (D3R Challenge Stage 1A) and cognate (Stage 1B) setup. Stage 1B submission was top ranked by mean and average RMSDs, even though no ligand knowledge was used in our simulations on this Stage. Furthermore, we demonstrate successful receptor conformational selection in Stage 1A, aided by the enhanced ‘4D’ multiple receptor conformation docking protocol with optimized scoring offsets. In the activity 3D QSAR modeling, predictivity of the BACE pKd model was modest, while for the second target (Cathepsin-S), leading performance was achieved. Difference in activity prediction performance between the targets is likely explained by the amount of available and relevant training data.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
D3R Grand Challenges
Computer aided drug design (CADD) methods are developing rapidly and CADD application in practical drug discovery and optimization projects is becoming routine. Yet it is often difficult to judge objectively how well modeling methods really perform: retrospective tests are inevitably biased as the algorithm development is influenced by the context of available 3D structures and activity data, and the design as well as composition of retrospective benchmarks only remotely reflect many real-life application scenarios. Published examples of practical applications suffer from publication bias [11, 19]—success stories are much more likely to be published, while failures mostly get known through word of mouth and eventually the common “modeling doesn’t work” general attitude. Drug design data resource (D3R) Grand Challenges provide the CADD developer community with an opportunity to test and compare methods head-to-head in context of a blinded evaluation versus yet unpublished data [13]. We participated in the bound pose prediction and the activity prediction challenges which test the two core tasks in CADD.
Macrocycle docking
D3R Grand Challenge 4 included prediction of the binding poses and activity for a series of macrocyclic inhibitors of Beta-Secretase 1 (otherwise known as beta-amyloid converting enzyme, BACE), presenting us with a possibility to test our methods in modeling of macrocycle ligand-receptor interactions. Unique utility of the macrocyclic compounds as drugs has long been appreciated in the medicinal chemistry, thanks to the numerous examples of macrocyclic natural products possessing exceptional potency, specificity and ability to modulate targets that are poorly druggable with typical ‘drug-like’ synthetic compounds (e.g. rapamycin [39], cyclosporin [29]). Cyclization can stabilize the molecule both chemically and conformationally. Chemical stability results in better bioavailability, and permeation was also shown to improve upon cyclization at least for certain classes of compounds [17]. From the standpoint of the ligand-receptor interaction thermodynamics, cyclic constraints restrict conformational space, sharply reducing entropic penalty and thus enhancing potency. At the same time, alternative conformations that might result in an off-target activity are eliminated. Larger, yet relatively rigid and still bioavailable macrocyclic ligands can efficiently interact with their targets over relatively flat extended interfaces where regular small-molecule ligands fail to achieve sufficient affinity. Many of the natural product macrocycles are formidably complex in terms of synthesis but the advances in synthetic chemistry recently led to increased exploration of the macrocyclic chemical space [41]. Recent approvals of a series of NS3-4A protease inhibitors for hepatitis C treatment (grazoprevir, simeprevir, paritaprevir and vaniprevir) [28] and a cancer drug lorlatinib [5] are the milestones paving way for broad application of fully synthetic macrocyclic drugs. Correspondingly, there is a growing demand for accurate modeling macrocyclic molecules to facilitate structure-based discovery and design of these ligands.
Presence of macrocycles represents a challenge for docking algorithms because the ligand conformational sampling, if any is included in the pose generation itself, is commonly geared towards performing simple bond torsion rotations rather than complex, coordinated motions of a macrocycle ring. To overcome this shortcoming, a number of protocols were proposed that couple a standard ligand docking algorithm with an external vigorous sampling stage (reviewed in [3]). Conformers generated by the external sampler are then individually docked to the target receptor, with the macrocyle now kept rigid. Another approach is to supplement the docking algorithm with a library of ring ‘templates’ [12], derived from extensive external sampling [30] and/or experimentally solved structures.
In ICM-Dock, sampling of flexible rings is directly incorporated into the docking procedure. However, we have not previously tested specifically the ability of our method to dock macrocycles. Therefore, to verify the accuracy of the algorithm and establish best practices of handling such systems, we compiled a large benchmark of experimentally solved macrocycle/protein complexes and performed basic cognate redocking tests.
Multiple receptor conformation and ligand-biased docking
Practical prospective applications of docking and blind tests such as D3R GC involve cross-docking, i.e. using conformations of the receptor that are not cognate to ligand(s) being docked (unlike retrospective benchmarking typically focused on cognate ligand/receptor structure pairs). Depending on the extent of receptor flexibility, cross-docking may require generation and/or selection of the receptor conformation that fits the ligand of interest. While unguided receptor sampling so far remains, in most cases, impractical, Multiple Receptor Conformation (MRC, or ensemble) docking has emerged as a viable alternative [37]. ICM-dock allows incorporation of receptor ensembles derived, for example, from multiple PDB structures, directly into docking procedure via ‘4D’ receptor grids [8]. Thus, receptor conformation optimally fitting the ligand of interest can be automatically selected during sampling.
Even optimal pairing of ligand pose to a non-cognate receptor conformation may not eliminate fit imperfections that impact scoring and overall ability of the procedure to identify near-native solutions. Knowledge derived from the experimentally determined bound ligand poses is another source of accuracy improvements, and various forms of docking templates have been in use [18, 31, 40]. In context of D3R GC2, we proposed a ligand-biased ensemble receptor docking (LigBEnD) hybrid ligand/receptor structure-based docking protocol. In this approach, each ligand was docked to each of the available protein structures with the co-crystallized ligand’s Atomic Property Field (APF) [33] bias [21]. In GC3, we combined the LigBEnD method with the 4D grid docking approach and introduced optimal receptor conformational ensemble selection [22]. For the present challenge, GC4, we further improved ‘4D’ sampling and LigBEnD protocol by introducing scoring/energy offsets that optimize selection of receptor conformers. We investigated whether the procedure selects receptor conformations close to the cognate structures. Finally APF 3D QSAR [33] models of ligand activity for the two GC4 targets were also developed and evaluated.
Methods
Flexible ring/macrocycle sampling in ICM-dock
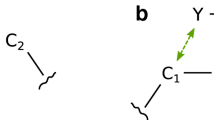
ICM Docking algorithm (vide infra) uses internal coordinates representation to sample the conformational space of the ligand efficiently [2, 35]. Representation of the acyclic molecules by ICM’s internal coordinate tree is straightforward—starting from any atom, the tree ‘grows’ along bonds and the position of each atom is determined in relation to the previously grown part of the tree by a triplet of internal coordinates—bond length, bond angle and torsion angle (or ‘phase’ angles at the branching points) (Fig. 1). Bond lengths, bond angles and ‘phase’ angles, generally, are kept constant (‘fixed’) when torsional sampling is performed, resulting in automatic preservation of the (near) ideal covalent bonding geometries. As a consequence, torsion space modeling normally does not require bond bending and stretching energy terms (it should be noted that in the ICM framework, when desired, all internal variables can be freed and appropriate terms added for full Cartesian space-like relaxation). However, the presence of rings in the molecular topology results in the internal coordinate trees where some of the bonds (one per ring) do not correspond to any tree edge. Therefore, for these bonds (which we term as ‘extra’ bonds), covalent geometry is not determined any longer by the locally corresponding internal variables but, rather, by a longer series of variables along the tree path surrounding the ring. If the ring can be considered rigid (e.g. a phenyl), the treatment is straightforward: for all tree edges/bonds belonging to the ring, not only bond lengths and angles but also torsions are considered constant/fixed. Thus, the entire ring becomes a rigid body and its orientation can be sampled highly efficiently, with all its atoms subject to the same common rotation/translation operators. However, when the ring system is substantially flexible (as is generally the case for macrocycles and even for the smaller saturated rings), maintaining proper ring closure at ‘extra’ bond by simply keeping all ring internal variables fixed is not possible if the ring flexing is to be modeled concurrently with all other conformational sampling (although one could, in principle, switch between a few discrete pre-defined ring conformations). The solution we use to allow continuous ring flexing, is to maintain ring closure dynamically by imposing a complete set of covalent geometry terms/restraints around the ‘extra’ bond. Because ICM-dock is based on the Monte-Carlo sampler coupled with gradient energy minimization after each random step, ring disruptions that arise due to the random perturbations are quickly ‘repaired’ by the gradient minimizer, guided by the forces created by covalent restraints.
ICM molecular internal coordinates tree. Internal coordinates bi, αi, Φi are ‘natural’ in that they directly correspond to local covalent geometry parameters, i.e. bond lengths, bond angles and bond rotors/torsions. However, tree representation of cyclic structures contains ‘extra’ bonds, for which covalent geometry is not directly determined by any specific bi, αi, Φi but rather by all variables along the path around the ring
Benchmark of macrocyclic ligand/receptor complexes
The benchmark set of experimentally resolved macrocycle/protein complexes was generated by processing the file components.cif from RCSB/PDB [7] (the set was originally derived from 2016 version of PDB). Chemical structures of ligands were extracted and largest ring sizes calculated using chemoinformatics functions in ICM (Molsoft, San Diego). Ligands with at least 9-member rings were retained as macrocycles. While some stricter definitions require at least 12 ring members, we felt that flexible 9–11 member rings are also of interest for algorithm testing purposes,in any case such compounds formed only a small subset (8 ligands). Resulting set of all PDB X-ray structures of macrocycle ligands was filtered first to remove lower resolution structures (> 2.5 Å). Next, redundant lower-resolution structures for each ligand/receptor pair were removed whenever multiple structures were available. Most structures were visually inspected and verified against available electron density maps. We removed structures with the following issues: large parts of the ligand did not correspond to electron density; large portions of the ligand had geometries inconsistent with the chemical structure, e.g. multiple sp2 atoms modeled with pyramidal geometries; ligands with macrocycles known to be essentially rigid, e.g. heme; a series of complexes with cyclic dinucleotide signaling molecules (e.g. c-di-GMP) that form ligand-ligand dimers. 3D structures of ligands in PDB entries were converted to flat 2D drawings using ICM cheminformatics functions. Chemical structures were spot-checked against the literature, errors in stereo-chemistry and bond typing were corrected. List of PDB entries, corresponding ligand IDs and Smiles strings for each ligand are provided as Supplementary Data. Distributions of macrocycle ring sizes, molecular weights, cLogP and flexible torsion counts (including flexible rings) in the final benchmark are shown in Supplementary Fig. 1. The set includes 10 covalent complexes, which were treated according to the standard ICM-dock covalent modeling procedure: appropriate reaction definitions were taken from ICM reaction library, ligand 2D drawings modified as needed to restore pre-covalent forms, and selection of the covalently modified residue was provided in the docking setup. Where needed to create complete binding sites, biological multimeric assemblies were rebuilt. Strongly protein-bound water molecules (≥ 3 hydrogen bonds to the receptor) were retained.
D3R target receptor conformations selection and processing for docking
All protein structures used came from the individual target’s corresponding Pocketome [20] entry. Pocketome is a large pocket-centric collection of protein–ligand complexes compiled from the PDB, each Pocketome entry is organized around a particular ligand pocket (i.e. PDB structures of the same protein may be present in different Pocketome entries if, for instance, ortho- and allo-steric pockets exist) from PDB entries of a single Uniprot entity (i.e. polypeptide gene product). Pockets are optimally pre-aligned/superimposed, making Pocketome entries a convenient starting point for ensemble docking. For BACE, only PDBs with co-crystallized ligands related to the compounds in GC4 set were retained: 2F3E, 2F3F, 3DUY, 3DV1, 3DV5, 3K5C, 3K5D, 3K5G, 4DPF, 4DPI, 4GMI, 4K8S, 4KE0, 4KE1. Similarly, for Cathepsin S, we only considered complexes of ligands related to the chemotypes in the challenge, leaving 24 pocket conformations from 13 PDBs: 3IEJ, 5QBU, 5QBX, 5QC1, 5QC3, 5QC4, 5QC5, 5QC6, 5QC8, 5QCA, 5QCD, 5QCE, 5QCJ (most of these entries contain two structurally non-identical copies of the protein in the crystallographic unit).
For each target, all proteins and their co-crystallized ligands were converted into an ICM object (i.e. prepared for use in ICM environment) using the standard ICM procedure [1]: The protein atoms were assigned to the correct atom types and charge based on a modified ECEPP/3 force field [26], the ligand atom types and charges were assigned based on the Merck molecular force field (MMFF94) [16]. Missing hydrogen atoms and zero-occupancy protein heavy atoms were added. Side chains with added atoms and polar hydrogen atoms, or side chains with multiple tautomeric or crystallographically ambiguous rotational conformations such as glutamine, asparagine, histidine, were sampled and optimized in the presence of the co-crystallized ligands. Mono-protonated state of the aspartic acid dyad is believed to be the most relevant for binding of most ligands [32], therefore for each BACE structure two alternative protonation states were considered, with one of the two catalytic aspartic acids residues Asp93 and Asp289 protonated (full Uniport entry sequence numbering).
To select minimal non-redundant set of receptor conformations for multiple receptor conformation (MRC) docking we applied our iterative approach based on ‘compatibility matrix’ [22]: receptor structures were added to the ensemble until compatibility with all experimentally resolved ligands was reached. For Cathepsin S, all co-crystallized ligands were compatible with one or both of the two pocket structures, 5QBU chain B and 5QC6 chain B, and these two structures were used for docking. For BACE, 7 PDBs were selected: 2F3E, 2F3F, 3K5C, 3K5D, 4DPF, 4DPI and 4GMI,each of these structures were represented in the ensemble by two entries corresponding to the alternative protonation states of two aspartates in the active site.
For docking Challenge 1b, organizers released cognate receptor structures. These included protein-bound and crystallographic water molecules. Per ICM-dock best practices, we attempted to detect tightly bound structural water. A single such water molecule was identified within the binding sites of 12 out of 20 receptor structures provided. This water molecule adjacent to Asn 294 is also seen in multiple PDB structures and was retained, although its impact on the docking accuracy proved minor overall (see “Results”).
Docking setup
For the macrocycle benchmark, 3D box was defined with margins of 5 Å around the co-crystallized ligand. For the CG4 ligand docking, the ligand-binding pocket was defined by protein residues within 5 Å of the co-crystallized ligand(s), and the box was defined around these residues (no additional margin). Standard ICM-dock grid potential maps within the box were calculated with a 0.5 Å grid spacing [27, 35]. Ensembles of receptor conformations were represented by ‘4D’ grids [8]. Each receptor conformation was associated with its corresponding receptor energy offset so that the final docking energy of each ligand was adjusted by receptor conformation energy offset.
Atomic property fields
APF is a representation of the 3D ligand by a set of seven fields representing spacial distributions of the physicochemical properties of its atoms [33]. These seven fields represent classic pharmacophore features as well as additional descriptors that together provide nuanced characterization of each atom type: hydrogen bond donor, hydrogen bond acceptor, sp2 hybridization, lipophilicity, size, formal charge, and electronegativity. The field of the molecule is normally generated as a total of Gaussian fields from each ligand atom and pre-calculated on grids. If another molecule is placed in the APF field, a score or pseudo-energy can be calculated as a sum of the dot products of atom/field property vectors. The score between any two poses of (identical or different) ligand(s) gives a topology-independent measure of chemical 3D similarity without the need of any specific atom-to-atom or bond-to-bond correspondence. Applications of the approach and its performance in ligand alignment, QSAR modeling, ligand based virtual screening and binding site comparison have been reported [14, 15, 33, 34].
Docking score offsets
During ‘4D’ ICM docking runs, the Monte-Carlo steps switch the sets of receptor interaction grids (and, when applicable, ligand-biasing APF potentials) corresponding to different receptor conformations. To correct for systematic errors in docking energies associated with different receptor conformations, we introduced a vector of energy offsets assigned to each receptor conformation. Optimal offsets to the ligand docking energy were determined as follows: All N co-crystallized ligands were docked to each of the M receptor conformations, generating matrices (MxN) of ligand RMSD, receptor Cα RMSD, and docking energies. The ligand RMSD was calculated using the docked pose versus the native pose of the ligand, whereas the receptor Cα RMSD was calculated between the receptor conformer selected in docking and the receptor structure cognate for the ligand. A vector of receptor docking energy offsets (with dimension M) was then added to the docking energies. Boltzmann weighted ligand RMSD (<RMSDlig > ) and receptor Cα RMSD (<RMSDrec > ) averages were calculated at 298 K using the docking energy with receptor offset. The receptor docking energy offsets were then optimized by Amoeba Simplex algorithm to minimize the objective function:
The function was designed to drive offset energy towards values that result in low-energy poses closest to native both on the ligand and receptor sides. Functional form with log() was chosen to attenuate impact of outliers with large RMSDs.
Ligand bias atomic property field (APF) grid maps preparation
The co-crystallized ligand poses were used, in the form of their APF [33] grid maps, to guide and accelerate the docking process. For Cathepin S, 24 co-crystallized ligands similar to the D3R challenge set were combined to calculate the ligand APF grid maps. For the BACE, 14 co-crystallized ligands were used. To prevent over-representation of certain chemotypes and heavy bias towards one single docking mode, the co-crystallized ligands were clustered using APF similarity, the contribution of each cluster of ligands to the ligand APF grids was normalized to the number of ligands in the cluster. For ‘4D’ docking simulations that sampled multiple receptor conformations, APF grids were also in 4D format and included, for each receptor conformation, only its ‘compatible’ ligands, according to compatibility matrix described above.
Ligand preparation for docking
Formal charges were set using ICM’s internal pKa prediction models at pH 5.5 for Cathepsin S and pH 4.5 for BACE (low pH conditions were indicated by organizers). Subsequent processing was done within standard ICM-dock protocol: hydrogen atoms added, atom types and partial charges were assigned as per MMFF94 force field [16], 2D structures converted to 3D, its rotational bonds sampled and all atoms minimized in the Cartesian coordinates in the absence of the receptor maps to generate the starting ensembles of ligand conformations for docking.
Ligand docking
ICM-dock protocol (in Molsoft ICM) was used throughout this study. Ligands were docked into the receptor potentials represented on grid maps and, where applicable, co-crystallized Ligand Template APF grid maps. ICM-dock [1] uses a multiple start biased probability Monte Carlo (BPMC) with local gradient minimization to optimize the ligand’s conformation and position within the binding site simultaneously [35]. The protocol first samples the free ligand with MC steps randomly changing flexible torsion angles (one at a time). Resulting low energy ligand conformers are next placed into the receptor interaction fields. Multiple MC trajectories are initiated from different initial ligand positions and conformations, now subjecting ligand torsions as well as its position and orientation to random steps. Special grid-switching MC steps also sample ensembles of protein conformations represented by 4D grid layers [8]. Small number (typically 3–10) of the lowest energy poses are retained and re-ranked in using ICM-VLS score with explicit receptor.
Ring flexibility
ICM uses internal coordinate representation of the simulated molecule, with bond angles and bond length variables kept constant in MC docking runs. Conformational space of the ligand is sampled via random changes and gradient minimizations of torsion angles. Flexible ring sampling mode (default in ICM-dock) includes torsion angles within rings such as macrocycles into sampling and gradient minimization (Fig. 1). Correct covalent geometry at ‘virtual’ bonds that are not part of ICM tree is maintained by a set of explicit bond bending and bond stretching constraints imposed during local gradient minimizations. Resulting forces restore ring closure after any disruption caused by MC steps altering ring torsions.
Docking ‘effort’ parameter, which defines the length of MC simulation and number of energy minimization steps, was set to 10. At the end of each independent docking simulation, 10 best conformations were retained according to the docking score combining its internal force-field energy, grid receptor interaction terms and ligand bias APF grid pseudo-energy. To ensure convergence, each ligand was docked in three independent simulations, generating 10 × 3 = 30 conformations. Poses were re-evaluated using ICM’s standard physics-based virtual ligand screening (VLS) score [36].
Post-docking processing and pose selection
ICM VLS docking score was combined with the APF similarity score SAPF of the docked pose to the co-crystallized ligands compatible with the corresponding protein conformation [21]. The resulting composite score SComp was used to rank poses and select the top scoring pose, without any manual intervention.
Training sets for activity prediction
To generate training sets, known ligands for Cathepsin S and BACE were extracted from ChEMBL version 24 [6]. Only compound records with measured IC50, Ki, or Kd were kept, compounds with remarks such as “inactive” or “inconclusive” were removed. The IC50, Ki and Kd values were treated interchangeably and converted into ‘pKd’ (i.e. −LogKd); while we are aware of potentially significant offsets between the different measures of activity, priority was given to maintaining the breadth of the data set. If a compound had multiple measured activities against a target, all the records of that compound were combined, and its top 20 percentile activity value was taken. For Cathespin S, ChEMBL data was supplemented with 135 compound activities from D3R GC3. The total number of ligands used for Cathepsin S was 430. For BACE, we filtered the training set to focus on the relevant data: 149 macrocyclic compounds were extracted; after docking, their APF similarities to the docking poses of the challenge set were calculated; only 103 compounds with high APF similarity (> 0.55) to the challenge set were used in training the model for sub-challenge 1a.
Three-fold clustered cross-validation procedure
Each training data set was split into three diverse sets for testing activity prediction protocols in the following procedure: The ligands were clustered using ligand 3D docked poses by APF method at a distance of 0.25. The clusters of ligands were sorted by cluster size, and sequentially assigned into one of three groups, ensuring that none of the groups is significantly larger in size than the other two groups, and that each group includes small and large clusters in similar proportion. During model training, one group of clusters would be set aside as validation set; the other two groups were used for training. The process was repeated for each group of clusters, so that Pearson correlation Q2 and RMSE value can be calculated for the whole set by this three-fold procedure.
Activity models
APF 3D-QSAR activity models were derived as previously described [22, 33]. APF + Physics (APF/P) hybrid models combine the APF 3D QSAR descriptors with ICM VLS score components. Poses for 103 training set compounds were generated following the same docking protocol as for Stage 1A pose prediction.
Pose-selection-optimized 3D QSAR
For Cathepsin S target, we attempted to make the APF 3D QSAR model that can automatically select optimal binding poses for prediction, i.e. given an ensemble of low-scoring docked poses for a ligand, 3D QSAR prediction can be made for each pose and the highest predicted pKd value will represent the final prediction for that ligand. Optimization was performed via iterative procedure that evolves the 3D QSAR model towards self-consistency: (1) first generation model is made using the set of training ligands’ best-scoring poses, as in our regular approach; (2) second and all consequent generation models are made by making pKd prediction for all training ligands’ poses (ensembles of 10 poses per ligand were used) and selecting top predicted pKd poses. Ten iterations of the procedure were performed (we observed that model accuracy did not change significantly after ~ 5 iterations).
Software and hardware
All calculations, including receptor and ligand preparation, grid potential map calculations, docking simulations, ICM VLS score terms, APF similarity score, ligand pose clustering, correlation and RMSD calculations, were carried out using ICM 3.8–7 (Molsoft LLC, San Diego, CA). The docking simulations and ICM VLS score calculations were performed on a Linux cluster of 20 eight-core (2xIntel Xeon E5620) compute nodes.
Results and discussion
Initial benchmarking of ICM-dock for macrocyclic ligands
As a preliminary test for the ability of ICM-dock to predict accurately bound poses and conformations of macrocycles, we assembled a retrospective benchmark of PDB structures comprising 246 complexes, including 198 unique ligands and 126 unique target proteins (see “Methods” and Supplementary Data). To our knowledge this is by far the largest such benchmark, with previously published sets comprising a few dozen complexes [4], [9]. The compounds in our set explore widely the so-called bRo5 (beyond the Rules of 5) [10] space, with 65% of ligands having MW > 500 Da. Macrocycle ring sizes ranged from 9 to 40 bonds with the median size of 16.
Results of ICM flexible ring docking on this benchmark confirmed that the method is capable of reproducing native-like structures for the large majority of complexes: among the top 5 solutions, a pose within 2 Å RMSD was found for 75% of complexes, and the top-scoring solution was within 2 Å for 62% complexes. Failures were more common among larger compounds. Indeed, if we consider half of the benchmark complexes comprising the smaller molecules (< 42 heavy atoms, or ≈ < 600 Da), success percentages rise to 86% and 76% (for best in top 5 and the top pose, respectively). Doubling the sampling ‘effort’ setting (from 5 to 10) was tested and resulted in a minor improvement (78% success for top 5 poses), therefore raising this parameter above 10 appears unnecessary, and setting of 5 may be an acceptable accuracy/speed compromise. When top 10 poses are considered in these longer simulations, success rate reaches 81%. Thus only a few near native solutions are found past the 5 top-ranked poses. We also investigated the effect of the grid box size, a factor that is often overlooked in benchmarks. Using narrower (± 3 Å) margins around the ligand to define the box, noticeably higher success percentages of 69% for the top scoring solution and 82% for the top 5 could be obtained (effort = 5), likely because some of the artifactual poses are eliminated. Median simulation time (at effort = 5, all stages, starting from 2D structure through rescoring of top poses) was 16 min. It should be noted that 81% of compounds in the set have > 15 flexible torsions (including the flexible rings), and therefore these ligands are considerably more challenging to sample adequately than most typical small molecule drug-like molecule.
No comparably large-scale macrocycle docking benchmarks (to our knowledge) have been previously reported. A binding mode prediction protocol consisting of the LowModeMD conformational search for the free ligand followed by ligand ensemble docking in MOE reproduced 52% of the 48 complex benchmark set with accuracy better than 2.5 Å [4]. In our benchmark, ICM-dock reproduced 67% of complexes at RMSD < 2.5 Å, although different benchmark sets may not be fully comparable. Of further interest, LowModeMD conformational search generated ensembles containing a conformer within 2.5 Å of the experimental bound conformation in 98% cases,our ICM-dock protocol includes integrated free ligand pre-sampling stage, which had the same percentage of success (again, difference in benchmark sets needs to be kept in mind). In another work, a set of 20 macrolides (i.e. a class of macrocyclic natural products) and related macrocycle complexes from PDB was used to test two versions of AutoDock (AD) [25], AD Vina [38], DOCK 6.0 [23] and Glide 6.6 [12], all coupled to a pre-sampling stage of MD/LLMod conformation generator (Castro-Alvarez et al. 2017. Most of the macrolide complexes (18 out of 20 overlapped with our benchmark and per-complex RMSD numbers were published, so that we were able to calculate directly comparable averages. Mean RMSD was 1.4/1.59/2.54/1.4/1.35 Å for ADVina/AD4.2/AD3.0/DOCK/Glide, respectively, versus 1.2 Å for ICM-Dock,and a median RMSD of 0.93/1.1/1.34/0.77/1.03 Å (same software order, versus 0.59 Å for ICM-Dock for the set of 18 complexes. Thus ICM-dock performance on macrocycles appears to compare favorably with published results for widely used docking software.
BACE pose prediction protocol optimization
Despite the overall success on the retrospective benchmark, considering a significant number of complexes for which near-native solution was found but not top-ranked, as well as the fact that cross-docking (i.e. docking into non-cognate receptor) context was likely to adversely impact performance, we used LigBEnD (ligand biased ensemble docking) approach for the Challenge 1A pose prediction. LigBEnD adds a knowledge-based bias to the receptor interaction potential grids during docking Monte-Carlo simulation and also into the rescoring step, while the 4D ensemble is used to take advantage of multiple available receptor X-ray structures for the receptor flexibility modeling.
In our experience, MRC docking in general can suffer from imbalances in scoring to different receptor conformers. Physically, receptor conformations may have substantially different internal energies—as an example, some conformers with deeper, more extensive and potentially stronger interacting binding pockets are likely to incur a certain internal energy penalty; such a penalty is not accounted for by the ligand-receptor interaction scoring functions and is also difficult to evaluate directly. Systematic errors in scoring function may also inaccurately favor certain receptor conformations. Furthermore, when the ligand APF bias is introduced, the issue is exacerbated by different strengths of APF fields: for example, “general” receptor conformations that are compatible with many co-crystallized ligands may have stronger ligand APF bias than “specialized” receptor conformations with one or few compatible ligands. Therefore, some receptor conformers in the MRC ensemble may produce artifactual low energies/scores for ligands that in reality would match better to a different conformer, if either the APF bias or physical interaction fields turn out to be excessively strong in comparison to the matching receptor conformer.
To counteract these effects, we introduced a vector of receptor energy offsets that are assigned to each receptor conformation, to adjust ‘prominence’ of an individual receptor conformation in MRC docking. Each 4D grid layer will have its specific energy offset. Offsets are optimized for the best recognition of cognate ligand/receptor conformer pairs on an ensemble of receptor conformations and ligand poses generated through exhaustive docking of all co-crystallized ligands to all receptor conformations as described in “Methods”.
Figure 2 shows the results of 4D grid redocking for the 14 co-crystallized ligands to 7 BACE receptor conformations from PDBs, each with 2 protonation states. Without receptor conformation offsets, two receptor conformers strongly dominated the top scoring solutions—all ligands are paired to one of these two conformers. Furthermore, one of the ligands was mis-docked (RMSD > 2.0 Å). With optimized offsets, all 14 ligands were redocked within 1.0 Å RMSD, and all 7 receptor conformations were utilized by some of the top docking poses. The receptor conformations selected by docking were also closer to the cognate structures, as reflected in the Cα RMSD. (note that in Fig. 2c, each of the 7 receptor conformations were present as two copies with different protonation states, therefore conformations #1 & #2 were from the same parent conformation, #3 & #4 the next one, and so on). Among the 7 ligands with cognate receptor conformations present in the 4D docking ensemble, matching receptor conformation was selected for 6 when receptor offsets were applied whereas only 2 matching pairs were identified without the offset. Thus, introduction of offsets improved accuracy of ligand poses as well as quality of conformational selection of receptor states in this retrospective test.
Docking 14 BACE Cc-crystallized ligands to the ‘4D’ ensemble of 7 selected receptor conformations, each represented by 2 protonation states. Blue: with receptor energy offset; red: without receptor energy offset. a Ligand RMSD. b Receptor Cα RMSD (within 5 Å of ligand). c Receptor conformations selected by docking. Note that consecutive pairs of receptor states correspond to the same conformer: for example, 9th and 10th state represent the same conformation and differ only in the protonation of the active site aspartic acid residues
Pose prediction accuracy for BACE
Ligand prediction accuracy
In challenge Stage 1a, we used 14 receptor conformations from 7 PDBs for docking. 14 co-crystallized ligands that are chemically related to challenge ligands were used as ligand APF bias to drive docking process towards binding modes similar to experimentally determined. At the end of simulation, 5 best poses for each ligand were selected according to a composite docking / ligand APF similarity score. Figure 3a shows the docking results of the 20 challenge compounds. The median and mean RMSD of the top rank poses is 0.7 Å and 0.92 Å respectively, with the least accurate ligand at 1.9 Å. The median and mean RMSDs of the best poses are 0.7 Å and 0.76 Å, with the least accurate ligand at 1.5 Å. For 12 out of the 20 ligands, our top ranked pose was also the lowest RMSD pose among the top five.
Docking pose prediction results for BACE in Challenge 1A. RMSDs of five top ranking poses submitted for each ligand. The lowest RMSD pose for each ligand is highlighted in red box. a Ligand RMSD; b receptor RMSD, heavy atoms within 10 Å of the ligand. Distribution of RMSDs for all seven receptor conformers in the ‘4D’ ensemble is shown as a min/mean/max bar. Pose #1 has receptor RMSD at the minimum or below the mean for 14 complexes, and lowest ligand RMSD pose for 15 complexes out of 20
Receptor prediction accuracy
In a realistic cross-docking scenario, both ligand and receptor in the docked complex model deviate from the experimental structure. Therefore, while it is common to report only ligand RMSD, the question of quality of the prediction is not restricted to the ligand pose, it is also important to consider how close the receptor conformation chosen (or generated) by the docking procedure is to the experimental cognate receptor structure. We are using MRC approach to account for receptor flexibility: ‘4D’ docking simulations switch on the fly from one conformer of the receptor to another within the 4D ensemble, which for BACE included 7 distinct conformers, and optimal energy offsets were derived (see previous section) to improve receptor conformer selection. Figure 3b shows that among best ligand poses, for half of the complexes (10 out of 20) our procedure selected receptor conformation closest to the cognate, and for most of the remaining complexes RMSD was below the MRC ensemble average, i.e. the receptor conformation selection was still better than random. Thus, our 4D docking simulations with receptor energy offsets are successfully mimicking receptor conformational selection mechanism of induced fit (within the ensemble).
For Challenge Stage 1B, D3R organizers released X-ray structures of the receptor for the 20 complexes (without the co-crystallized ligands) for re-docking of the ligands to their cognate receptor conformations. Given the encouraging results in the macrocycle re-docking benchmark, we chose to perform this exercise without using any ligand APF bias. Figure 4a summarizes the results. Top ranking poses have a mean/median RMSD of 0.61/0.56 Å with a maximum of 1.2 Å; the best poses out of 5 have a mean/median RMSD of 0.58/0.56 Å with maximum of 1.0 Å; for 19 out of 20 ligands, the top ranking pose is the best pose.
For the challenge submission, we included a single tightly-bound structural water molecule for 12 out of 20 complexes. Post-challenge, we decided to investigate the impact of including or omitting this water on the accuracy, and repeated calculations without any water molecules included. The results (Fig. 4b) were overall very similar, with median RMSDs unchanged. Some loss of accuracy was noted for two ligands: top pose for compounds #17 and #18 went from RMSDs of 0.5 Å and 0.6 Å to 1.6 Å and 1.5 Å, respectively, although sub-angstrom pose was still present for #18 at rank 2. Visual inspection revealed formation of a water-mediated hydrogen bond for these ligands, explaining greater deviations from native pose when the water molecule is omitted (Supplementary Fig. 2)
Overall, our blind cross-docking (Stage 1A) results were among the top ranked submission and cognate re-docking (Stage 1B) results were the top ranked submission in this challenge. The latter result is also remarkable in that no ligand data was used for Stage 1B prediction.
Activity prediction
APF 3D QSAR predictions for BACE
To select optimal approach for the blind challenge, two prediction methods were first tested using a threefold clustered cross-validation method: pure APF 3D QSAR PLS model, and a hybrid APF/P 3D QSAR PLS model, in which 6 energy terms were introduced as additional, physics-based descriptors. In the D3R GC4 the latter approach led to improved models across multiple kinase targets [22]. Pure APF 3D QSAR gave cross-validated RMSE of 0.90 pKd unit, and a Pearson and Kendall’s τ-b correlation of 0.74 and 0.51, respectively. The hybrid model achieved an RMSE of 0.91 pKd unit, and a Pearson and Kendall’s τ-b correlation of 0.72 and 0.49, respectively. Thus, no significant differences in performance between models were observed in cross-validation on the training set, and we decided to opt for the simpler APF model for the challenge submission. Unfortunately, the model accuracy in the prediction of 154 BACE challenge set turned out to be significantly below our cross-validation results: RMSE of 1.15 pKd unit, Pearson and Kendall’s τ-b correlation of 0.35 and 0.21, respectively.
It should be noted that some of the submissions for this Challenge did achieve higher correlations, up to 0.38 τ-b. We decided to investigate whether any simple base-line activity correlates may produce similar results. Several physical properties that frequently correlate with activity were calculated (using Molsoft ICM cheminformatics module): molecular weight, cLogP, polar surface area, molecular surface area, and molecular volume, as shown in Table 1. Also included was the hydrophobic term from the ICM scoring function. Remarkably, for the Challenge set ligands, pKd values were significantly correlated with several physical properties as well as the hydrophobic term in ICM-score. Using molecular volume alone as an activity predictor would result in a τ-b value of 0.38, on par with the top result among all submissions. However, these correlations are much weaker already on our focused training sets and essentially disappear when the calculation is done on the full set of 7124 compounds with available BACE inhibition data in ChEMBL—as one might expect, relatively strong size correlations seem to be restricted to the specific chemotypes. Nevertheless, the implication of these findings is that a model with a strong size and/or lipophilicity bias would be able to generate Challenge set predictions with a τ-b value in the range of 0.31–0.38.
APF 3D QSAR predictions for Cathepsin S
For this target the challenge did not include any docking accuracy component, only the predictions of binding affinity for ligands. When building APF 3D QSAR model, we attempted to make the activity (pKd) model that could also optimally select bound ligand poses in an ensemble generated by docking using a procedure resembling “autoshimming” approach in Ref. [24]. In a series of model ‘generations’, we would use the previous generation model to predict pKd for multiple poses of each ligand and select highest predicted pKd poses as a basis for the next generation QSAR. Initial set of poses for the first generation QSAR model was based on the regular scoring. We expected that the procedure could result in a better 3D QSAR model because pose ranking/selection becomes consistent with pKd prediction, as it should be in terms of ligand binding physics. In the three-fold clustered cross-validation this ‘pose-ranking-consistent’ (PRC) 3D QSAR model achieved a pKd RMSE, Pearson and Kendall’s τ-b correlations of 0.65, 0.60 and 0.45, respectively, as compared to 0.69, 0.50 and 0.38 for the base-line model using initial selection of top-ranking docking poses. Marked improvement across all accuracy measures in cross-validation led us to use the PRC approach for our GC4 submission. On the blind test set, the submitted model achieved 0.4 RMSE, 0.75 Pearson and 0.54 Kendall τ-b metrics, exceeding our expectations based on cross-validation accuracy, and sharing top rank with one more submission among all models submitted to this Challenge. It should be noted that in the post-challenge evaluation, comparison of the PRC model results with the base-line model using top-ranking docking poses revealed that on the GC4 set the models performed essentially identically (Table 2), despite the apparent advantage of the PRC approach in cross-validation and minor but marked differences in poses selected by the two methods (Supplementary Fig. 3). More extensive testing will be needed to further validate the benefits of PRC model.
Why were we able to build a 3D QSAR models with good predictivity for this target, while similar approach did not yield a meaningfully predictive model for BACE, and none of the other methods could do better than a simple baseline physical property correlation? A possible explanation transpires if we consider the distribution of the Tanimoto distances (i.e. chemical dissimilarity) from each challenge compound to a closest available training set compound (Supplementary Fig. 4a, b). Median Tanimoto distance to the training set among BACE compounds is 0.25, while among Cathepsin S compounds it is only 0.03. Thus, good coverage of activity data for the closely related compounds appears to be the critical factor.
Conclusions
ICM-dock shows accurate predictions for macrocyclic ligand complexes both, on a broad retrospective benchmark and the BACE target of the prospective blind Challenge. In the cross-docking scenario of Stage 1A, we introduced receptor conformation energy offsets to improve accuracy of ‘4D’ receptor ensemble docking and demonstrated that receptor conformational selection during 4D sampling identifies near-cognate receptor conformations in the ensemble.
Good quality predictive 3D QSAR model was constructed using APF approach for Cathepsin S but not for BACE. Proximity of challenge set to the training compounds with available experimental data appears to be crucial for the prediction accuracy.
Abbreviations
- PDB:
-
Protein data bank
- APF:
-
Atomic property field
References
Abagyan R (2017). ICM user manual. https://www.molsoft.com/
Abagyan R, Totrov M, Kuznetsov D (1994) ICM-A new method for protein modeling and design: applications to. J Comp Chem 15(5):488–506
Allen SE, Dokholyan NV, Bowers AA (2016) Dynamic docking of conformationally constrained macrocycles: methods and applications. ACS Chem Biol 11(1):10–24
Anighoro A, de Leon AD, Bajorath J (2016) Predicting bioactive conformations and binding modes of macrocycles. J Comput Aided Mol Des 30(10):841–849
Basit S, Ashraf Z, Lee K, Latif M (2017) First macrocyclic 3(rd)-generation ALK inhibitor for treatment of ALK/ROS1 cancer: clinical and designing strategy update of lorlatinib. Eur J Med Chem 134:348–356
Bento AP, Gaulton A, Hersey A, Bellis LJ, Chambers J, Davies M, Kruger FA, Light Y, Mak L, McGlinchey S, Nowotka M, Papadatos G, Santos R, Overington JP (2014) The ChEMBL bioactivity database: an update. Nucleic Acids Res 42:D1083–1090
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28(1):235–242
Bottegoni G, Kufareva I, Totrov M, Abagyan R (2009) Four-dimensional docking: a fast and accurate account of discrete receptor flexibility in ligand docking. J Med Chem 52(2):397–406
Castro-Alvarez A, Costa AM, Vilarrasa J (2017) The performance of several docking programs at reproducing protein-macrolide-like crystal structures. Molecules 22(1):136
Doak BC, Over B, Giordanetto F, Kihlberg J (2014) Oral druggable space beyond the rule of 5: insights from drugs and clinical candidates. Chem Biol 21(9):1115–1142
Easterbrook PJ, Berlin JA, Gopalan R, Matthews DR (1991) Publication bias in clinical research. Lancet 337(8746):867–872
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47(7):1739–1749
Gaieb Z, Parks CD, Chiu M, Yang H, Shao C, Walters WP, Lambert MH, Nevins N, Bembenek SD, Ameriks MK, Mirzadegan T, Burley SK, Amaro RE, Gilson MK (2019) D3R Grand Challenge 3: blind prediction of protein-ligand poses and affinity rankings. J Comput Aided Mol Des 33(1):1–18
Giganti D, Guillemain H, Spadoni J-L, Nilges M, Zagury J-F, Montes M (2010) Comparative evaluation of 3D virtual ligand screening methods: impact of the molecular alignment on enrichment. J Chem Inf Model 50(6):992–1004
Grigoryan AV, Kufareva I, Totrov M, Abagyan RA (2010) Spatial chemical distance based on atomic property fields. J Computer-Aided Mol Design 24(3):173–182
Halgren TA (1996) Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J Comp Chem 17(5–6):490–519
Hewitt WM, Leung SS, Pye CR, Ponkey AR, Bednarek M, Jacobson MP, Lokey RS (2015) Cell-permeable cyclic peptides from synthetic libraries inspired by natural products. J Am Chem Soc 137(2):715–721
Husby J, Bottegoni G, Kufareva I, Abagyan R, Cavalli A (2015) Structure-based predictions of activity cliffs. J Chem Inf Model 55(5):1062–1076
Ioannidis JP (2005) Why most published research findings are false. PLoS Med 2(8):e124
Kufareva I, Ilatovskiy AV, Abagyan R (2012) Pocketome: an encyclopedia of small-molecule binding sites in 4D. Nucleic Acids Res 40:D535–540
Lam PC, Abagyan R, Totrov M (2018) Ligand-biased ensemble receptor docking (LigBEnD): a hybrid ligand/receptor structure-based approach. J Comput Aided Mol Des 32(1):187–198
Lam PC, Abagyan R, Totrov M (2019) Hybrid receptor structure/ligand-based docking and activity prediction in ICM: development and evaluation in D3R Grand Challenge 3. J Comput Aided Mol Des 33(1):35–46
Lang PT, Brozell SR, Mukherjee S, Pettersen EF, Meng EC, Thomas V, Rizzo RC, Case DA, James TL, Kuntz ID (2009) DOCK 6: combining techniques to model RNA-small molecule complexes. RNA 15(6):1219–1230
Martin EJ, Sullivan DC (2008) AutoShim: empirically corrected scoring functions for quantitative docking with a crystal structure and IC50 training data. J Chem Inf Model 48(4):861–872
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30(16):2785–2791
Némethy G, Gibson KD, Palmer KA, Yoon CN, Paterlini G, Zagari A, Rumsey S, Scheraga HA (1992) J Phys Chem 96:6472
Neves MA, Totrov M, Abagyan R (2012) Docking and scoring with ICM: the benchmarking results and strategies for improvement. J Comput Aided Mol Des 26(6):675–686
Pillaiyar T, Namasivayam V, Manickam M (2016) Macrocyclic hepatitis C virus NS3/4A protease inhibitors: an overview of medicinal chemistry. Curr Med Chem 23(29):3404–3447
Ruegger A, Kuhn M, Lichti H, Loosli HR, Huguenin R, Quiquerez C, von Wartburg A (1976) Cyclosporin A, a peptide metabolite from trichoderma polysporum (Link ex Pers.) Rifai, with a remarkable immunosuppressive activity. Helv Chim Acta 59(4):1075–1092
Sindhikara D, Spronk SA, Day T, Borrelli K, Cheney DL, Posy SL (2017) Improving accuracy, diversity, and speed with prime macrocycle conformational sampling. J Chem Inf Model 57(8):1881–1894
Slynko I, Da Silva F, Bret G, Rognan D (2016) Docking pose selection by interaction pattern graph similarity: application to the D3R grand challenge 2015. J Comput Aided Mol Des 30(9):669–683
Sussman F, Villaverde MC, Dominguez JL, Danielson UH (2013) On the active site protonation state in aspartic proteases: implications for drug design. Curr Pharm Des 19(23):4257–4275
Totrov M (2008) Atomic property fields: generalized 3D pharmacophoric potential for automated ligand superposition, pharmacophore elucidation and 3D QSAR. Chem Biol Drug Des 71(1):15–27
Totrov M (2011) Ligand binding site superposition and comparison based on atomic property fields: identification of distant homologues, convergent evolution and PDB-wide clustering of binding sites. BMC Bioinform 12(Suppl 1):S35
Totrov M, Abagyan R (1997) Flexible protein-ligand docking by global energy optimization in internal coordinates. Proteins 1:215–220
Totrov M, Abagyan, R (1999) Derivation of sensitive discrimination potential for virtual ligand screening. In: Proceedings of the third annual international conference on computational molecular biology, pp. 312–320
Totrov M, Abagyan R (2008) Flexible ligand docking to multiple receptor conformations: a practical alternative. Curr Opin Struct Biol 18(2):178–184
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455–461
Vezina C, Kudelski A, Sehgal SN (1975) Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J Antibiot (Tokyo) 28(10):721–726
Xu X, Ma Z, Duan R, Zou X (2019) Predicting protein-ligand binding modes for CELPP and GC3: workflows and insight. J Comput Aided Mol Des 33(3):367–374
Yu X, Sun D (2013) Macrocyclic drugs and synthetic methodologies toward macrocycles. Molecules 18(6):6230–6268
Acknowledgements
The authors thank D3R organizers for coordinating the challenge. We also thank Eugene Raush for technical assistance, and Andrew Orry for proofreading of this manuscript.
Author information
Authors and Affiliations
Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing financial interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lam, P.CH., Abagyan, R. & Totrov, M. Macrocycle modeling in ICM: benchmarking and evaluation in D3R Grand Challenge 4. J Comput Aided Mol Des 33, 1057–1069 (2019). https://doi.org/10.1007/s10822-019-00225-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-019-00225-9